Abstract
We report here the identification of a new shared human melanoma antigen recognized by a human leukocyte antigen (HLA)-A*68011–restricted cytotoxic T lymphocyte clone (CTL 128). The cDNA encoding this antigen is composed of a partially spliced form of the melanocyte differentiation antigen tyrosinase-related protein (TRP)-2, containing exons 1–4 with retention of intron 2 and part of intron 4 (TRP-2–INT2). The sequence coding for the antigenic epitope is located at the 5′ end of intron 2 and is available for translation in the same open reading frame of the fully spliced TRP-2 mRNA. This peptide is also recognized by CTL 128 when presented by the HLA-A*3301, a member of the HLA-A3–like supertype that includes the HLA-A*68011. Quantitative reverse transcription PCR analysis carried out on total and/or cytoplasmic mRNA demonstrated that, in contrast to the fully spliced TRP-2 mRNA expressed in melanomas, normal skin melanocytes, and retina, the TRP-2–INT2 mRNA could be detected at significant levels in melanomas but not in normal cells of the melanocytic lineage. Instead, in these normal samples, both the spliced and the unspliced transcript of gp100 were expressed at high levels. Absence of endogenous TRP-2–INT2 expression in melanocytes was also confirmed by lack of recognition of HLA-A*68011–transduced, TRP-2+ melanocyte lines by CTL 128. These results indicate that a partially spliced form of a differentiation antigen mRNA, present in the cytoplasmic compartment of neoplastic but not normal cells of the melanocytic lineage, can be the source of a melanoma-restricted T cell epitope.
Keywords: melanoma, antigen, T cell, specificity, intron-encoded epitope
Several studies have demonstrated the existence of an MHC class I–restricted CTL response specifically directed against human tumors (1). The role of such tumor-specific effectors in mediating the regression of metastatic melanomas has been suggested by the results of several clinical trials of both adoptive immunotherapy (2, 3) and vaccination (4–6). The determinants recognized by such CTLs have been studied extensively on melanoma cells, and several antigens have been characterized at the molecular level (1). According to the pattern of gene expression in normal and neoplastic cells, these antigens can be divided into three classes which may have different clinical relevance.
The first class includes antigens that are shared between melanomas and tumors of other histological type, but not by normal tissues with the exception of testis and placenta (1, 7, 8). However, these tissues, since they are negative for expression of MHC class I molecules (9) needed for antigen presentation, cannot be targeted by T cells. Clinical trials based on the use of these antigens are in progress in patients affected by melanoma and other neoplastic diseases, and no relevant side-effects have been detected in the treated patients (1, 4).
The second class contains tissue-specific antigens expressed in normal and neoplastic cells of the melanocytic lineage (1, 10–12). Recently, the treatment of HLA-A2 melanoma patients with a gp100 peptide, modified to increase its binding to this HLA allele, in IFA plus IL-2, was shown to be associated with a significant clinical response rate (5). Even though no significant side-effects were observed in that or other similar clinical studies, the development of cross-reacting responses against normal tissues (i.e., skin melanocytes and pigmented retinal cells) must be carefully considered when self-differentiation antigens are used as vaccines (13).
The third class includes antigens expressed only by the tumor cells from which they have been isolated (1), since these antigenic epitopes are usually generated by a point mutation occurring in otherwise ubiquitously expressed proteins. The lack of natural tolerance against these antigens may allow the induction of a strong immune response, while avoiding the development of potential autoimmune reactions. However, until a panel of widely shared mutations has been discovered, the clinical application of such antigens is limited to the treatment of those few patients whose tumor carries the given mutation.
The existence of a fourth set of tumor antigens has been recently suggested based on the pattern of reactivity of a panel of CTL clones able to recognize the autologous and HLA-matched allogeneic melanomas, but not melanocytes or other targets of different histological origin (14). Here we report the characterization of a melanoma antigen belonging to this group and found to be encoded by a retained intron in a partially spliced transcript of the tyrosine-related protein 2 (TRP-2)1 differentiation antigen.
The features of this new shared melanoma-specific antigen may allow the development of safer and more efficacious immunotherapy trials.
Materials and Methods
Cell Lines.
The melanoma cell line Me18732 was established from a metastatic lesion obtained from a surgical specimen of patient 18732, admitted for surgery to the Istituto Nazionale Tumori (Milan, Italy). PBLs of this patient were typed serologically as HLA-A2/28, -B44/51, -C2/5. The human metastatic (Me17697, Me2559/1, Me12657, Me17088/1, Me4023) and primary (Me20842) melanoma cell lines, established in our laboratory, the carcinoma lines CALU3, SKBR3, and SKOV3 (American Type Culture Collection, Rockville, MD), and the WEHI-164.13 cells (15) were cultured in 10% FCS/RPMI 1640. The melanoma lines LB33, MZ2-MEL, and SK23-MEL, and the COS-7 cell line were provided by Prof. T. Boon (Ludwig Institute for Cancer Research, Brussels, Belgium) and maintained in 10% FCS/RPMI 1640 (melanomas) or 10% FCS/DMEM (COS-7). Normal human epidermal melanocyte lines (NHEM)-2489, -2311, and -4528 (Clonetics, San Diego, CA), and DL-1 melanocytes, the gift of Dr. M. De Luca (Istituto Dermopatico dell'Immacolata, Rome, Italy), were kept in melanocyte growth medium (MGM-2; Clonetics). The C1R-A*03301 transfectant, the homozygous EBV-transformed lymphoblastoid cell line (LCL) SCHU (HLA-A*0301, B*0702, C7), AMA-1 (HLA-A*6802, -B*5301, -C4), and WT-100-bis (HLA-A11, B35, C4) were provided by Dr. Soo Young Yang (Memorial Sloan-Kettering Cancer Center, New York); the EBV-LCL JHAF (HLA-A*31011, -B51, -C8) and LB (HLA-A*68011, -B*40011, -C2/3) were obtained from American Type Culture Collection. EBV-LCL cells were maintained in 10% FCS/RPMI 1640. CTL 128 (CD3+, CD4−, CD8+, TCR αβ+) was derived and grown as described previously (14). Its lytic activity was tested in a standard chromium- release assay (14) in the presence or absence of HLA-specific mAbs.
Subcloning of the HLA-A*68011 Allele.
cDNA obtained from Me18732 total RNA was amplified by PCR using a primer pair suitable for amplification and directional cloning of the HLA-A full-length coding region (gift of Dr. Soo Young Yang). The 1.1-kb PCR product was subcloned into the eukaryotic expression vector pcDNA3 (Invitrogen Corp., Abingdon, Oxon, UK). A plasmid clone (pcDNA3/A*6801) encoding the HLA-A*68011 allele was identified by diagnostic restriction mapping and sequencing.
Construction and Screening of the cDNA Library.
Poly(A)+ RNA was isolated from Me18732 cells using the FastTrack mRNA extraction kit (Invitrogen Corp.). The library was constructed converting 5 μg of poly(A)+ RNA to cDNA with the SuperScript Choice System kit (GIBCO BRL, Gaithersburg, MD) using an oligo-dT primer containing a NotI site at its 5′ end. cDNAs were then ligated to BstXI adapters (Invitrogen Corp.) and digested with NotI. After size fractionation, cDNAs were unidirectionally cloned into the BstXI/NotI site of the mammalian expression vector pcDNA3.1 (Invitrogen Corp.). Recombinant plasmids were electroporated into DH5-α Escherichia coli and selected with ampicillin (50 μg/ml). The library was divided into 1,300 pools of ∼100 cDNA clones. Each pool of bacteria was amplified to saturation, and plasmid DNA was extracted and transfected (100 ng) together with pcDNA3/A*6801 (100 ng) into COS-7 cells by the DEAE-dextran-chloroquine method (16). Using the same technique, in other experiments COS-7 cells were cotransfected with 100 ng of pcDNA3/A*6801 vector and 100 ng of expression vectors encoding one of the following melanoma antigens: Melan-A/MART-1, tyrosinase, gp100, MAGE-1, -2, -3, -4, -12, BAGE-1, -2, GAGE-1, -2, -3, -4, -5, -6 (provided by Dr. P. van der Bruggen, Ludwig Institute for Cancer Research), TRP-1 (provided by Dr. R.S. Wang, National Cancer Institute, National Institutes of Health, Bethesda, MD). Full-length TRP-2 cDNA was amplified by reverse transcription (RT)-PCR using specific primers located in the 5′-untranslated region (UTR; PRIT-1b, 5′-GGAGAAAAGTACGACAG-3′) and at the end of exon 8 (TRP-2L, 5′-ACCCTAGGCTTCTTCTGTGTATCTCTT-3′), cloned into pcDNA3, and sequenced to verify correspondence to the published cDNA sequence (17). Transfected COS-7 cells or stimulating cell lines were tested for their ability to induce TNF release by CTL 128, as described previously (18).
