Abstract
To characterize gene expression in activated mast cells more comprehensively than heretofore, we surveyed the changes in genetic transcripts by the method of serial analysis of gene expression in the RBL-2H3 line of rat mast cells before and after they were stimulated through their receptors with high affinity for immunoglobulin E (FcεRI). A total of 40,759 transcripts derived from 11,300 genes were analyzed. Among the diverse genes that had not been previously associated with mast cells and that were constitutively expressed were those for the cytokine macrophage migration inhibitory factor neurohormone receptors such as growth hormone- releasing factor and melatonin and components of the exocytotic machinery. In addition, several dozen transcripts were differentially expressed in response to antigen-induced clustering of the FcεRI. Included among these were the genes for preprorelaxin, mitogen-activated protein kinase kinase 3, and the dual specificity protein phosphatase, rVH6. Significantly, the majority of genes differentially expressed in this well-studied model of mast cell activation have not been identified before this analysis.
Keywords: receptor aggregation, signal transduction, exocytosis, cell differentiation, allergy
ast cells perform a significant role in host defense against parasitic and some bacterial infections (1). They are also widely distributed in neural and endocrine tissues and have been referred to as the “emergency kit” for the response to stress (2). As part of their armamentarium for host defense, they, and the closely related circulating basophils, express large numbers of receptors having a high affinity for IgE (FcεRI) (3). A signaling cascade initiated by aggregation of the FcεRI leads to the rapid release of preformed, granule-associated mediators, such as histamine, serotonin, and proteases, and to the secretion of membrane-derived arachidonic acid metabolites such as leukotriene C4 and prostaglandin D2 (4). These rapid responses are followed by changes in gene expression and release of multiple cytokines, chemokines, and other immunomodulatory molecules (5). Allergic reactions, mediated in part by mast cells, involve an apparent aberration of the protective role played by IgE and FcεRI. In the allergic individual, harmless environmental antigens evoke an anomalous IgE response and the interaction of the IgE with the FcεRI “sensitizes” the cells so that subsequent exposure to the antigen can lead to a dramatic, sometimes even fatal systemic anaphylaxis.
Earlier studies of such events were largely concerned with examining functional or biochemical phenomena; more recently many genes coding for molecules thought to be involved in the activation of mast cells have been identified and in a few instances manipulated to determine the role of the proteins they code for. The aim of the present investigation was to define comprehensively the profiles of expressed genes in resting and activated mast cells in order to discover unsuspected molecular participants in the cellular events.
We chose to use serial analysis of gene expression (SAGE)1 for this purpose because it has proved to be useful for a high throughput survey of which genes are being transcribed (6, 7). The principles of this method are as follows: (a) when derived from a defined location of a transcript, a sequence of ∼10 bp is sufficient to provide a unique “tag” for any gene from even a large, e.g., mammalian, genome. (b) By ligating many such tags together along with a restriction enzyme recognition sequence that acts as a linker, a large number of such tags can be efficiently sequenced and identified. Specifically what is done is to convert the desired mRNA into double stranded cDNA. These cDNAs are then digested with an endonuclease that recognizes only the four nucleotide sequence CATG. Consequently an average random sequence of 256 bp should contain one such site. Separate aliquots are then ligated with distinctive linkers, and cleaved with a “tagging” endonuclease that will leave a short sequence of ∼14 bp: the CATG and a unique 10-bp sequence. The cDNAs containing the discrete linkers are then ligated and such “ditags” amplified by PCR. Many such ditags are then ligated to form concatamers and the concatamers are inserted into a plasmid and sequenced, the terminal CATG acting as commas between successive ditags. Any single messenger species should be represented by a corresponding single species of tag at a multiplicity proportional to the multiplicity of that messenger species. The cells we used, RBL-2H3 cells (8), were derived from a rat mucosal mast cell line (9–11), and have been widely used for studying mast cell biology.
Materials and Methods
Cells and Cell Culture.
Rat basophilic leukemia cells (RBL-2H3) were maintained as described previously (8). When stimulated cells were desired, the RBL-2H3 cells were sensitized overnight with mouse anti-DNP IgE (12) at 2 μg/ml, washed three times, and stimulated in culture medium with DNP18-BSA at a concentration of 100 ng/ml for varying times. HMC-1 cells (13) were grown in Iscove's medium supplemented with 10% FCS and 10−5 M monothioglycerol. UK812 cells (14) were cultured in RPMI 1640 and 15% FCS. Peritoneal mast cells and macrophages were purified as described (15, 16). In brief, peritoneal cells (3 × 108) from ten Sprague Dawley rats were harvested using Tyrode's buffer (135 mM NaCl, 5 mM KCl, 10 mM Hepes, and 5.6 mM glucose) with 0.1% BSA. The cells were sedimented by centrifugation at 100 g for 5 min. The cell pellets were pooled and resuspended at a concentration of 5 × 107 cells/ml. A 1-ml suspension of cells was layered on a 2-ml cushion of 22.5% wt/ vol metrizamide and centrifuged at 400 g for 15 min. The cells remaining at the buffer-metrizamide interface were aspirated and saved for preparation of macrophages. The cells in the pellet were pooled, washed twice in Tyrode's buffer, and determined to be 95% pure mast cells as measured by staining with 0.1% toluidine blue. The cells collected at the interface were seeded in tissue culture flasks and incubated at 37°C for 3 h. After washing off nonadherent cells, the adherent cells (macrophages) were collected. Both mast cells and macrophages were used for preparing RNA immediately after purification.
RNA and SAGE.
Total RNA was prepared from peritoneal mast cells and macrophages with Trizol (GIBCO BRL, Gaithersburg, MD) following the manufacturer's protocol. The isolated total RNA from peritoneal mast cells was treated with heparinase I (Sigma Chemical Co., St. Louis, MO) as previously described (17) before performing reverse transcription (RT) PCR. Messenger RNAs were purified from resting and stimulated RBL-2H3 cells with the MessageMaker Kit (GIBCO BRL) following the manufacturer's instruction. Messenger RNAs from unstimulated cells and cells stimulated for 3.5 h (above) were used for generating the “resting” and “activated” SAGE libraries. SAGE was performed as described (18). Detailed protocols and sequence analysis software were provided by Dr. Kenneth W. Kinzler (Johns Hopkins Oncology Center, Baltimore, MD). DNA sequencing of 1,600 clones containing concatenated tags was done using −21M13 dye primer fluorescent sequencing chemistry (PE Applied Biosystems, Foster City, CA). Sequencing reactions were run on a 377 ABI automated sequencer. All electropherograms were reanalyzed by visual inspection to check for ambiguous base calls and to correct misreads.
Statistical Analysis.
The data were analyzed as indicated in the Appendix.
RT-PCR, Northern Blot and Library Screening.
RT-PCR was carried out following the manufacturer's protocol using an RT-PCR kit (Stratagene, La Jolla, CA). The primers we used for amplification of the corresponding gene transcripts were: macrophage migration inhibitory factor (MIF)(5′-CGGAATTCGGGTCACGTAGTCAGGTCCCAGACT-3′, 5′-GCTCTAGAGGTCTCAAACCATTTATTTCTCCCG-3′); preprorelaxin (5′-CGGAATTCTGAACCGCCCAGGAGCACCGCCCA-3′, 5′-GCTCTAGATTA-TTAACAGAACTACAACAATGCA-3′); Hrs-2 (5′-TTG-GAGACAGACTGGAGTCCATT-3′, 5′-TCCCAGCCTCCACAGA-CCAGCAAC-3′); vesicle-associated membrane protein (VAMP)-2 (5′-TGGGAGCTTCCAACATGTTGATTA-3′, 5′-CCACAGA-ATTGGGGGCATGGGGCA-3′); annexin V (5′-ACAGCAT-CTGGCTCTCGAGGCA-3′, 5′-GTTCACAATTTCATGGCTAAGGTTCC-3′); gonadotropin-releasing hormone (GnRH) receptor (5′-AGTGGCATTTGGCACCTCCTTTGT-3′, 5′-CCT-AATAATCTGTGCTGTTGGTCTATA-3′); melatonin receptor (5′-TACGTGTTCCTGATATGGACACTG-3′, 5′-GGCCGGATCTGAGGCCACAATAAG-3′); RMCP-5 (5′-AGATCATCGGGGGCACGGAGTGCA-3′, 5′-AGGAACTTTCTG-TGACTTGTAAGAACC-3′); TNF-α (5′-CAAGGAGGAG-AAGTTCCCAA-3′, 5′-CGGACTCCGTGATGTCTAAG-3′); IL-3 (5′-GTATGCTGCTCCCGCTCCTGATG-3′, 5′-CATTCCACGGTCATAGGGCGAAAG-3′); β-actin (5′-GTGGGGCGCCCCAGGCACCA-3′, 5′-GTCCTTAATGTCACGCACG-ATTTC-3′).