DNA Sequencing.
Sequencing was performed with an automated DNA sequencer (ABI Prism 377; Perkin-Elmer Corp., Norwalk, CT) using the Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer Corp.). The computer search for the sequence homology was done with the program FASTA EMBL-Heidelberg.
Production of Subfragments of cDNA 131.
Subfragments of cDNA 131 (5′-end 1500, 5′-end 800, 200, and 3′ end) were obtained by digestion of the plasmid with HindIII and BstUI and cloned into the pcDNAI plasmid (Invitrogen Corp.). From the 5′-end 1500, the smaller fragments INT2-434, -166, and -107 were generated by PCR amplification. The sense primer KS-INT2 (5′-GCCGCCATGTATTCTGTTAGAGATACA-3′) was used for amplification of all fragments. This primer generates an ATG start codon (underlined), with the appropriate Kozak consensus sequence, in the same open reading frame (ORF) as TRP-2. The antisense primers used for amplification of the INT2-434, -166, and -107 fragments, respectively, were SP6 (5′-ATTTAGGTGACACTATAG-3′) located in the pcDNAI plasmid, INT2-as1 (5′-ATCCGCTCGAGCATGAAATTACTTCCC-3′), and INT2-as2 (5′-ATCCGCTCGAGGATAATTCTACGAC-3′), located in intron 2 of TRP-2. The antisense primers INT2-as1 and -as2 contained an XhoI restriction site (underlined) for directional cloning.
Retroviral HLA-A*68011 Gene Transfer into Melanocytes.
A retroviral vector encoding the full-length HLA-A*68011 cDNA, under the control of the viral LTR, and the truncated form of the human low-affinity nerve growth factor receptor (ΔLNGFR) driven by the SV40 promoter was constructed as described previously (19). An amphotropic packaging cell line was established by infection of the GP+env Am 12 line and immunoselection for ΔLNGFR expression by the use of magnetic beads (Dynabeads M-450; Dynal A.S., Oslo, Norway) coated with the LNGFR-specific mAb 20.4 (American Type Culture Collection). Transduction of melanocytes and SK23-MEL was performed by cultivation with retrovirus-containing supernatant in the presence of polybrene (0.8 mg/ml). Three rounds of infection of at least 4 h were performed.
Separation of Nuclear and Cytoplasmic RNA.
Cells (2–3 × 106) were harvested, washed with PBS, and then carefully resuspended in 400 μl of buffer A (10 mM Hepes, pH 8.0, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol). After 10 min of incubation on ice, cytoplasmic membranes were disrupted by the addition of 20 μl NP-40 (10%), and the suspension was vigorously vortexed for 10 s and then centrifuged for 30 s at 15,000 rev/min. The supernatant containing the cytoplasmic RNA was transferred to a new tube. The pellet containing the intact nuclei was washed with buffer A to eliminate residual cytoplasmic RNA. Cytoplasmic and nuclear RNAs were purified from the cell fractions using the RNeasy mini kit (QIAGEN Inc., Chatsworth, CA). The absence of contaminating nuclear material (i.e., DNA and nuclear mRNA) in the cytoplasmic compartment was checked with specific primers flanking at least one intron of MAGE-3 (20) or complementary to intron–exon junctions flanking exon 8 of p53 (21).
Expression of TRP-2 and Its Partially Spliced Form TRP-2–INT2.
cDNA corresponding to 150 ng of total RNA from melanoma lines and fresh samples (tumors, skin, and retina) was amplified by PCR in 25 μl of water containing 200 μM of each dNTP, 0.6 μM of each primer, 1× PCR buffer, 1 U of Taq-Gold (all reagents from Perkin-Elmer Corp.). TRP-2 cDNA was amplified using the sense primer PR3 located in exon 2 (5′-TTCGGCAGAACATCCATTCC-3′) and the TRP-2L antisense primer, originating an amplicon of 1186 bp. TRP-2–INT2 was amplified using the sense primer PRIT-1b and the antisense primer INT2-1260 (5′-ACCTCACCAACTCACATCTT-3′), located in the 5′-UTR and in intron 2, respectively, and giving an amplification product of 977 bp. PCR amplification was performed for 30 cycles: 1 min at 94°C, 1 min at 55°C (58°C for TRP-2), and 1 min at 72°C. Primer positions were chosen in order to avoid contamination from genomic DNA. Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) cDNA was amplified with specific primers (22).
Evaluation of Primer Pair Sensitivity.
Plasmids containing TRP-2–INT2, TRP-2, and gp100 cDNAs were suspended to 10 pg/μl, a concentration equivalent to 1,026,119, 1,193,338, and 1,251,907 molecules/μl, respectively, serially twofold diluted, and amplified by PCR under the conditions described above. To mimic the target complexity of “true” cDNA sample, cDNA corresponding to 50 ng of total RNA from the carcinoma line SKBR3, negative for expression of TRP-2, TRP-2–INT2, and gp100, was added. TRP-2 was amplified with primers PR3 and TRP-2L. For TRP-2–INT2, in addition to PRIT-1b and INT2-1260, we also tested the sense primer PR15 (5′-AATTGGCAGTTCCAGTGTCC-3′), located in intron 2, and the antisense primer PR16 (5′-ATTGCAGGTACAGGAGCCAT-3′), located in intron 4, giving an amplicon of 906 bp. To amplify gp100, a previously described primer pair giving an amplicon of 361 bp was used (23). After an initial incubation at 94°C for 10 min and a final incubation at 72°C for 7 min, temperature cycling was conducted, with minor modifications, according to the original stepdown PCR parameters (24). Cycling was repeated 37 times, divided into 6 groups, with a denaturation of 20 s at 94°C and an elongation of 1 min 15 s at 72°C. The six groups consisted of three, six, six, six, nine, and seven cycles. Annealing temperatures lasted 40 s and decreased by 3°C from one group of cycles to the next, starting at 64°C in the first three cycles, 61°C in the following six, etc., and ending with 50°C in the last seven cycles. For gp100, extension time at 72°C lasted for 1 min in all cycles. Amplification products were size fractionated on agarose gels in the presence of ethidium bromide, digitally acquired under UV transillumination (Eagle Eye II, Still Video System; Stratagene Inc., La Jolla, CA), and analyzed densitometrically (ImageQuant version 3.3 software; Molecular Dynamics, Sunnyvale, CA). For each primer pair, the OD of gel bands (expressed in arbitrary units on a linear scale) was plotted against the initial concentration (expressed in picograms per microliter on a logarithmic scale) of target plasmid present in the sample. The minimum concentration of target plasmid detectable by each primer pair was indicated by the lowest point in the linear amplification range of the curve. The mean of this value, evaluated in three experiments and expressed as number of molecules, was considered as the sensitivity of each primer pair.