Northern blot analyses were performed as described (19). The RBL-2H3 cDNA library was screened with 32P-labeled oligonucleotides derived from clone 1 and 9 tag sequences as described (18).
Rabbit Anti–Rat MIF Antiserum.
A full-length cDNA for MIF was amplified from resting RBL-2H3 cell mRNA by RT-PCR using the following primers: 5′-CGGAATTCATGCCTATGTTCATCTGAACA-3′ and 5′-CCGCTCGAGTCAAGCGAAGGTGGAACCGTTC-3′. After digestion with EcoRI and XhoI, the PCR fragment was cloned into the pGEX4T-1 vector (Pharmacia, Piscataway, NJ). A glutathione-S-transferase-MIF fusion protein was expressed in Escherichia coli and purified as described (19). Rabbit anti–rat MIF antiserum was generated by immunizing rabbits with the purified fusion protein following standard protocols.
Immunoprecipitation and Western Blots.
RBL-2H3, HMC-1, and KU812 cell lysates were prepared as previously described (20). 4 μg of goat anti–human MIF antibody (R&D Systems, Minneapolis, MN) and 25 μl of protein G beads were added to 1 ml of lysate (3 × 106 cell equivalents) or culture supernatants or fresh culture medium. After incubation for 3 h, the beads were recovered by centrifugation, and the immunoprecipitates washed twice. Bound proteins were released by boiling in SDS sample buffer, separated by electrophoresis in 4–20% gradient SDS-PAGE gels, and transferred to nitrocellulose membranes. MIF immunoprecipitates from HMC-1 and KU812 samples were blotted with monoclonal anti-human MIF (R&D Systems) at 0.5 μg/ml, whereas MIF immunoprecipitates from RBL-2H3 samples were blotted with rabbit anti-rat MIF serum at 1:2,000 dilution. To detect mitogen-activated protein kinase kinase (MAPKK3) protein, cell lysates were prepared as above from resting and activated RBL-2H3 cells at various times after stimulation, and separated on 10% SDS-PAGE gels. Isoform-specific rabbit anti-MAPKK3 antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was used as the primary antibody at 0.5 μg/ml.
Results
Methodological Aspects of SAGE Analysis and General Results.
We decided to compare resting cells with cells stimulated through their FcεRI using conditions that were previously known to give a robust response and at times where prior studies had demonstrated active transcription of newly activated genes (5). Messenger RNA was purified from the activated RBL-2H3 cells and from an equivalent number of unstimulated cells, and analyzed by SAGE. A total of 40,759 tags were generated; approximately half were from the population of resting cells and half from the activated cells.
We extended some of the results and validated the fidelity of the SAGE analysis using RT-PCR, or Northern blotting (with or without Western analysis). In all 16 instances in which this was done, the genes identified by the expression of the tags analyzed in this study were confirmed.
Approximately 7,000 distinct tags were identified in each of the SAGE libraries separately prepared from the resting and activated transcripts (Table 1) with the vast majority of the distinct tags likely to correspond to unique genes. The characteristics of the frequency distribution of expressed genes are strikingly similar in the resting and activated cells: the most abundant transcripts, whose levels are equal to or >0.1%, are derived from <2% of the genes analyzed but constitute >34% of the total transcripts. Correspondingly, 65% of the genes account for those transcripts present at low abundance, with an average of 0.005%, and contribute only 23% of the total transcripts (Table 1).
Table 1.
Summary of SAGE Data
| Frequency distribution* | ≥20 | 19 to 5 | 4 to 2 | =1 | ||||
|---|---|---|---|---|---|---|---|---|
| Resting cells | ||||||||
| No. unique genes identified | 131 (1.8)‡ | 487 (6.8) | 1,850 (25.9) | 4,673 (65.4) | ||||
| No. tags sequenced§ | 7,015 (34.8) | 3,822 (18.9) | 4,667 (23.1) | 4,673 (23.2) | ||||
| Activated cells | ||||||||
| No. unique genes identified | 132 (1.9) | 451 (6.4) | 1,647 (23.3) | 4,830 (68.4) | ||||
| No. tags sequenced | 7,938 (38.6) | 3,667 (17.8) | 4,147 (20.1) | 4,830 (23.5) |
The frequency distribution of a given tag identified in this analysis was calculated within a total population of 20,177 tags sequenced from resting cells and 20,582 tags sequenced from stimulated RBL-2H3 cells.
Percentage in frequency group.
The total number of tags identified that are derived from genes with a copy number indicated above are shown.
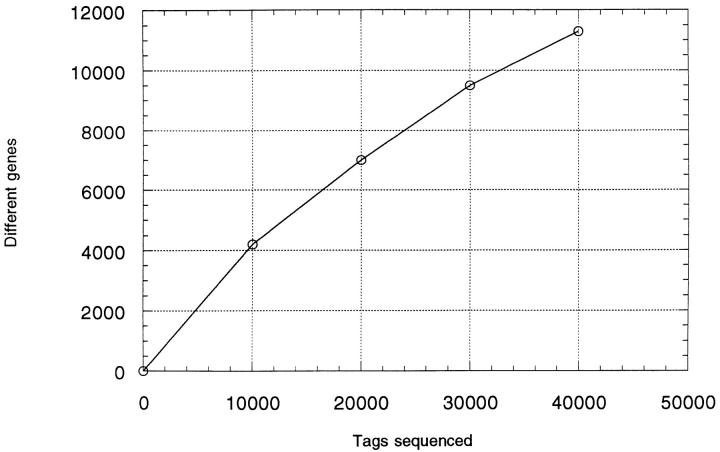
As expected, the number of unique genes identified was directly proportional to the number of tags sequenced, with a total of 11,300 unique genes identified in the combined data sets of the two libraries (Fig. 1). Statistical analysis (Appendix) reveals that there was a 95% chance of detecting a transcript expressed at an average abundance of >0.0075% in the RNA samples from the two populations.
Figure 1.
Results of the combined analysis of two SAGE tag libraries.
Results from Unactivated Cells.
Consistent with a study in yeast (21), housekeeping genes such as those for cytoskeletal structures and enzymes involved in protein synthesis and energy metabolism, constitute the majority of highly expressed genes in RBL-2H3 cells. It is noteworthy that each of the 50 most abundant housekeeping genes we identified was represented by a unique tag thus confirming that the vast majority of the unique tags represent discrete transcripts. However, in one instance of a very highly expressed endogenous retrovirus-like gene, we observed a low number of two tags that had been generated from internal sequences of the same transcript (below).
Many genes and their corresponding proteins characteristic of mast cells, such as Fc receptors, mediators and their synthetic enzymes, have been studied, but no systematic quantitation of the levels of gene transcripts for these components has been published. Table 2 lists the mRNA levels of some representative genes characteristic of mast cells in unstimulated RBL-2H3 cells. Consistent with a previous study, RBL-2H3 cells, like normal mast cells, express low-affinity Fc receptors for IgG (i.e., FcγRIIb2 and FcγRIII; reference 22). In fact, our analysis indicates that the mRNA levels for these two α chains are comparable to the level of FcεRI α (Table 2).
Table 2.
Expression of Transcripts Characteristic of Mast Cells Before and After Stimulation by FcεRI
| Tag sequence* | Resting cells‡ | Activated cells§ | Gene | |||
|---|---|---|---|---|---|---|
| Mediators | ||||||
| AAACAATGTG | 49 | 28 | Proteoglycan peptide core protein | |||
| AAGTCCTGCA | 25 | 11 | Carboxypeptidase A | |||
| TGGGATCTGG | 7 | 0 | Rat mast cell protease-5 | |||
| TCCGAAGACT | 2 | 2 | Rat mast cell protease-10 | |||
| Mediator synthetases | ||||||
| CTTCAGAGGG | 11 | 19 | Histidine decarboxylase | |||
| CTACTTCTTC | 5 | 2 | 5-Lipoxygenase | |||
| CCCACATAGT | 3 | 4 | S-glutathione-S-transferase | |||
| GGCAGAGGAA | 1 | 2 | Prostaglandin D2 synthetase | |||
| CGAAATAAAT | 1 | 1 | c-Phospholipase-A2 | |||
| TGTGTCTCCA | 1 | 1 | Aromatic l-amino acid decarboxylase | |||
| Receptors | ||||||
| TTGGTTTGGG | 17 | 10 | FcεRI β | |||
| CAATAAATAA | 11 | 7 | FcεRI α | |||
| ATGCTCTGGA | 9 | 5 | FcγRIII α | |||
| CTAAGGAGAT | 6 | 3 | FcγRIIb2 α | |||
| CTAGGCTCTA | 4 | 2 | FcεRI γ |
In this and the succeeding tables, the tag sequences represent the 10-bp SAGE tags 3′ of the NlaIII site.
In this and subsequent tables the copy numbers are from 20,177 tags from the resting cells.
In this and subsequent tables the copy numbers are from 20,582 tags from the activated cells.
Transcripts for constituents of mast cell granules were also identified: The proteoglycan peptide core protein serves as a packing site to store mediators, such as histamine, serotonin, and proteases, and transcripts for this protein were found to be expressed at a high level. Of the mRNA coding for proteases, that for carboxypeptidase A was present at a much higher level than others. This is notable since the older, nonsecreting RBL-1 line of these cells has been reported to lack transcripts coding for this protease (23). The number of gene transcripts coding for histidine decarboxylase, which converts histidine to histamine (24), was tenfold higher than that for aromatic l-amino acid decarboxylase, the enzyme that converts tryptophan to serotonin (see Fig. 7 in reference 16). Transcripts encoding enzymes regulating arachidonic acid metabolism (25) such as phospholipase A2 (PLA2), which generates arachidonic acid from phospholipids, 5-lipoxygenase, a synthetase for the 5-HPETE intermediate for leukotrienes, glutathione- S-transferase, responsible for the production of the leukotriene LTC4, and prostaglandin D2 synthetase, were expressed at low to intermediate levels.
The variation in mRNA levels for such mast cell-specific components was notable. For example, transcripts for the proteoglycan peptide core protein were observed 49 times, which can be calculated to correspond to ∼735 copies/ cell, assuming 300,000 total transcripts per cell (Materials and Methods). In contrast, transcripts for PLA2 and for aromatic l-amino acid decarboxylase were observed only once, corresponding to ∼15 copies/cell.
Diverse gene transcripts for proteins not previously associated with mast cells, were found to be constitutively expressed in RBL-2H3 cells. Some of these are listed in Table 3. Among them was the macrophage migration inhibitory factor, MIF, a proinflammatory cytokine primarily produced constitutively by the anterior pituitary and by macrophages, and released in response to endotoxin (26). More than 520 copies/cell were expressed in the RBL-2H3 cells. Using RT-PCR, we observed that normal rat peritoneal mast cells, like peritoneal macrophages, express MIF (Fig. 2 A). These results suggest that mast cells are a major source of production of MIF. We did not observe the release of MIF from resting (Fig. 2 B) or FcεRI-activated RBL-2H3 cells (data not shown), whereas the human immature mast cell line, HMC-1, and the basophil cell line, KU812, constitutively express and secrete MIF, as shown by Western blotting (Fig. 2 B).
Table 3.
Transcripts of Known Genes Not Previously Described in Mast Cells
| Tag sequence | Resting cells | Activated cells | Gene | |||
|---|---|---|---|---|---|---|
| Cytokine | ||||||
| AACGCAGCCA | 39 | 34 | MIF* | |||
| Exocytotic machinery | ||||||
| CACAGCAAGG | 3 | 4 | Hrs-2‡ | |||
| TTCAAGAAGT | 4 | 3 | Complexin II | |||
| AACGTTGGGA | 2 | 2 | Munc-18-2 | |||
| CAAGCGAGCC | 2 | 1 | rSec6 | |||
| CCCCCAATTC | 2 | 3 | Vamp-2‡ | |||
| AAAATAAACT | 1 | 1 | Synaptotagmin-2 | |||
| CCTCTACGAA | 1 | 2 | α-SNAP | |||
| PLA2 inhibitors | ||||||
| AGACACTTCC | 5 | 3 | Annexin II | |||
| AATGAAATCA | 3 | 5 | Annexin I | |||
| GCTAAGTGGC | 1 | 3 | Annexin VI | |||
| AAATTGTGAA | 1 | 2 | Annexin V‡ | |||
| Receptors | ||||||
| TGAAAAAAAA | 2 | 4 | GnRH receptor‡ | |||
| GCCCCCAGGA | 1 | 2 | Melatonin receptor‡ |
Confirmed by Northern blotting, cloning, sequencing, and Western blotting.
Confirmed by RT-PCR.
Figure 2.
Characterization of transcripts and their products. (A) MIF detected by RT-PCR. Total RNAs were prepared from rat peritoneal mast cells and macrophages and used for RT-PCR using specific primers for rat MIF. The marker (leftmost lane) shows the position of a cDNA product having the expected size of 550 bp. (B) MIF detected by Western blot. Immunoprecipitates were prepared by reacting goat polyclonal anti-human MIF antibodies with the material indicated below, separated on polyacrylamide gels, and blotted either with a mouse monoclonal anti-human MIF antibody (lanes 1–6), or with polyclonal rabbit anti–rat MIF antibodies (lanes 7–9). Lanes 1, 4, and 7 were loaded with lysates from 3 × 106 HMC-1, KU812, and RBL-2H3 cells, respectively; lanes 2, 5, and 8 with 1 ml of culture supernatants from HMC-1, KU812, and RBL-2H3 cells; lanes 3, 6, and 9 with 1 ml of the medium used to culture HMC-1, KU812, and RBL-2H3 cells, as a control. (C) MAPKK3 detected by Western blot. Lysates from 2 × 105 RBL-2H3 cells were blotted with anti-MAPKK3 antibody. Lane 1, Unstimulated cells. Lanes 2–5, Cells stimulated for 0.5, 3.5, 7, and 16 h.
At least seven proteins previously shown to mediate secretory events in other systems, are also expressed in RBL-2H3 cells suggesting that these components are parts of the exocytotic machinery in mast cells as well. VAMP-2, synaptotagmin-2 and Hrs-2 have been described as being primarily expressed in neuronal systems and are implicated in synaptic vesicle:plasma membrane fusion (27, 28). Both VAMP-2 and synaptotagmin-2 (also known as v-SNAREs) are located on vesicle membranes. The former is a component of the minimal machinery for vesicle docking and fusion, whereas the latter may function as the Ca2+ sensor for triggering synaptic vesicle fusion (29). Hrs-2 (which interacts with SNAP-25), Munc-18-2 (which interacts with syntaxins; reference 30), and complexin II (which interacts with the SNARE complex; reference 31) have been proposed to regulate synaptic vesicle exocytosis through interacting with the components of SNARE complex.
Annexins are a family of structurally related proteins and of the 13 known isoforms (32), we observed in RBL-2H3 cells transcripts for four: annexins I, II, V, and VI. New potential functions of annexins are still being found (33), and some of these may be especially relevant in mast cells. One such is that each of these four isoforms can inhibit PLA2 activity in vitro, and like PLA2, exhibit Ca2+-dependent binding of phospholipid in vivo. Consequently, they are thought to act as Ca2+-dependent, nonspecific inhibitors of PLA2 by depleting substrate. Possibly, they act as counterbalance after activation of PLA2 in stimulated mast cells. Some annexins also mediate membrane fusion. For example annexin I and II have been implicated in exocytosis in neutrophil and chromaffin cells (32). In addition it has recently been reported that based on lipid sequestration, annexin V can act as an inhibitor of protein kinase C (34).
Finally, we observed transcripts for the receptors for two hormones in the RBL-2H3 cells: for GnRH and for melatonin. These neurohormones have direct immunomodulatory effects, and receptors for both have been observed on many immune cells including T cells (35–37) but not previously on mast cells. It will be interesting to see whether they are in fact expressed on mast cells and if so, what their physiological function may be.
Results from Activated Cells: Upregulated Genes.
When gene expression profiles between resting and activated RBL-2H3 cells were compared, we observed that <1% of the genes we analyzed were expressed at detectably different levels in the unstimulated and stimulated cells. However, the expression levels of several dozen genes were found to be dramatically different between the two populations. Table 4 lists 34 tags displaying the most significant increases after stimulation. We estimate (Appendix) that the probabilities are between 54 and 100% that the expression levels for these genes differ by at least a factor of three for the samples taken from the resting and activated states.
Table 4.