Quantitative RT-PCR Analysis.
cDNA samples prepared from total or cytoplasmic RNA of Me18732 were used as a reference, i.e., a sample whose level of expression of a given transcript is arbitrarily defined as 100%. GAPDH expression was used to normalize experimental results. The linear amplification range of GAPDH, TRP-2, gp100, TRP-2–INT2, and gp100-in4 was determined experimentally for the reference Me18732 cDNAs, starting from 2 μl cDNA, a volume corresponding to 100 ng total RNA. For each primer pair, the appropriate number of cycles to attain exponential amplification was found for a certain range of dilutions, varying the number of the final cycles of the stepdown PCR, performed as described above. The total number of cycles for GAPDH was 22 (groups of 3, 6, 6, and 7 cycles), and for gp100, TRP-2–INT2, TRP-2, and gp100-in4 was 30, 33, 35, and 40, respectively, according to groups of 3, 6, 6, 6, 9 plus n cycles. TRP-2 was amplified by primer pair PR3 plus TRP-2L; TRP-2–INT2 by PR15 plus PR16; and gp100, gp100-in4, and GAPDH by a pair of published primers (10, 22, 23). Linear regression analysis of experimental OD values allowed construction of standard curves for Me18732. Serial dilutions of cDNAs from all samples to be quantified were analyzed in these cycling conditions. Their OD values were compared with the appropriate standard curve always included in each PCR run, and normalized to the expression level of the GAPDH gene.
The lowest point in the linear amplification range of the calibration curve of a given primer pair, corresponding to a dilution expressed as a percentage of the first dilution of Me18732 cDNA, arbitrarily taken as 100%, was considered the minimum sensitivity threshold for that primer pair. The absence of a detectable amplification product, or OD values falling below this threshold, were reported as less than this threshold value. For primer pair PR3 plus TRP-2L (TRP-2) the threshold was 0.01%, and for PR15 plus PR16 (TRP-2–INT2) 0.1%. To quantify the level of TRP-2–INT2 expression in fresh specimens (skin and retina samples) in which the proportion of melanocytic cells is unknown and variable, the method described was modified as follows. The same cDNA dilutions from Me18732 total RNA were amplified using 31 (for TRP-2) and 33 (for TRP-2–INT2) cycles of stepdown PCR; after densitometric analysis, the OD results were expressed in arbitrary units. cDNA dilutions of skin and retina samples were amplified with TRP-2 primers for n number of cycles (ranging from 40 to 45) in order to obtain, after amplification, an OD of the gel band within the range covered by the Me18732 cDNA. For TRP-2–INT2, the same quantities of starting material were amplified for n plus at least 2 cycles to reflect the difference in the number of cycles (31–33) needed to detect TRP-2 and TRP-2–INT2, respectively, in the reference melanoma.
Peptides and CTL Assays.
The TRP-2–INT2 sequence was analyzed at a Web site of the National Institutes of Health (http: //bimas.dcrt.nih.gov:80/molbio/hla_bind/) for the presence of potential HLA-A*68011–binding peptides. Lyophilized peptides, 90% pure as indicated by analytical HPLC, were purchased from Primm Srl (Milan, Italy), dissolved at 10 mM concentration in DMSO, and stored at −80°C. Various concentrations of the peptides were added to 51Cr-labeled target cells. After 1 h of incubation at room temperature, CTL 128 was added at an E/T ratio of 20:1. Lysis was measured 4 h later. Alternatively, stimulator cells were incubated for 1 h at room temperature with a fixed concentration of peptides; after extensive washing, CTL 128 was added, and the TNF release was evaluated 18–20 h later on WEHI-164.13 cells.
Results
Identification of HLA-A*68011 as Restriction Element for CTL 128.
Melanoma cell line Me18732 was established from a metastatic lesion of patient 18732. Lymphocytes of the patient were typed as HLA-A2 and HLA-A28 by serological methods and then as HLA-A*0201 and HLA-A*68011 (hereafter referred to as HLA-A*6801) by sequence-specific oligonucleotide probe (SSOP) subtyping (25). HLA-restricted antitumor CTL clones were obtained from mixed lymphocyte tumor culture, as described (14). In particular, CTL clone 128 recognized the autologous tumor, and its lytic activity was inhibited by an mAb recognizing HLA-A2 and -A28, but not by an anti–HLA-A2 and -A69 mAb, indicating that HLA-A28 (A*6801) was the restricting molecule for CTL 128 (data not shown).
Antigen Specificity of CTL 128.
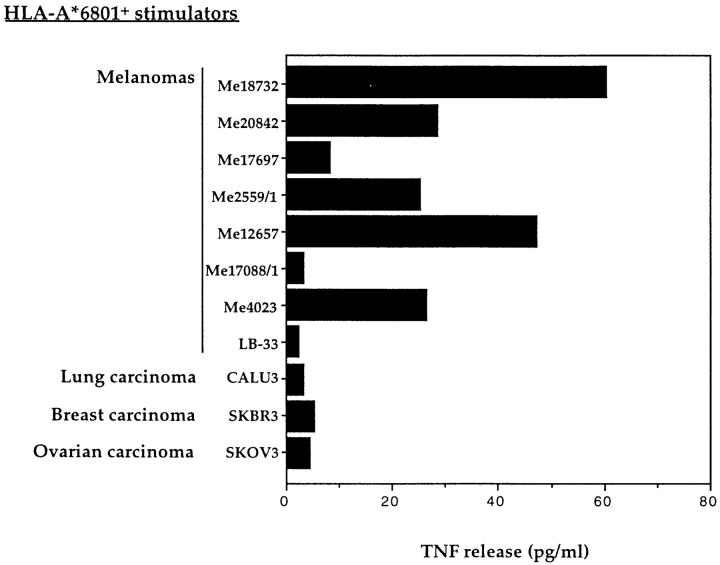
To evaluate the frequency of expression of the antigen recognized by CTL 128, a panel of HLA-A*6801 tumor lines was tested in a CTL stimulation assay. Five out of eight melanoma cell lines induced TNF release by CTL 128 (Fig. 1). However, no reactivity was observed with three HLA-A*6801 carcinoma lines of different histological origin (Fig. 1). The pattern of reactivity displayed by CTL 128 towards the melanoma lines tested did not correlate with expression of previously described melanoma antigens in these cell lines, as assessed by RT-PCR (data not shown). This was confirmed by the lack of TNF release by CTL 128 in the presence of COS-7 cells cotransfected with plasmid pcDNA3/A*6801, together with most of the cDNAs known to encode shared melanoma antigens (see Materials and Methods; data not shown). These results suggested that CTL 128 recognized a new antigen shared by a number of melanomas.
Figure 1.
Recognition by CTL 128 of HLA-A*6801+ melanoma and carcinoma cell lines. 1.5 × 103 CTLs were added to 2.5 × 104 stimulator cells, and the TNF content of the supernatant was tested 20 h later on WEHI-164.13 cells.
Cloning of a cDNA Encoding the Melanoma Antigen Recognized by CTL 128.