Transcripts Increased in Mast Cells after FcεRI Stimulation
| Tag Sequences |
Cells | Gene |
Probability |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Resting | Activated | |||||||||
| Known to be induced after stimulation | % ‡ | |||||||||
| AATACTAGAA | 2 | 43 | MCP-1 § | 100 | ||||||
| GGTGACAGGG | 1 | 8 | M-CSF | 61 | ||||||
| ATCGTGCGCT | 1 | 7 | TGF-β | 54 | ||||||
| Not known to be induced after stimulation | ||||||||||
| Tag No. | ||||||||||
| 1 | GTTCAGGGTC | 43 | 410 | Endogenous retrovirus-like | 100 | |||||
| 2 | GAGCTGACAC | 5 | 85 | Endogenous retrovirus-like | 100 | |||||
| 3 | GTGGACAGCT | 9 | 80 | Endogenous retrovirus-like | 100 | |||||
| 4 | AAACTGTTTG | 9 | 63 | No match | 99 | |||||
| 5 | GATCCGGAGG | 3 | 53 | No match | 100 | |||||
| 6 | CAGGCAGAAA | 4 | 41 | No match | 99 | |||||
| 7 | TTGATCCGGA | 0 | 29 | No match | 100 | |||||
| 8 | CTTTGGGTAC | 2 | 16 | Cyclin G | 81 | |||||
| 9 | GAAGCTAATA | 1 | 16 | MAPKK3 | 91 | |||||
| 10 | AGTTCTGCTT | 1 | 14 | No match | 86 | |||||
| 11 | GTGGCTTACA | 2 | 13 | No match | 69 | |||||
| 12 | AGCAAGAATT | 0 | 10 | Adrenodoxin | 87 | |||||
| 13 | CTTGCTGGTC | 1 | 10 | No match | 72 | |||||
| 14 | TGTCTCAGCC | 0 | 10 | No match | 87 | |||||
| 15 | ACCTTGTCCT | 1 | 9 | No match | 67 | |||||
| 16 | GACTGCGTCG | 1 | 9 | No match | 67 | |||||
| 17 | AGCCTCCCTT | 1 | 8 | No match | 61 | |||||
| 18 | GCGGGCGTCT | 0 | 8 | No match | 80 | |||||
| 19 | GCTGGGCTCC | 1 | 8 | No match | 61 | |||||
| 20 | GTCTCTCTTC | 1 | 8 | No match | 61 | |||||
| 21 | TATTTATTCC | 1 | 8 | No match | 61 | |||||
| 22 | TCATCATTCT | 1 | 8 | No match | 61 | |||||
| 23 | AAACATTGGG | 1 | 7 | No match | 54 | |||||
| 24 | ACACCTCCTG | 0 | 7 | No match | 76 | |||||
| 25 | ACTCACAGGA | 0 | 7 | No match | 76 | |||||
| 26 | CCTGATTGGG | 1 | 7 | No match | 54 | |||||
| 27 | CTCGAGTTTC | 0 | 7 | No match | 76 | |||||
| 28 | GGGCAATAAC | 0 | 7 | No match | 76 | |||||
| 29 | GTTCATTTTG | 1 | 7 | No match | 54 | |||||
| 30 | TGTTCCTTAG | 0 | 5 | rVH6, dsPTP | 63 | |||||
| 31 | GATCGAAGTC | 0 | 4 | preprorelaxin | 56 | |||||
The 34 tags displaying the largest increase after antigen stimulation are shown.
In this and the following table the percentage refers to the probability that there is at least a threefold difference in expression of the transcript in the activated cells.
In this and the following table, genes in boldface were confirmed by Northern blot analysis and/or cloning and sequencing.
Of the 34 tags that were found to be expressed at higher levels in the stimulated cells, three matched the genes encoding monocyte chemotactic protein (MCP)-1, TGF-β, and M-CSF, in the database. The first two are among many chemokines and cytokines previously shown to be upregulated in mast cells and basophils in response to the aggregation of cell surface IgE receptors (Table 11.2 in reference 5); expression of the third was constitutive in the human cell line HMC-1 and up-regulated after stimulation with phorbol ester and ionophore (38). The remaining 31 tags represent newly identified inducible genes. Four of them have matches in the database: cyclin G (39), adrenodoxin (40), rVH6 (41), and preprorelaxin (42).
The sequences of the additional 27 differentially expressed tags were not identical to sequences currently in the rat gene data bank (the last search having been conducted in early July, 1998). Because even a single nucleotide difference will cause a sequence to be labeled as no match, unidentified sequences could represent species variants rather than novel genes. However, because the tags are so short (10 specific bp) and allelic variations so infrequent, it is unlikely that many of the “no matches” simply represent allelic variants of known genes.
Further Study of Differentially Expressed Genes.
It was of obvious interest to explore further some of the differentially expressed genes to see whether some of those not represented in the sequence database (no match) could be identified.
Tag 1 (Table 4) was chosen because it is highly expressed in both unstimulated and stimulated cells and increases tenfold upon stimulation. Tag 9 was chosen somewhat at random as typical of a sequence differentially expressed at an intermediate level. Labeled oligos representing these tags were used to probe Northern blots as well as an RBL-2H3 cDNA library from resting cells.
The probe from tag 1 hybridized to a 6-kb transcript on Northern blots (data not shown) and identified a 1.5-kb cDNA clone during screening of the library. It is likely that this cDNA clone is derived from an endogenous retrovirus-like DNA sequence since it shows significant similarity to retroviral sequences in the database. For example it has 84% homology to a rat endogenous retroviral RAL element (43), 83% similarity to proviral MO-VL30 in the tpl-2 locus (44) and 78% homology to the VL30 element (45). Sequence analysis of the cDNA identified sequences of tags 2 and 3 (Table 4) just upstream of a short (8–6 bp, respectively) polyadenine stretch. It is plausible that a small amount of internal oligo dT-priming occurred at these sites.
The cDNA cloned with a tag 9 probe is a 2.5-kb fragment and represents the full transcript as confirmed by Northern blotting (Fig. 3). It codes for rat MAPKK, a protein that had not previously been characterized in rats but which has an amino acid sequence 99% identical to the murine MAPKK3 protein. MAPKK3 is a member of the MAPKK supergene family and phosphorylates and activates p38, a member of the MAPK family (46). The original prototypes of the MAPK family are ERK1 and 2, which are activated by MAPKK1 and 2 through phosphorylations on tyrosine and threonine, and inactivated via dephosphorylation of either of the phosphorylated residues by MAPK phosphatases such as CT100 and PAC1. A variety of extracellular stimuli promote expression of these enzymes (47).
Figure 3.
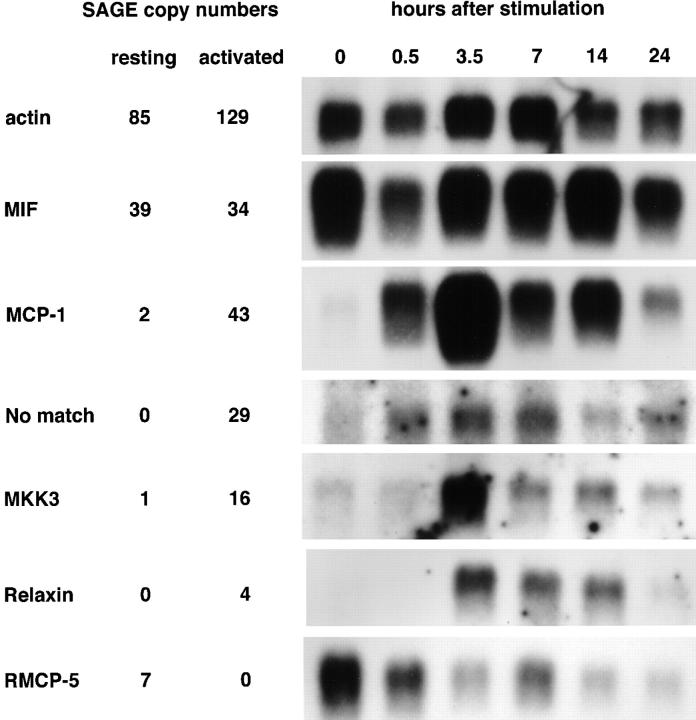
Northern blots of selected genes. Messenger RNAs were prepared and used for Northern blotting with end-labeled 15–40-bp oligonucleotides in all cases except for RMCP-5; for the latter a randomly labeled PCR-amplified cDNA was used. The β-actin blot is a loading control. The columns to the left of the blots list the copy numbers of the tags observed in the resting and activated SAGE libraries for the indicated genes. MKK3 refers to MAPKK3.
Other Differentially Expressed Genes.
Transcription of the recently identified ubiquitous MAPK phosphatase, rVH6, is induced during nerve growth factor-mediated differentiation in PC12 cells (41). SAGE analysis revealed its expression in RBL-2H3 cells in response to stimulation through FcεRI (Table 4). It is interesting to speculate that this is linked to the contemporaneous induction of MAPKK3.