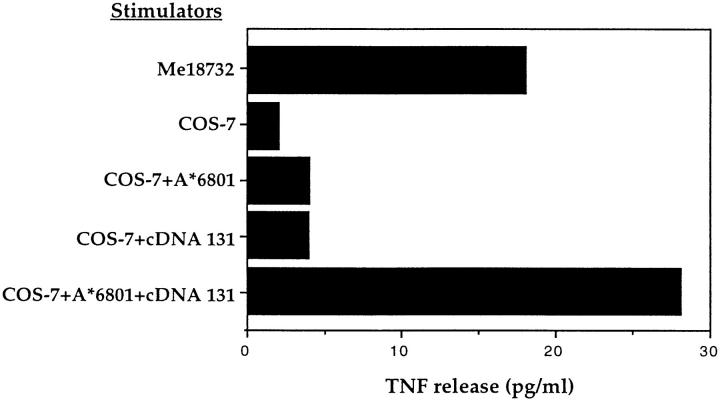
A cDNA library was constructed in expression vector pcDNA3.1 with poly(A)+ RNA extracted from Me18732 cells. Plasmid DNA from pools of 100 recombinant clones was cotransfected into COS-7 cells together with plasmid pcDNA3/A*6801. The DNA of one pool induced the production of a high level of TNF by CTL 128 (data not shown). By subcloning the bacteria of this pool, we obtained several clones that were able to transfer expression of the antigen. The results obtained with one of these clones, cDNA 131, are shown in Fig. 2. Recognition of the antigen was independent of the high, artificial expression level achieved in COS-7 cells. Indeed, an HLA-A*6801+ melanoma cell line that was not recognized by CTL 128 acquired, when transfected with cDNA 131, the ability to induce TNF release, and became sensitive to lysis by CTL 128 (data not shown). The sequence of the cDNA 131 proved to be 3535 bp long (Fig. 3), with nucleotides (nt) 1–994 and 3081–3347 identical to two noncontiguous regions of TRP-2 cDNA (17), which has been recently identified as a melanoma antigen of the melanocyte lineage (12). The first region contained exon 1, whose 5′-UTR lacked the first 15 bases, and exon 2, whereas the second region encompassed exons 3 and 4, of the TRP-2 gene, respectively. The sequences between nt 995 and 3080 and that downstream of nt 3347 showed no significant homology with any sequence recorded in the databanks, and were likely to be retained intron sequences. Indeed, a stretch of 10 amino acids flanking the exonic portions matched the described sequences at exon–intron junctions of the TRP-2 gene (26). The length of sequence 995–3080 (2086 nt) was comparable with intron 2 of TRP-2, as deduced from the published genomic map, and the sequence downstream of nt 3347 of cDNA 131 presented a 5′ donor splice site sequence identical to that of intron 4 of TRP-2 (26). Since it lacks the 3′ acceptor splice site sequence and its length is considerably shorter than that estimated from the published genomic map, we concluded that this was the sequence of intron 4 of TRP-2, truncated at nt 176. Sequencing of intron 4, amplified by PCR from genomic DNA of Me18732, revealed the presence of a polyadenine stretch of 10 nt, starting from nt 177 (data not shown). This finding supports the hypothesis of a fortuitous priming of this sequence by the oligo-dT used for construction of the cDNA library. Thus, cDNA 131 appeared to be composed of a partially spliced form of the melanocyte differentiation antigen TRP-2, containing exons 1–4 with retention of intron 2 and the initial portion of intron 4.
Figure 2.
Stimulation of CTL 128 by COS-7 cells transfected with cDNA 131 and HLA-A*6801 cDNA. COS-7 cells were transfected with HLA-A*6801 and with cDNA 131. The production of TNF by CTL 128 was measured after 24 h of coculture with the transfected cells, using the TNF-sensitive cell line WEHI-164.13. As control, COS-7 cells were transfected with cDNA 131 or HLA-A*6801 alone.
Figure 3.

Sequence of cDNA 131. The ATG start codon of the TRP-2 cDNA, and the first stop codon (TAG) in the same ORF, are doubly underlined. Exonic and intronic sequences are uppercase and lowercase, respectively. Positions of the primers used in the study for amplification of TRP-2–INT2 are indicated by horizontal arrows. The nucleotide sequence coding for the antigenic peptide (bold) recognized by CTL 128 is translated in the corresponding amino acid sequence using the single letter amino acid code. The nucleotide sequence is available under GenBank accession no. AJ000503.
Identification of the Sequence Encoding the Peptide Recognized by CTL 128.
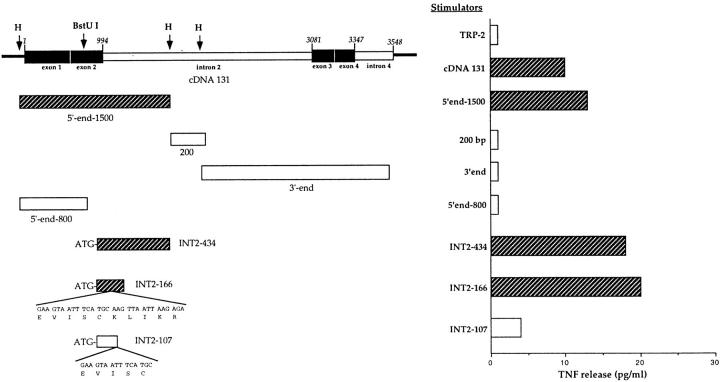
Three subfragments of ∼1.5, 0.2, and 2 kb, respectively (5′-end 1500, 200, and 3′ end; Fig. 4), were obtained by digesting cDNA 131 with HindIII, then cloned into pcDNAI vector and transfected into COS-7 cells along with pcDNA3/A*6801. At this step, the presence in the 200 and 3′-end fragments of start codons regulating their translation was not investigated. The level of TNF released by CTL 128 in the presence of COS-7 cells transfected with the 5′-end 1500 fragment was comparable to that stimulated by cDNA 131 (Fig. 4), indicating that the antigenic peptide was encoded within this region. Since CTL 128 was not stimulated by COS-7 cells cotransfected with both the fully spliced TRP-2 cDNA and HLA-A*6801 (Fig. 4), we hypothesized that the sequence coding for the antigen recognized by CTL 128 could be entirely or partially located in the intronic portion of the TRP-2 gene present in cDNA 131. This notion received support by the lack of recognition of COS-7 cells transfected with the 5′-end 800 cDNA fragment (Fig. 4), derived by truncation of the 5′-end 1500 fragment at a BstUI site and comprising exon 1 and half of exon 2. The intronic localization of the sequence encoding the antigenic peptide was confirmed by the ability of a PCR-amplified fragment encompassing the first 434 bp of intron 2 (INT2-434; see Materials and Methods) to convey expression of the antigen (Fig. 4). A sequence that coded for a decapeptide (EVISCKLIKR; Fig. 3) that possessed anchor residues (positions 2, 9, and 10) corresponding to the HLA-A*6801 peptide-binding motif (27) was present in the same ORF of the previous exons.
Figure 4.
Identification of the sequence coding for the antigenic peptide recognized by CTL 128. Top left, Exon–intron organization of cDNA 131. Exons (black boxes) and introns (white boxes) are indicated; the horizontal line at the extremities represents pcDNA3.1 vector; the numbering of the sequence is relative to the 5′ end of cDNA 131. Subfragments derived from cDNA 131 and PCR products, shown below cDNA 131, were cloned into expression vectors and transfected into COS-7 cells with the HLA-A*6801 cDNA, and those recognized by CTL 128 (striped boxes) are indicated. The peptide-encoding sequences present in the PCR fragments INT2-166 and INT2-107 are shown (bottom left). The spliced full-length form of TRP-2 cDNA was obtained by PCR as described (see Materials and Methods). TNF release by CTL 128 was evaluated on WEHI-164.13 cells (right). Striped bars, Recognition of cDNA 131 and its subfragments.
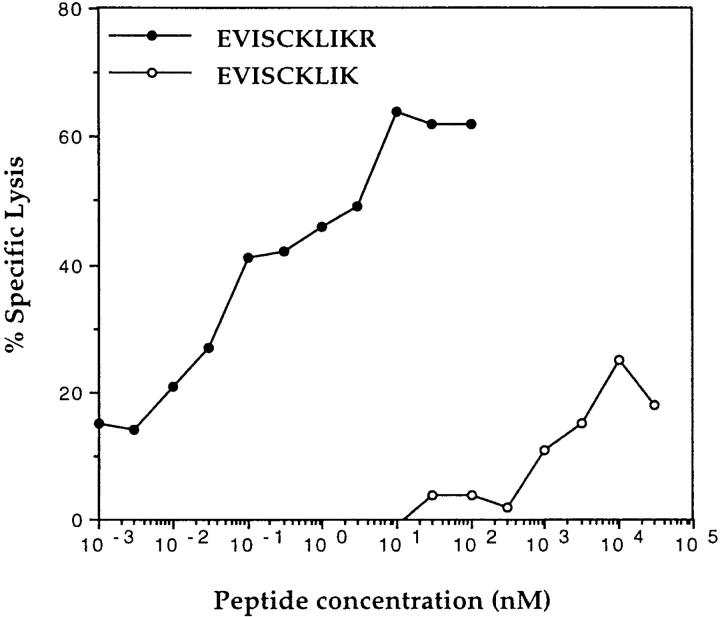
The sense primer KS-INT2 (which generates an ATG start site with the appropriate Kozak consensus sequence) and the reverse primer INT2-as1 or INT2-as2 (Fig. 3) were used to generate fragments including (INT2-166) or not (INT2-107) the entire putative peptide-encoding sequence. Only fragment INT2-166 was able to stimulate TNF release by CTL 128 (Fig. 4). Both 10- and 9-mer peptides, EVISCKLIKR and EVISCKLIK, were then synthesized and incubated with the HLA-A*6801 homozygous LCL line LB. The decapeptide was able to sensitize LB cells to lysis by CTL 128, with half-maximum lysis obtained at a concentration of ∼100 pM, whereas the nonapeptide had a very low efficiency (Fig. 5). A control peptide, WTGPNCERKK, derived from exon 2 of the fully spliced TRP-2 gene (17) able to bind to HLA-A*6801, was not recognized (data not shown). Given the localization of the antigenic epitope within intron 2 of TRP-2, the antigen was named TRP-2–INT2 and the epitope TRP-2–INT2222-231.