A gene whose upregulation was unanticipated was that for preprorelaxin (Table 4). This preprohormone structurally belongs to the preproinsulin family and it is primarily produced by corpora lutea (48). Relaxin's well characterized function is to inhibit uterine contraction and to contribute to the remodeling and softening of tissues in the birth canal during pregnancy (49, 50). Among its other biological activities, it has been reported to inhibit the release of histamine by mast cells in vitro (51), and to alleviate experimental allergic asthma in vivo (52). Since we have now shown it to be upregulated in mast cells after stimulation (Table 4 and Fig. 3), it will be particularly interesting to explore its role in mast cell physiology.
Table 5 lists the nine tags whose expression showed the largest decrease when the RBL cells were stimulated. The only sequence that matched with a citation in the rat database was for RMCP-5. Its decreased expression after stimulation was confirmed by Northern blotting (Fig. 3).
Table 5.
Transcripts Decreased in Mast Cells after FcεRI Stimulation*
| Tag sequence |
Cells | Gene |
Probability |
|||||
|---|---|---|---|---|---|---|---|---|
| Resting | Activated | |||||||
| TGATCTTTTT | 11 | 1 | No match |
%76 |
||||
| TTGTATAATA | 10 | 1 | '' | 72 | ||||
| TTCATTCATG | 9 | 1 | '' | 67 | ||||
| TGCAGTCATC | 8 | 0 | '' | 80 | ||||
| TGGAAGGACC | 8 | 0 | '' | '' | ||||
| GAGACTTGCT | 7 | 0 | '' | 76 | ||||
| GCCAAATTTG | 7 | 0 | '' | '' | ||||
| GGGGTAAGAG | 7 | 0 | '' | '' | ||||
| TGGGATCTGG | 7 | 0 | RMCP-5 | '' | ||||
The nine tags displaying the largest decrease in expression after stimulation as shown.
Some genes or gene products that were previously shown to be upregulated in activated mast cells were not found in either of our SAGE libraries. There are several possible explanations for this apparent discrepancy: (a) since in various published studies a variety of mast cell lines, cultured mast cells, and different tissue mast cells were examined, any individual cell line may not display the full heterogeneity that characterizes the different cell preparations; (b) different studies employed different times and doses of antigen than used here; (c) if the expression of a gene was low we may not have observed it since in order to detect a transcript with 95% probability, there would have had to be on the average, 23 copies of it per cell in either population (Appendix); (d) some unanticipated deficiency in the generation of the SAGE libraries.
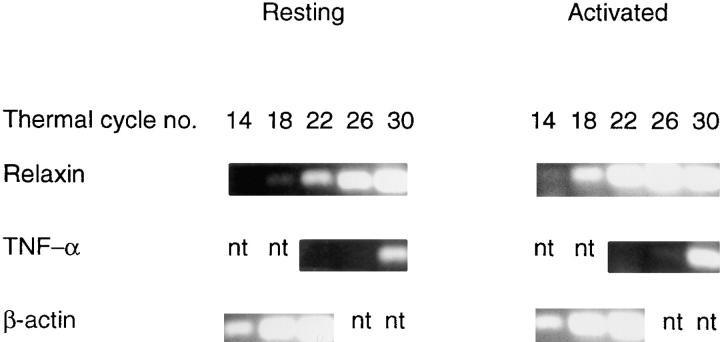
To distinguish between possibilities c and d, we used RT-PCR to compare with preprorelaxin, the expression of two representative transcripts (TNF-α and IL-3) that were not revealed in our SAGE libraries. PCR amplification for each transcript was performed for a variable number of thermal cycles. To assure that spurious differences in amplification were not generated, β-actin was used as a control. Fig. 4 shows that the β-actin transcript was observed after only 14 cycles and that there was no appreciable difference between the resting and activated cells. Transcripts of preprorelaxin were readily observed after 18 cycles and were clearly upregulated in the stimulated cells. TNF-α has previously been shown to be constitutively present and upregulated, but at the time we chose to prepare the library from stimulated cells (3.5 h), the observed enhancement was modest consistent with our RT-PCR results (compare our Fig. 4 with Fig. 3 in reference 53). However, it took 30 cycles for the TNF-α transcript to reach the level attained by preprorelaxin after only 22 cycles.
Figure 4.
Results of RT-PCR amplification of mRNAs from resting and activated cells using primers specific for selected transcripts. Items shown as “nt” were not tested.
The prior study showed that the level of IL-3 transcripts was enhanced after receptor-mediated activation but was on the decline when probed at 4 h; in our study, the level of PCR product observed on the photograph of the gel after 30 cycles was so low that it would not have reproduced adequately, and we saw no enhancement in the PCR product prepared from the mRNA of the cells activated for 3.5 h (data not shown). This result indicates that the level of expression of some upregulated genes may have been below the threshold necessary for us to have detected it by the SAGE analysis.
Discussion
The term transcriptome has been coined to describe the identity and multiplicity of mRNAs expressed by a defined population of cells (21). The object of the work described in this paper was to begin to define such a profile for mast cells before and after stimulation of the cells by one of their important cell surface receptors, FcεRI. To get sufficient amounts of material (mRNA) we used a rodent mucosal mast cell line, RBL-2H3, even though we recognize that findings on the RBL-2H3 cells will ultimately need to be extended explicitly to their normal counterparts and to human cells. Clearly, this extension can be carried out much more incisively on the basis of the initial work on the rat cell line as has already proven to be the case with respect to FcεRI and many other molecules and phenomena first studied in this cell line.
We obtained a partial transcriptome of RBL-2H3 cells by generating >40,000 tags derived from transcripts of resting and activated cells using the SAGE method. The results of the analysis provided a quantitative overview of the expression of 11,300 genes in the RBL-2H3 cells. Although the more limited sequences in the rat database may have precluded immediate identification of many of these tags, this is clearly a temporary situation and the genes encoding a significant number of the tags have already been identified. In this section we discuss briefly several of our new findings that we think are of special interest.
MIF.
The observation that mRNA for MIF, a cytokine not previously associated with mast cells, is expressed constitutively in high amounts is interesting. MIF was first identified 30 yr ago as a soluble factor that is produced by activated lymphocytes (T cells) and that inhibits the random migration of cultured macrophages. Recent studies have identified macrophages and cells of the anterior pituitary as additional important sources of production of MIF (26). In both cases, MIF is found in preformed granules and is released in response to inflammatory stimuli such as lipopolysaccharide (54, 55). We found that like macrophages and pituitary cells, RBL-2H3 cells, normal peritoneal mast cells, the human mast cell line HMC-1, and the human basophil cell line KU812, constitutively express MIF at high levels.
The high level expression of MIF in mast cells provides further evidence that host defense is a major function of these cells and suggests that MIF is one of the mediators. Direct evidence that mast cells play an important role in host defense comes from two studies using mast cell-deficient W/Wv mice. The mortality for mast cell-deficient W/Wv mice after challenge with virulent bacteria is significantly higher than their litter mate controls (56, 57) and their reconstitution with cultured mast cells restores their resistance to infection.
Mast cell-derived TNF-α is a crucial mediator in this response by recruiting bactericidal leukocytes, but other factor(s) may contribute since exogenous TNF-α only partially restores the host protection mediated by mast cells. Potentially, MIF fills this role. It activates macrophages to produce TNF-α and nitrous oxide (58), and thus may contribute to the development of an inflammatory response, a critical element of innate immunity. It would be interesting to explore to what extent simultaneous administration of exogenous MIF and TNF-α into mast cell–deficient W/Wv mice would restore host protection against infection to a level equivalent to that provided by mast cells.
It is noteworthy that hyperplasia of mast cells and high levels of MIF have been observed in synovium from patients with rheumatoid arthritis and that administration of antibodies capable of neutralizing MIF delayed the onset of arthritis in a mouse model (59–61). These observations may relate to the central role MIF is thought to play in delayed-type hypersensitivity (DTH). Thus, MIF mRNA and protein are expressed in DTH lesions and neutralizing anti-MIF antibodies significantly inhibit the development of DTH (62).
Mast cells have been long been implicated in DTH. The best evidence is defective elicitation of DTH in W/Wv and SI/SId mast cell–deficient mice, and restoration of the DTH response after adoptive transfer of cultured mast cells into these mice (63). Our finding that both mast cells and macrophages are important sources of MIF, may relate to the two types of DTH that have been observed: mast cell– dependent DTH as in footpad DTH, and macrophage- dependent DTH as in classic tuberculin DTH (64).
Exocytotic Machinery.
Aggregation of FcεRI causes an increase in the intracellular concentration of Ca2+ and an activation of protein kinase C—two events that are required for mediator release (65). Although several low molecular weight GTPases have been implicated in the process (66, 67), many of the molecular components of the mast cell exocytotic machinery remain unknown.
In this study, we found that RBL-2H3 cells express at least seven proteins known to be involved in exocytosis in other systems, particularly at nerve terminals. According to the SNARE hypothesis (27), four proteins constitute the core exocytotic machinery at the nerve terminal: synaptotagmin and VAMP (v-SNAREs) on the vesicle membrane, SNAP-25, and syntaxin (t-SNAREs) on the target membrane. Two cytosolic proteins (NSF and SNAP) are involved in the recycling of vesicles after fusion. The process is believed to be regulated by other proteins such as Hrs-2, complex II, and Munc-18 due to their ability to interact with the core complex. Finding that these proteins are common to neurons and mast cells (see Results) suggest they play similar roles in the two systems.
Neurohormone Receptors.
There is increasing evidence for an integrated bidirectionally-regulated interaction between the neuroendocrine and immune systems (68). For example, both systems express and respond to a variety of common regulatory molecules (neuropeptides, cytokines, and steroids). In this study, we uncovered another example for such an interaction by finding that mast cells, like cells of the pituitary, express MIF, as well as mRNA for the receptors for GnRH and melatonin. By regulating gonadal steroids the hypothalamic hormone GnRH modulates immune responses indirectly but it can also influence such responses directly by stimulating proliferation of T and B cells through their receptors for GnRH (35, 69, 70). Similarly, melatonin, produced in the pineal gland of the hypothalamus, has immunoregulatory properties: it can activate monocytes and neutrophils (36), and induce proliferation of T cells and their release of cytokines (37).
Other Differentially Expressed Genes.
We anticipated that the genes for MCP-1, M-CSF, and TGF-β would be induced in the activated cells (5). Of these, MCP-1 deserves special attention because treatment with anti-MCP-1 antibody reduces recruitment of monocytes by 65% during the development of a late phase allergic reaction in humans (5). The high level of expression (∼650 copies/cell) of MCP-1 mRNA in activated RBL-2H3 cells, and its persistence (at least 24 h), is consistent with its having an important role in the delayed reaction.
Among the inducible genes, MAPKK3 is also particularly interesting. Because they are involved in the regulation of such a diverse array of cellular process, MAPK cascades have attracted considerable attention in the past 10 yr (46). The kinases are themselves regulated by phosphorylation/dephosphorylation. In the RBL cells, both the mRNA and protein for MAPKK3 are increased more than tenfold after activation. This response is specific and other members of the MAPK family, such as MAPKK 1, 2, and 5 were not differentially expressed in the resting and activated states in RBL-2H3 cells (data not shown). It will be interesting to determine the physiological consequence of increases in the MAPKK3 protein after activation.
In recent years, the changes in expression of genes in activated mast cells have been actively pursued. The standard methods such as Northern blotting (71), RT-PCR (53), and subtraction libraries (72) have been used in such studies. Although several dozens of cytokines, chemokines, and other immunomodulatory molecules have been shown to be upregulated after activation (5), such methods can analyze only limited numbers of genes in a single study and the quantitative analysis of the transcription of individual genes is difficult. We have found that the SAGE method is particularly useful for such studies and in this initial investigation have already uncovered a rich trove of expressed genes, exploration of whose function is likely to give further insight into the physiology of mast cells.
Acknowledgments
We are grateful to Dr. Kenneth W. Kinzler for his generous advice and assistance, and to him and Dr. Juan Rivera for careful review of the manuscript.
Abbreviations used in this paper
- DTH
delayed-type hypersensitivity
- GnRH
gonadotropin releasing hormone
- MAPKK
mitogen-activated protein kinase kinase
- MCP
monocyte chemotactic protein
- MIF
macrophage migration inhibitory factor
- PLA2
phospholipase A2
- RT
reverse transcription
- SAGE
serial analysis of gene expression
- VAMP
vesicle- associated membrane protein
Appendix
Detecting a Transcript.
Assume that a single cell contains 300,000 transcripts, that each transcript has an equal chance of detection, and that 20,000 tags are generated for each of two cell lines. Then a specific mRNA with an average (mean) of N transcripts in each cell line will generate on average a total of T = N/7.5 tags. Assuming a Poisson distribution on the total number of tags actually generated, the probability that no tags are seen is e-T, which is ∼5% when T = 3. Thus a transcript that appears a mean of 22.5 times in two cell lines, i.e., at an average abundance of 0.0075%, has a 95% chance of being detected.
Changes in Levels of Expression.
In activated and resting cells respectively, assume that A and B copies are found of a tag corresponding to a particular mRNA species. We wish to calculate the probability that the expression of this mRNA has increased by a factor of at least F. This problem is best approached through Bayesian statistical analysis (chapter 8 in reference 73). Assume that given the actual concentration of the mRNA in the activated and resting cells, the expected numbers of tags generated by the experiment are Y and Z, respectively, and let x = Y/(Y+Z). The variable x takes on a value between 0 and 1, and we need to assume a prior probability distribution (i.e., before observing A and B) for x. For mathematical convenience, it is easiest to choose this distribution from among the two-parameter family of beta distributions (chapter 6 in reference 73). The beta distribution with parameters a and b has a probability density function
 |
1 |
where a and b are both positive real numbers, Γ is the gamma function and x ranges as stated from 0 to 1. With beta (a, b) as the prior, Bayesian analysis (details omitted) implies that after observing A and B tags respectively from activated and resting cells, the posterior probability density for x is simply beta (A + a, B + b). Assuming we generated an approximately equal number of tags for both resting and activated cells, for the mRNA expression to be increased by a factor of at least F, x must fall between F/(F + 1) and 1. We may calculate this probability simply by integrating beta (A + a, B + b) from F/(F + 1) to 1.
For a specific experiment, we need to choose appropriate values for the parameters a and b. So-called “uninformative” priors use a = b = 1, for which the beta distribution reduces to the uniform distribution over the interval [0,1]. However, in this experiment our data suggest the expression levels of most genes do not differ greatly in resting and activated cells, similar to what has been observed when normal and cancerous cells were compared (7, 74). Thus our prior distribution should not be uniform but should be peaked near 0.05. This corresponds to a beta distribution with parameters a and b equal, and greater than 1. The larger the value of the parameters a and b, the greater the concentration of the prior probability for x near 0.5, and the more evidence that is needed to infer a given change in the level of expression. For detectable mRNA species, the complete set of ordered pairs representing the number of tags observed in resting and activated cells (data not shown), implies a reasonable choice for the parameters a and b is 2.0.
We apply this analysis to calculate the probability P that the level of expression of a given mRNA species has been increased by at least threefold. Assuming the prior distribution beta (2,2), and noting that for positive integral values of n, Γ (n) = (n − 1)!, we have
 |
2 |
This equation is used to produce the probabilities in Tables 4 and 5.
References
- 1.Abraham SN, Malaviya R. Mast cells in infection and immunity. Infect Immun. 1997;65:3501–3508. doi: 10.1128/iai.65.9.3501-3508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selye, H. 1965. The Mast Cells. Butterworth Inc., Washington DC. 1–498.
- 3.Rigby, L., M.D. Hulett, R.I. Brinkworth, and P.M. Hogarth. 1998. The structural basis of the interaction of IgE and FcεRI. In IgE Receptor (FcεRI) Function in Mast Cells and Basophils. M.M. Hamawy, editor. R.G. Landes Company, Austin, TX. 7–32.
- 4.Hamawy, M.M., and W.D. Swaim. 1997. FcεRI-mediated cell degranulation, proliferation and adhesion. In IgE Receptor (FcεRI) Function in Mast Cells and Basophils. M.M. Hamawy, editor. R.G. Landes Company, Austin, TX. 173–180.
- 5.Gordon, J.R. 1997. FcεRI-induced cytokine production and gene expression. In IgE Receptor (FcεRI) Function in Mast Cells and Basophils. M.M. Hamawy, editor. R.G. Landes Company, Austin, TX. 209–242.