Figure 5.
Lysis by CTL 128 of HLA-A*6801 cells pulsed with the synthetic peptide EVISCKLIKR (TRP-2–INT2222-231). 51Cr-labeled HLA-A*6801 LCL line LB was pulsed for 2 h at room temperature with the synthetic peptides EVISCKLIKR (filled circles) and EVISCKLIK (open circles) and incubated with CTL 128 at an E/T ratio of 20:1, at the concentration indicated. 51Cr release was measured after 4 h of coincubation.
To exclude the possibility that TRP-2–INT2222-231 may have been generated by a mutation that occurred in the tumor, a 2152-bp fragment spanning the entire intron 2 (nt 995–3080 in cDNA 131) was amplified by PCR from genomic DNA of CTL 128 and from a different melanoma, MZ2-MEL. The PCR products were cloned, sequenced, and transfected together with pcDNA3/A*6801 into COS-7 cells. All clones had the expected sequences and were able to transfer expression of the antigen (data not shown).
Presentation of the Antigenic Peptide by Alleles of the HLA-A3–like Supertype.
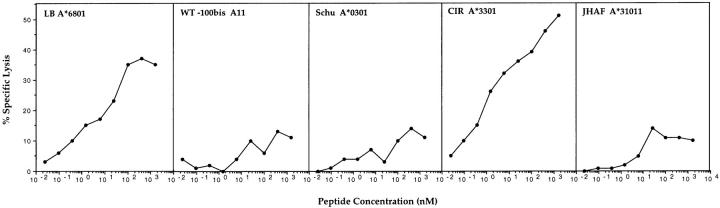
HLA-A*6801, along with A3, A11, A31, and A*3301, belongs to an A3-like supertype of HLA-A alleles with similar peptide-binding characteristics (28). To investigate whether the TRP-2–INT2222-231 peptide can be presented to CTL 128 by HLA alleles of the A3-like supertype, EBV LCL cells expressing such alleles were used as targets for CTL 128 after pulsing with the peptide. Among the A3-like supertype alleles, only A*3301 was able to present TRP-2–INT2222-231 peptide with the same efficiency as A*6801 (Fig. 6).
Figure 6.
Recognition by CTL 128 of the TRP-2–INT2221-231 peptide when presented by HLA alleles of the A3-like supertype. 51Cr-labeled LCL cells were incubated with CTL 128 at an E/T ratio of 20:1, in the presence of peptide TRP-2–INT2222-231 at different concentrations. 51Cr release was measured after 4 h of coincubation.
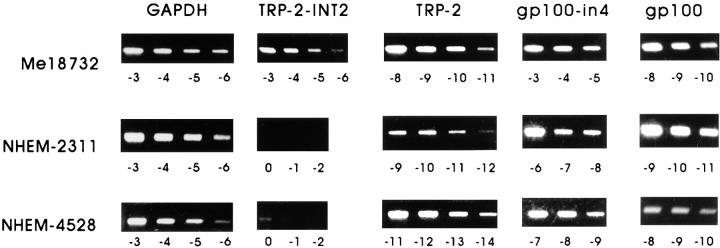
Expression of the Sequence Encoding TRP-2–INT2 in Tumors and Normal Tissues.
Expression of transcripts retaining intron 2 was analyzed by RT-PCR with the PRIT-1b plus INT2-1260 primer pair. The completely spliced TRP-2 messenger was detected with the PR3 plus TRP-2L primer pair. Among the tumors tested, only melanomas proved positive for TRP-2 and TRP-2–INT2 expression (Table 1), whereas 17 carcinomas (4 ovary, 4 breast, 4 lung, 2 pancreas, and 3 bladder) were negative (data not shown). 84% (16 out of 19) and 68% (13 out of 19) of melanoma cell lines analyzed expressed TRP-2 and TRP-2–INT2, respectively. In fresh melanoma samples, the normal form of TRP-2 was expressed in 69% (9 out of 13) of the analyzed samples and TRP-2–INT2 in 54% (7 out of 13; Table 1). Expression of TRP-2–INT2 in the absence of TRP-2 was never observed. This result is in agreement with the notion that TRP-2 is a melanocytic differentiation antigen (12) and that the same promoter drives the synthesis of a common messenger from which both antigens arise. Normal tissues in which TRP-2 is known to be present were analyzed for TRP-2–INT2 expression. 4 melanocyte cell lines, 13 skin samples, and 1 retina tested were all negative in the RT-PCR assay (Table 1).
Table 1.
Expression of TRP-2–INT2 and TRP-2 mRNA in Tumors and Normal Tissues
| No. of samples with antigen expression/no. samples tested* | ||||
|---|---|---|---|---|
| TRP-2–INT2 | TRP-2 | |||
| Melanoma cell lines | 13/19 (68%)‡ | 16/19 (84%) | ||
| Melanocyte cell lines | 0/4 | 4/4 | ||
| Fresh melanoma samples | 7/13 (54%)‡ | 9/13 (69%) | ||
| Skin | 0/13 | 13/13 | ||
| Retina | 0/1 | 1/1 | ||
| Carcinomas | 0/17 | ND | ||
cDNA synthesis and PCR were performed as described in Materials and Methods.
TRP-2–INT2 expression was detected only in samples positive for TRP-2.
Quantitative RT-PCR Analysis for TRP-2–INT2 Expression.
To quantitate levels of expression of TRP-2 and TRP-2–INT2 in both neoplastic and normal cells, primer pair sensitivity (i.e., the minimum number of target molecules detectable) as well as amplification efficiency had to be comparable. As detailed in Materials and Methods, serially diluted plasmids containing TRP-2, TRP-2–INT2, and gp100 cDNA were amplified for 37 cycles with specific primers. After densitometric analysis, the sensitivity of the PR3 plus TRP-2L primer pair was determined in the order of 34 molecules for TRP-2, whereas 19 was the lowest number of molecules of gp100 detectable with the previously described primers (23). For TRP-2–INT2, the primer pair PR15 plus PR16, which displays an even higher sensitivity (four molecules), was chosen for this analysis. All primer pairs showed a nearly identical amplification efficiency, as indicated by parallelism of regression lines in the linear amplification range of the curves (data not shown).
Therefore, levels of expression of TRP-2 and TRP-2–INT2 in nine melanoma cell lines were determined by quantitative RT-PCR on cDNA derived from total RNA. Me18732 was arbitrarily considered to have 100% expression of these genes and was used as the reference standard (Table 2). All measurements were normalized on GAPDH expression to take into account differences in template integrity. Expression of TRP-2–INT2 in the absence of TRP-2 was never observed. The six tumor lines that stimulated TNF release by CTL 128 expressed both TRP-2 and TRP-2–INT2. The level of expression of TRP-2–INT2 mRNA ranged from 6 to 187% of that present in Me18732, and that of TRP-2 from 5 to 35%. Among the three melanoma lines not recognized by CTL 128, LB33 was below the threshold of sensitivity of the primer pair used for expression of both transcripts, whereas Me17088/1 expressed low levels (0.1%) of TRP-2 mRNA only; MZ2-MEL*6801, a melanoma cell line stably transfected with HLA-A*6801, expressed low levels of both TRP-2 (0.5%) and TRP-2–INT2 (0.4%). Since RT-PCR analysis performed on total RNA does not distinguish the cytoplasmic, partially spliced TRP-2–INT2, from which the antigenic peptide is generated, from pre–TRP-2 mRNA of nuclear origin, RNA was extracted from both the nucleus and the cytosol of MZ2–MEL-A*6801. A strong PCR signal was obtained from nuclear RNA, whereas the TRP-2–INT2 message in the cytoplasm accounted for 0.2% of that present in the same compartment of Me18732. The absence of TNF release by CTL 128 in the presence of MZ2–MEL-A*6801 demonstrates that cytoplasmic TRP-2–INT2 transcript levels of at least >0.2% of that found in Me18732 are needed for functional recognition of target cells.
Table 2.
Quantitative RT-PCR of TRP-2 and TRP-2–INT2 mRNA Expression and Recognition by CTL Clone 128
| Melanoma cell lines | Recognition by CTL 128 | TRP-2–INT2* | TRP-2* | |||
|---|---|---|---|---|---|---|
| Me18732 | + | 100 | 100 | |||
| Me12657 | + | 187 | 34 | |||
| Me20842 | + | 28 | 17 | |||
| Me2559/1 | + | 6 | 5 | |||
| Me4023 | + | 55 | 22 | |||
| SK23–MEL-A*6801‡ | + | 15 | 35 | |||
| MZ2–MEL-A*6801‡ | − | 0.4 | 0.5 | |||
| Me17088/1 | − | <0.1 | 0.1 | |||
| LB33 | − | <0.1 | <0.01 |
Expression of TRP-2–INT2 and TRP-2 mRNA in the different melanoma samples was assessed by quantitative RT-PCR. Me18732 was arbitrarily considered to have 100% expression of these genes, and the limit of detection for TRP-2–INT2 and TRP-2 was 0.1 and 0.01%, respectively.
Cell line obtained by calcium-phosphate transfection of pcDNA3/ A
*6801 and selection in G418. Expression of the transfected HLA- A
*6801 allele was verified by flow cytometry with specific mAbs (data not shown).
To exclude the possibility of even low contamination of pre–TRP-2 nuclear message, quantitative analysis of TRP-2 and TRP-2–INT2 expression in four melanocyte cell lines at their first in vitro passages was performed with cDNA derived from cytoplasmic RNA. Expression levels of gp100 and its partially spliced form gp100-in4 mRNAs, containing sequences coding for CTL-defined antigens expressed by melanomas and melanocytes (1, 10), were also included in the analysis. As reported in Table 3, TRP-2–INT2 mRNA of the cultured skin melanocytes was either undetectable (3/4) or at the lower limit of detection (1/4), whereas TRP-2 mRNA ranged from 15 to 614% of that expressed by Me18732. However, all melanocyte lines expressed gp100 and particularly gp100-in4 at high levels, from 79 to 1,613% and from 776 to 5,000%, respectively, of that found in Me18732. Fig. 7 shows the gel bands used for densitometric analysis.
Table 3.
Quantitative RT-PCR of TRP-2–INT2, TRP-2, gp100, and gp100-in4 mRNAs in the Cytoplasm of Human Melanocytes
| Cytoplasmic mRNA | TRP-2–INT2 | TRP-2 | gp100-in4 | gp100 | ||||
|---|---|---|---|---|---|---|---|---|
| % | ||||||||
| Me18732 | 100 | 100 | 100 | 100 | ||||
| NHEM-2489 | <0.1 | 21 | 776 | 79 | ||||
| NHEM-2311 | <0.1 | 133 | 1039 | 360 | ||||
| NHEM-4528 | 0.1 | 614 | 2363 | 155 | ||||
| DL-1 | <0.1 | 15 | 5000 | 1613 | ||||
Expression of the different transcripts was measured by quantitative RT-PCR. Me18732 was arbitrarily considered to have 100% expression of these genes. The limit of detection for TRP-2–INT2 was 0.1%.
Figure 7.
Quantitative RT-PCR analysis. Serial twofold dilutions of cDNA derived from cytoplasmic RNA of DL-1 and NHEM-4528 melanocytes were amplified and simultaneously compared with the reference Me18732, as described in Materials and Methods. Depending on the sample and on the gene tested, starting dilutions ranged from 0 to −11 (i.e., 20 = undiluted, 2−11 = 1/2,048). Results obtained after densitometric scanning of the amplified products are shown in Table 3.
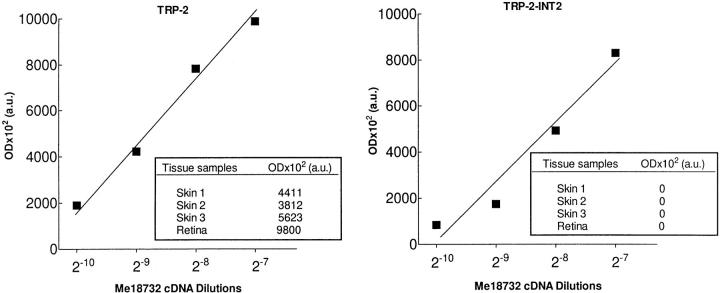
We then examined samples of three TRP-2 + normal skin tissue and one retina. Since in fresh specimens the proportion of melanocytic cells is variable and generally very low, we used a slightly modified protocol, detailed in Materials and Methods. Total RNA was analyzed, since attempts to separate nuclei from cytoplasmic compartments were unsuccessful, as demonstrated by a consistent level of nuclear contamination in cytoplasm (data not shown), due to the high level of cellular damage and/or deaths resulting from the enzymatic treatment used to prepare single cell suspensions. Results are reported in the original OD values and are shown in Fig. 8. An amplification product for TRP-2 was obtained in each of the four samples (three skin tissue and one retina) analyzed, whereas TRP-2–INT2 was undetectable. In this case too, both gp100 and gp100-in4 were expressed at high levels (data not shown).
Figure 8.
Expression of TRP-2–INT2 and TRP-2 on normal tissues. Expression was tested by quantitative RT-PCR on cDNA derived from total RNA. Results are expressed in arbitrary OD units (a.u.) after densitometric analysis of the amplified products, as described in Materials and Methods. For Me18732, the plotted OD points for TRP-2 and TRP-2–INT2 correspond to four identical and consecutive serial dilutions, chosen in order to obtain, with the lowest one, a still detectable amplification signal for TRP-2–INT2. Amplification of TRP-2 from normal samples was performed in conditions (i.e., cDNA amount and cycle number) allowing the attainment of an OD value included in the range covered by Me18732. TRP-2–INT2 was consequently amplified using comparable parameters (see Materials and Methods). (Insets) OD arbitrary units (a.u.) are reported for normal samples.
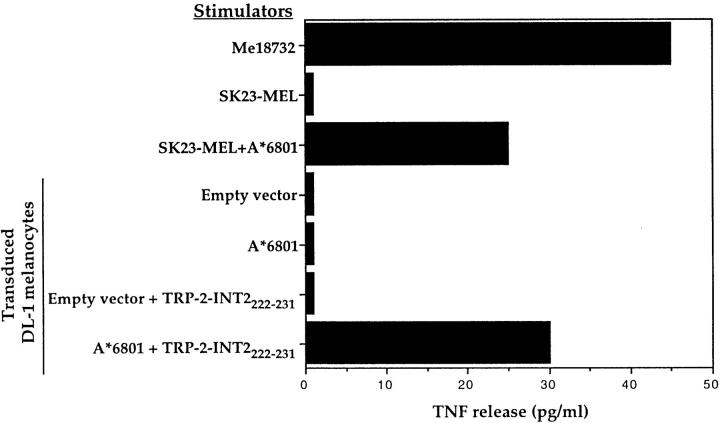
The absence of expression of endogenous TRP-2–INT2 in normal melanocytic cells was also confirmed by the lack of recognition by CTL 128 of melanocyte lines transduced with HLA-A*6801 (data for the DL-1 line are shown in Fig. 9). The transduced HLA-A*6801 molecule was able to present the endogenous TRP-2–INT2 product as shown by recognition of the HLA-A*6801–transduced SK23-MEL. Moreover, expression of HLA-A*6801 by the transduced melanocytes was confirmed by their ability to functionally present the exogenously provided TRP-2–INT2222-231 peptide to CTL 128.