- 6.Madden SL, Galella EA, Zhu J, Bertelsen AH, Beaudry GA. SAGE transcript profiles for p53-dependent growth regulation. Oncogene. 1997;15:1079–1085. doi: 10.1038/sj.onc.1201091. [DOI] [PubMed] [Google Scholar]
- 7.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis [see comments] Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 8.Barsumian EL, Isersky C, Petrino MG, Siraganian RP. IgE-induced histamine release from rat basophilic leukemia cell lines: isolation of releasing and nonreleasing clones. Eur J Immunol. 1981;11:317–323. doi: 10.1002/eji.1830110410. [DOI] [PubMed] [Google Scholar]
- 9.Eccleston E, Leonard BJ, Lowe JS, Welford HJ. Basophilic leukaemia in the albino rat and a demonstration of the basopoietin. Nature New Biol. 1973;244:73–76. doi: 10.1038/newbio244073b0. [DOI] [PubMed] [Google Scholar]
- 10.Kulczycki A, Jr, Isersky C, Metzger H. The interaction of IgE with rat basophilic leukemia cells. I. Evidence for specific binding of IgE. J Exp Med. 1974;139:600–616. doi: 10.1084/jem.139.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seldin DC, Adelman S, Austen KF, Stevens RL, Hein A, Caufield JP, Woodbury RG. Homology of the rat basophilic leukemia cell and the rat mucosal mast cell. Proc Natl Acad Sci USA. 1985;82:3871–3874. doi: 10.1073/pnas.82.11.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu FT, Bohn JW, Ferry EL, Yamamoto H, Molinaro CA, Sherman LA, Klinman NR, Katz DH. Monoclonal dinitrophenyl-specific murine IgE antibody: preparation, isolation, and characterization. J Immunol. 1980;124:2728–2737. [PubMed] [Google Scholar]
- 13.Butterfield JH, Weiler D, Dewald G, Gleich GJ. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leukocyte Res. 1988;12:345–355. doi: 10.1016/0145-2126(88)90050-1. [DOI] [PubMed] [Google Scholar]
- 14.Kishi K. A new leukemia cell line with Philadelphia chromosome characterized as basophil precursors. Leukocyte Res. 1985;9:381–390. doi: 10.1016/0145-2126(85)90060-8. [DOI] [PubMed] [Google Scholar]
- 15.Holgate ST, Lewis RA, Austen KF. 3′,5′-Cyclic adenosine monophosphate-dependent protein kinase of the rat serosal mast cell and its immunologic activation. J Immunol. 1980;124:2093–2099. [PubMed] [Google Scholar]
- 16.Kaplan JE, Cardarelli PM, Rourke FJ, Weston LK, Moon DG, Blumenstock FA. Fibronectin augments binding of fibrin to macrophages. J Lab Clin Med. 1989;113:168–176. [PubMed] [Google Scholar]
- 17.Gilchrist M, Macdonald AJ, Neverova I, Ritchie B, Befus AD. Optimization of the isolation and effective use of mRNA from rat mast cells. J Immunol Methods. 1997;201:207–214. doi: 10.1016/s0022-1759(96)00230-x. [DOI] [PubMed] [Google Scholar]
- 18.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression [see comments] Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 19.Anonymous. 1994. Current Protocols in Molecular Biology. J. Wiley & Sons, New York.
- 20.Wofsy C, Kent UM, Mao S-Y, Metzger H, Goldstein B. Kinetics of tyrosine phosphorylation when IgE dimers bind to Fc receptors on rat basophilic leukemia cells. J Biol Chem. 1995;270:20264–20272. doi: 10.1074/jbc.270.35.20264. [DOI] [PubMed] [Google Scholar]
- 21.Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Jr, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 22.Bocek P, Jr, Dráberová L, Dráber P, Pecht I. Characterization of Fcγ receptors on rat mucosal mast cells using a mutant FcεRI-deficient rat basophilic leukemia line. Eur J Immunol. 1995;25:2948–2955. doi: 10.1002/eji.1830251035. [DOI] [PubMed] [Google Scholar]
- 23.Lutzelschwab C, Pejler G, Aveskogh M, Hellman L. Secretory granule proteases in rat mast cells. Cloning of 10 different serine proteases and a carboxypeptidase A from various rat mast cell populations. J Exp Med. 1997;185:13–29. doi: 10.1084/jem.185.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schayer RW. Histidine decarboxylase in mast cells. Annu NY Acad Sci. 1963;103:164–172. doi: 10.1111/j.1749-6632.1963.tb53696.x. [DOI] [PubMed] [Google Scholar]
- 25.Gilfillan, A.M. 1997. Signal transduction pathways regulating arachidonic acid metabolite generation following FcεRI aggregation. In IgE Receptor (FcεRI) Function in Mast Cells and Basophils. M.M. Hamawy, editor. R.G. Landes Company, Austin, TX. 181–208.
- 26.Donnelly SC, Bucala R. Macrophage migration inhibitory factor: a regulator of glucocorticoid activity with a critical role in inflammatory disease. Mol Med Today. 1997;3:502–507. doi: 10.1016/S1357-4310(97)01133-7. [DOI] [PubMed] [Google Scholar]
- 27.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 28.Bean AJ, Seifert R, Chen YA, Sacks R, Scheller RH. Hrs-2 is an ATPase implicated in calcium-regulated secretion. Nature. 1997;385:826–829. doi: 10.1038/385826a0. [DOI] [PubMed] [Google Scholar]
- 29.Goda Y. SNAREs and regulated vesicle exocytosis [comment] Proc Natl Acad Sci USA. 1997;94:769–772. doi: 10.1073/pnas.94.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hata Y, Sudhof TC. A novel ubiquitous form of Munc-18 interacts with multiple syntaxins. Use of the yeast two-hybrid system to study interactions between proteins involved in membrane traffic. J Biol Chem. 1995;270:13022–13028. doi: 10.1074/jbc.270.22.13022. [DOI] [PubMed] [Google Scholar]
- 31.McMahon HT, Missler M, Li C, Sudhof TC. Complexins: cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83:111–119. doi: 10.1016/0092-8674(95)90239-2. [DOI] [PubMed] [Google Scholar]
- 32.Raynal P, Pollard HB. Annexins: the problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim Biophys Acta. 1994;1197:63–93. doi: 10.1016/0304-4157(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 33.Ishitsuka R, Kojima K, Utsumi H, Ogawa H, Matsumoto I. Glycosaminoglycan binding properties of annexin IV,V, and VI. J Biol Chem. 1998;273:9935–9941. doi: 10.1074/jbc.273.16.9935. [DOI] [PubMed] [Google Scholar]
- 34.Dubois T, Mira JP, Feliers D, Solito E, Russo-Marie F, Oudinet JP. Annexin V inhibits protein kinase C activity via a mechanism of phospholipid sequestration. Biochem J. 1998;330:1277–1282. doi: 10.1042/bj3301277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobson JD, Crofford LJ, Sun L, Wilder RL. Cyclical expression of GnRH and GnRH receptor mRNA in lymphoid organs. Neuroendocrinology. 1998;67:117–125. doi: 10.1159/000054306. [DOI] [PubMed] [Google Scholar]
- 36.Barjavel MJ, Mamdouh Z, Raghbate N, Bakouche O. Differential expression of the melatonin receptor in human monocytes. J Immunol. 1998;160:1191–1197. [PubMed] [Google Scholar]
- 37.Maestroni GJ. T-helper-2 lymphocytes as a peripheral target of melatonin. J Pineal Res. 1995;18:84–89. doi: 10.1111/j.1600-079x.1995.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson G, Svensson V, Nilsson K. Constitutive and inducible cytokine mRNA expression in the human mast cell line HMC-1. Scand J Immunol. 1995;42:76–81. doi: 10.1111/j.1365-3083.1995.tb03628.x. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K, Kanaoka Y, Jinno S, Nagata A, Ogiso Y, Shimizu K, Hayakawa T, Nojima H, Okayama H. Cyclin G: a new mammalian cyclin with homology to fission yeast Cig1. Oncogene. 1993;8:2113–2118. [PubMed] [Google Scholar]
- 40.Sagara Y, Watanabe Y, Kawamura K, Yubisui T. Cloning and sequence analysis of a full-length cDNA of rat adrenodoxin. Biol Pharm Bull. 1996;19:39–41. doi: 10.1248/bpb.19.39. [DOI] [PubMed] [Google Scholar]
- 41.Mourey RJ, Vega QC, Campbell JS, Wenderoth MP, Hauschka SD, Krebs EG, Dixon JE. A novel cytoplasmic dual specificity protein tyrosine phosphatase implicated in muscle and neuronal differentiation. J Biol Chem. 1996;271:3795–3802. doi: 10.1074/jbc.271.7.3795. [DOI] [PubMed] [Google Scholar]
- 42.Bani D. Relaxin: a pleiotropic hormone. Gen Pharmacol. 1997;28:13–22. doi: 10.1016/s0306-3623(96)00171-1. [DOI] [PubMed] [Google Scholar]