Figure 9.
Lack of recognition by CTL 128 of HLA-A*6801+–transduced melanocytes. DL-1 melanocytes and the SK23-MEL lines were transduced by a retroviral vector encoding HLA-A*6801. 1.5 × 103 CTLs were added to 104 transduced and untransduced stimulator cells, and the TNF content of the supernatant was tested 20 h later on WEHI-164.13 cells. Efficiency of transduction was evaluated by the ability of the transduced cells to present the TRP-2–INT2222-231 peptide.
Taken together, these results indicate that the presence of a transcript corresponding to the unspliced TRP-2 gene accompanies TRP-2 mRNA expression in the cytoplasmic compartment of the majority of melanoma cells. This phenomenon does not occur in normal cells, despite the presence of comparable or even higher levels of TRP-2, allowing a partially spliced form of a differentiation antigen encoding mRNA to become the source of a melanoma-restricted T cell epitope.
Discussion
This study reports the molecular identification of an antigen (TRP-2–INT2) belonging to a new class of shared melanoma antigens absent from melanocytes. The epitope recognized by HLA-A*6801–restricted CTL 128 is encoded within intron 2 of a partially spliced mRNA of the TRP-2 gene. In its completely spliced form, TRP-2 is a member of the tyrosinase-related gene family encoding an enzyme acting in the melanogenic pathway and containing an epitope recognized in an HLA-restricted fashion by T cells on melanomas and melanocytes (12). The sequence coding for the peptide recognized by CTL 128 is located at the 5′ end of intron 2, which is available for translation because of a failure in the splicing pathway. An increasing body of evidence suggests the immunological significance of unconventionally produced epitopes. Indeed, epitopes encoded in alternative ORFs (29), within introns (10, 16), under the putative control of a cryptic promoter (30), or governed by a variant of conventional translation mechanisms (31) have been described. It has also been proposed that the increased expression in splice variants of pigment genes that occurs in melanoma compared with normal melanocytes may contribute to the pool of melanoma-specific antigenic peptides (32).
TRP-2–INT2 expression was not observed in melanomas lacking TRP-2. Moreover, no TRP-2–INT2 was found when TRP-2 expression was 0.1% of that found in Me18732. Such a low level of TRP-2 transcription, detected by the very sensitive quantitative RT-PCR, could not be evidenced in Northern blots (data not shown) and probably accounts for the percentage of TRP-2 + fresh melanomas that fail to express TRP-2–INT2. In all normal tissues containing TRP-2 + cells of melanocytic origin (i.e., skin and retina) and in three out of four melanocyte samples that had levels of TRP-2 ranging from 21 to 133% of that expressed by Me18732, TRP-2–INT2 was undetectable (Table 3, and Figs. 7 and 8). Furthermore, TRP-2–INT2 was still at the lower threshold of detectability (i.e., 0.1%) in a melanocyte line that displayed more than a sixfold increase (614%) of TRP-2 compared with Me18732 (Table 3, and Fig. 7). Transduction of melanocyte lines with the HLA-A*6801 allele confirmed the absence of endogenous TRP-2–INT2222-231 expression at levels allowing CTL 128 recognition (Fig. 9).
Northern blot analysis has indicated that TRP-2 and, in general, pigment gene expression are low in human melanoma compared with the average expression in melanocytes (33). This finding was confirmed by our quantitative PCR analysis in which two out of four melanocyte lines had increased levels of TRP-2 over that of Me18732, one of the tumors that displayed the highest level of expression for this gene (Tables 2 and 3). We had similar findings with gp100, whose levels of expression in three out of four melanocyte lines were 1.3–16 times higher than those of Me18732. Given the lower expression of TRP-2 in transformed compared with normal cells, these results rule out the possibility that the presence of the unspliced form of the TRP-2 gene in melanoma is simply related to high levels of gene transcription. Rather, our findings suggest that the splicing pathway that involves this gene is operating in normal melanocytes but is altered in melanoma cells, allowing the transport of certain intron-containing transcripts into the cytoplasm. As also observed for gp100-in4 by Robbins et al., the intron-encoded epitope is expressed after transfection into COS-7 cells (10). Lack of adjacent regulatory sequences might be responsible for the splicing failure in COS-7 cells (34), since our cDNA clone is probably truncated at its 3′ end by a fortuitous priming of a polyadenine stretch in intron 4. However, the presence of unspliced mRNAs in the cytoplasm cannot be considered a distinguishing feature of transformed cells, but appears to be gene specific. Indeed, weak consensus sequences at intron–exon boundaries may determine a very low splicing efficiency and ubiquitous retention of intervening sequences in mRNA even in normal cells (35). Moreover, although the mechanism has not been investigated, expression of the unspliced form of the differentiation antigen gp100 (gp100-in4) has been found in normal melanocytes at levels comparable to or even higher than that found in melanomas (10), a finding confirmed by our quantitative PCR analysis (Table 3).
TRP-2–INT2 codes for a putative protein of 237 amino acids that runs, using the same ORF of TRP-2, from the start codon in position 400 to the terminator site (nt 1111) located in intron 2, just 18 nt downstream of the peptide-coding region. Wild-type TRP-2 protein (519 amino acids) was identified as the DOPAchrome tautomerase involved in melanin biosynthesis (17, 36). The TRP-2–INT2 translation product, devoid of the transmembrane domain, is expected to have a different localization from TRP-2, which exists as a melanosome membrane-bound form. Therefore, the functional role, if any, of TRP-2–INT2 in the tumor cells needs to be addressed.
Although the minimum level of TRP-2–INT2 mRNA expression allowing CTL-mediated recognition has not been established, an HLA-A*6801 melanoma line expressing in its cytoplasmic compartment 0.2% of the level found in Me18732 failed to induce TNF production by CTL 128. However, a melanoma line (Me 2559/1) expressing TRP-2–INT2 at levels equal to 6% of that found in Me18732 (Fig. 1, and Table 2) was recognized. On the basis of the frequency of hybridizing cDNA colonies, Me18732 appears to express ∼20 TRP-2–INT2 mRNA molecules per cell (data not shown). Accordingly, 6% expression corresponds to one molecule per cell and could be considered the minimum level of TRP-2–INT2 that allows CTL 128– mediated recognition. This level of expression is lower than that required for CTL recognition of most of the other melanoma antigens (37, 38), suggesting that CTL 128 is a high-affinity effector. The HLA-binding affinity of the TRP-2–INT2222-231 peptide is unknown, but the very low concentration (100 pM) required to sensitize cells to lysis (see Fig. 5) supports this hypothesis. Detection of high-affinity effectors specific for epitopes encoded by intron 2 suggests that this sequence may be a source of “non-self” antigenic epitopes. Indeed, CTLs recognizing autoantigens have been shown to escape intrathymic clonal deletion only when expressing low-affinity receptors, while high-affinity CTLs against autoantigens are deleted (39). Furthermore, in peptide sensitization assays, many differentiation antigen–derived peptides are active only at concentrations higher than those of the tumor-specific peptides (40). Taken together, these observations strongly indicate that TRP-2–INT2 codes for a new type of tumor antigen shared among melanomas, but absent in normal tissues.
Analysis of the structural similarities in the antigen-binding groove of HLA molecules and the identification of cross-reactive peptides have allowed grouping of HLA alleles into supertypes (41). Based on its peptide-binding characteristics, HLA-A*6801 belongs to the HLA-A3–like supertype that also comprises A3, A11, A31, and A*3301 (28). Among the alleles of the A3-like supertype, recognition of TRP-2–INT2222-231 peptide was also observed when it was presented by A*3301, which, along with A*6801, is expressed by 13% of the world's population (28). It has been previously reported that identical peptides derived from viral proteins (HBcAg from hepatitis B) or from tumor antigens (TRP-2) can be recognized in the context of different HLA alleles, HLA-A31 and A68 or HLA-A31 and A33, respectively, by specific and differentially HLA-restricted CTL clones (42, 43). Moreover, in agreement with our findings, single T cells have been shown to recognize a TRP-2 melanoma–associated peptide when presented by both HLA-A31 and A*3301 (43).