- 43.Nakamuta M, Furuich M, Takahashi K, Suzuki N, Endo H, Yamamoto M. Isolation and characterization of a family of rat endogenous retroviral sequences. Virus Genes. 1989;3:69–83. doi: 10.1007/BF00301988. [DOI] [PubMed] [Google Scholar]
- 44.Makris A, Patriotis C, Bear SE, Tsichlis PN. Structure of a Moloney murine leukemia virus-virus-like 30 recombinant: implications for transduction of the c-Ha-ras proto-oncogene. J Virol. 1993;67:1286–1291. doi: 10.1128/jvi.67.3.1286-1291.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Firulli BA, Anderson GR, Stoler DL, Estes SD. Anoxia-inducible rat VL30 elements and their relationship to ras-containing sarcoma viruses. J Virol. 1993;67:6857–6862. doi: 10.1128/jvi.67.11.6857-6862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su B, Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr Opin Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 47.Keyse SM. An emerging family of dual specificity MAP kinase phosphatases. Biochim Biophys Acta. 1995;1265:152–160. doi: 10.1016/0167-4889(94)00211-v. [DOI] [PubMed] [Google Scholar]
- 48.Hudson P, Haley J, Cronk M, Shine J, Niall H. Molecular cloning and characterization of cDNA sequences coding for rat relaxin. Nature. 1981;291:127–131. doi: 10.1038/291127a0. [DOI] [PubMed] [Google Scholar]
- 49.Zhao S, Kuenzi MJ, Sherwood OD. Monoclonal antibodies specific for rat relaxin. IX. Evidence that endogenous relaxin promotes growth of the vagina during the second half of pregnancy in rats. Endocrinology. 1996;137:425–430. doi: 10.1210/endo.137.2.8593785. [DOI] [PubMed] [Google Scholar]
- 50.Hwang JJ, Sherwood OD. Monoclonal antibodies specific for rat relaxin. III. Passive immunization with monoclonal antibodies throughout the second half of pregnancy reduces cervical growth and extensibility in intact rats. Endocrinology. 1988;123:2486–2490. doi: 10.1210/endo-123-5-2486. [DOI] [PubMed] [Google Scholar]
- 51.Masini E, Bani D, Bigazzi M, Mannaioni PF, Bani-Sacchi T. Effects of relaxin on mast cells. In vitro and in vivo studies in rats and guinea pigs. J Clin Invest. 1994;94:1974–1980. doi: 10.1172/JCI117549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bani D, Ballati L, Masini E, Bigazzi M, Sacchi TB. Relaxin counteracts asthma-like reaction induced by inhaled antigen in sensitized guinea pigs. Endocrinology. 1997;138:1909–1915. doi: 10.1210/endo.138.5.5147. [DOI] [PubMed] [Google Scholar]
- 53.Chang EY, Szallasi Z, Acs P, Raizada V, Wolfe PC, Fewtrell C, Blumberg PM, Rivera J. Functional effects of overexpression of protein kinase C-alpha, -beta, -delta, -epsilon, and -eta in the mast cell line RBL-2H3. J Immunol. 1997;159:2624–2632. [PubMed] [Google Scholar]
- 54.Nishino T, Bernhagen J, Shiiki H, Calandra T, Dohi K, Bucala R. Localization of macrophage migration inhibitory factor (MIF) to secretory granules within the corticotrophic and thyrotrophic cells of the pituitary gland. Mol Med. 1995;1:781–788. [PMC free article] [PubMed] [Google Scholar]
- 55.Calandra T, Bernhagen J, Mitchell RA, Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Echtenacher B, Mannel DN, Hultner L. Critical protective role of mast cells in a model of acute septic peritonitis. Nature. 1996;381:75–77. doi: 10.1038/381075a0. [DOI] [PubMed] [Google Scholar]
- 57.Malaviya R, Ikeda T, Ross E, Abraham SN. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature. 1996;381:77–80. doi: 10.1038/381077a0. [DOI] [PubMed] [Google Scholar]
- 58.Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF) Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 59.Kopicky-Burd JA, Kagey-Sobotka A, Peters SP, Dvorak AM, Lennox DW, Lichtenstein LM, Wigley FM. Characterization of human synovial mast cells. J Rheumatol. 1988;15:1326–1333. [PubMed] [Google Scholar]
- 60.Metz CN, Bucala R. Role of macrophage migration inhibitory factor in the regulation of the immune response. Adv Immunol. 1997;66:197–223. doi: 10.1016/s0065-2776(08)60598-2. [DOI] [PubMed] [Google Scholar]
- 61.Mikulowska A, Metz CN, Bucala R, Holmdahl R. Macrophage migration inhibitory factor is involved in the pathogenesis of collagen type II-induced arthritis in mice. J Immunol. 1997;158:5514–5517. [PubMed] [Google Scholar]
- 62.Bernhagen J, Bacher M, Calandra T, Metz CN, Doty SB, Donnelly T, Bucala R. An essential role for macrophage migration inhibitory factor in the tuberculin delayed-type hypersensitivity reaction. J Exp Med. 1996;183:277–282. doi: 10.1084/jem.183.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Askenase PW, Van Loveren H, Kraeuter-Kops S, Ron Y, Meade R, Theoharides TC, Nordlund JJ, Scovern H, Gerhson MD, Ptak W. Defective elicitation of delayed-type hypersensitivity in W/Wv and SI/SId mast cell-deficient mice. J Immunol. 1983;131:2687–2694. [PubMed] [Google Scholar]
- 64.Torii I, Morikawa S, Harada T, Kitamura Y. Two distinct types of cellular mechanisms in the development of delayed hypersensitivity in mice: requirement of either mast cells or macrophages for elicitation of the response. Immunology. 1993;78:482–490. [PMC free article] [PubMed] [Google Scholar]
- 65.Beaven, M.A., and T. Kassessinoff. 1997. Role of phospholipases, protein kinases and calcium in FcεRI-induced secretion. In IgE Receptor (FcεRI) Function in Mast Cells and Basophils. M.M. Hamawy, editor. R.G. Landes Company, Austin, TX. 55–73.
- 66.Smith J, Thompson N, Thompson J, Armstrong J, Hayes B, Crofts A, Squire J, Teahan C, Upton L, Solari R. Rat basophilic leukaemia (RBL) cells overexpressing Rab3a have a reversible block in antigen-stimulated exocytosis. Biochem J. 1997;323:321–328. doi: 10.1042/bj3230321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roa M, Paumet F, Le Mao J, David B, Blank U. Involvement of the ras-like GTPase rab3d in RBL-2H3 mast cell exocytosis following stimulation via high affinity IgE receptors (Fc epsilonRI) J Immunol. 1997;159:2815–2823. [PubMed] [Google Scholar]
- 68.Wilder RL. Neuroendocrine-immune system interactions and autoimmunity. Annu Rev Immunol. 1995;13:307–338. doi: 10.1146/annurev.iy.13.040195.001515. [DOI] [PubMed] [Google Scholar]
- 69.Marchetti B, Guarcello V, Morale MC, Bartoloni G, Raiti F, Palumbo G, Jr, Farinella Z, Cordaro S, Scapagnini U. Luteinizing hormone-releasing hormone (LHRH) agonist restoration of age-associated decline of thymus weight, thymic LHRH receptors, and thymocyte proliferative capacity. Endocrinology. 1989;125:1037–1045. doi: 10.1210/endo-125-2-1037. [DOI] [PubMed] [Google Scholar]
- 70.Morale MC, Batticane N, Bartoloni G, Guarcello V, Farinella Z, Galasso MG, Marchetti B. Blockade of central and peripheral luteinizing hormone-releasing hormone (LHRH) receptors in neonatal rats with a potent LHRH-antagonist inhibits the morphofunctional development of the thymus and maturation of the cell-mediated and humoral immune responses. Endocrinology. 1991;128:1073–1085. doi: 10.1210/endo-128-2-1073. [DOI] [PubMed] [Google Scholar]
- 71.Burd PR, Rogers HW, Sundararajan J, Wilson SD, Dvorak AM, Galli SJ, Dorf ME. Interleukin 3-dependent and -independent mast cells stimulated with IgE and antigen express multiple cytokines. J Exp Med. 1989;170:245–257. doi: 10.1084/jem.170.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho JJ, Vliagoftis H, Rumsaeng V, Metcalfe DD, Oh CK. Identification and categorization of inducible mast cell genes in a subtraction library. Biochem Biophys Res Commun. 1998;242:226–230. doi: 10.1006/bbrc.1997.7644. [DOI] [PubMed] [Google Scholar]
- 73.Kendall, M. and A. Stuart. 1977. The Advanced Theory of Statistics. Vol. 1: Distribution Theory. Macmillan Publishing Co., Inc., New York.
- 74.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]