Apart from the TRP-2–INT2222-231 peptide, screening introns and intron–exon junctions of TRP-2–INT2 for the presence of known motifs revealed peptide sequences that are predicted to bind with high efficiency several HLA-A and -B alleles, including HLA-A2, frequently present in all major ethnic groups. This suggests that other tumor antigens with the same degree of lineage and tumor specificity may be expressed by melanoma cells in the context of more frequent HLA-A and -B alleles. Furthermore, it is possible that other specific antigens can be generated in melanoma and other tumors by mechanism(s) of altered splicing of lineage-related genes.
Acknowledgments
We thank Dr. K. Fleischhauer (Istituto Scientifico San Raffaele) for DNA typing of the HLA-A locus, and Dr. S. Battocchio (Division of Pathology, Istituto Nazionale Tumori) for providing mammary skin tissue samples. We are also indebted to Ms. M.T. Radice for oligonucleotide synthesis, Ms. D. Penso for help with the automated DNA sequencer, Ms. A. Molla for technical assistance, and Mr. M. Azzini and Ms. G. Barp for editing.
Abbreviations used in this paper
- LCL
lymphoblastoid cell line
- LNGFR
low-affinity nerve growth factor receptor
- nt
nucleotide(s)
- ORF
open reading frame
- RT
reverse transcription
- TRP
tyrosinase-related protein
- UTR
untranslated region
Footnotes
R. Lupetti is the recipient of a fellowship from the Italian Association for Cancer Research (AIRC), Milan. This work was supported in part by grants from the AIRC and from the European Community Biomed2 Program (BMH4-CT95-1627).
References
- 1.Van den Eynde BJ, van der Bruggen P. T cell defined tumor antigens. Curr Opin Immunol. 1997;9:684–693. doi: 10.1016/s0952-7915(97)80050-7. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Yannelli JR, Yang JC, Topalian SL, Schwartzentruber DJ, Weber JS, Parkinson DR, Seipp CM, Einhorn JH, White DE. Treatment of patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and interleukin-2. J Natl Cancer Inst. 1994;86:1159–1166. doi: 10.1093/jnci/86.15.1159. [DOI] [PubMed] [Google Scholar]
- 3.Arienti F, Belli F, Rivoltini L, Gambacorti-Passerini C, Furlan L, Mascheroni L, Prada A, Rizzi M, Marchesi E, Vaglini M, et al. Adoptive immunotherapy of advanced melanoma patients with interleukin-2 (IL-2) and tumor-infiltrating lymphocytes selected in vitro with low doses of IL-2. Cancer Immunol Immunother. 1993;36:315–322. doi: 10.1007/BF01741170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchand M, Weynants P, Rankin E, Arienti F, Belli F, Parmiani G, Cascinelli N, Bourlond A, Vanwijck R, Humblet Y, et al. Tumor regression responses in melanoma patients treated with a peptide encoded by gene MAGE-3. Int J Cancer. 1995;63:883–885. doi: 10.1002/ijc.2910630622. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Dudley ME, Schwarz SL, Spiess PJ, et al. Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat Med. 1998;4:321–327. doi: 10.1038/nm0398-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nestle FO, Alijagic S, Gilliet M, Sun Y, Grabbe S, Dummer R, Burg G, Schadendorf D. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat Med. 1998;4:328–333. doi: 10.1038/nm0398-328. [DOI] [PubMed] [Google Scholar]
- 7.Jäger E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jäger D, Arand M, Wada H, Noguchi Y, Stockert E, et al. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human-histocompatibility leukocyte antigen (HLA)-A2– binding peptide epitopes. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K, Shichijo S, Noguchi M, Hirohata M, Itoh K. Identification of MAGE-1 and MAGE-4 proteins in spermatogonia and primary spermatocytes of testis. Cancer Res. 1995;55:3478–3482. [PubMed] [Google Scholar]
- 9.Haas GG, Jr, D'Cruz OJ, De Bault LE. Distribution of human leukocyte antigen-ABC and -D/DR antigens in the unfixed human testis. Am J Reprod Immunol Microbiol. 1988;18:47–51. doi: 10.1111/j.1600-0897.1988.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 10.Robbins PF, El-Gamil M, Li YF, Fitzgerald EB, Kawakami Y, Rosenberg SA. The intronic region of an incompletely spliced gp100 gene transcript encodes an epitope recognized by melanoma-reactive tumor infiltrating lymphocytes. J Immunol. 1997;159:303–308. [PubMed] [Google Scholar]
- 11.Wang RF, Robbins PF, Kawakami Y, Kang X, Rosenberg SA. Identification of a gene encoding a melanoma tumor antigen recognized by HLA-A31–restricted tumor-infiltrating lymphocytes. J Exp Med. 1995;181:799–804. doi: 10.1084/jem.181.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang RF, Appella E, Kawakami Y, Kang X, Rosenberg SA. Identification of TRP-2 as a human tumor antigen recognized by cytotoxic T lymphocytes. J Exp Med. 1996;184:2207–2216. doi: 10.1084/jem.184.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugita S, Sagawa K, Mochizuchi M, Shichijo S, Itoh K. Melanocyte lysis by cytotoxic T lymphocytes recognizing the MART-1 melanoma antigen in HLA-A2 patients with Vogt-Koyanagi-Harada disease. Int Immunol. 1996;8:799–803. doi: 10.1093/intimm/8.5.799. [DOI] [PubMed] [Google Scholar]
- 14.Anichini A, Mortarini R, Maccalli C, Squarcina P, Fleischhauer K, Mascheroni L, Parmiani G. Cytotoxic T cells directed to tumor antigens not expressed on normal melanocytes dominate HLA-A2.1-restricted immune repertoire to melanoma. J Immunol. 1996;156:208–217. [PubMed] [Google Scholar]
- 15.Espevik T, Nissen-Meyer J. A highly sensitive cell line, WEHI 164 clone 13, for measuring cytotoxic factor/tumor necrosis factor from human monocytes. J Immunol Methods. 1986;95:99–105. doi: 10.1016/0022-1759(86)90322-4. [DOI] [PubMed] [Google Scholar]
- 16.Coulie PG, Lehmann F, Lethé B, Herman J, Lurquin C, Andrawiss M, Boon T. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci USA. 1995;92:7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama K, Suzuki H, Yasumoto K, Tomita Y, Shibahara S. Molecular cloning and functional analysis of a cDNA coding for human DOPAchrome tautomerase/tyrosinase-related protein-2. Biochim Biophys Acta. 1994;1217:317–321. doi: 10.1016/0167-4781(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 18.Traversari C, van der Bruggen P, Van den Eynde B, Hainaut P, Lemoine C, Ohta N, Old L, Boon T. Transfection and expression of a gene coding for a human melanoma antigen recognized by autologous cytolytic T lymphocytes. Immunogenetics. 1992;35:145–152. doi: 10.1007/BF00185107. [DOI] [PubMed] [Google Scholar]
- 19.Fleischhauer K, Tanzarella S, Russo V, Sensi ML, van der Bruggen P, Bordignon C, Traversari C. Functional heterogeneity of HLA-A*02 subtypes revealed by presentation of a MAGE-3-encoded peptide to cytotoxic T cell clones. J Immunol. 1997;159:2513–2521. [PubMed] [Google Scholar]
- 20.Weynants P, Lethé B, Brasseur F, Marchand M, Boon T. Expression of MAGE genes by non-small-cell lung carcinomas. Int J Cancer. 1994;56:826–829. doi: 10.1002/ijc.2910560612. [DOI] [PubMed] [Google Scholar]
- 21.Donghi R, Longoni A, Pilotti S, Michieli P, Della G, Porta, Pierotti M. Gene p53 mutations are restricted to poorly differentiated and undifferentiated carcinomas of the thyroid gland. J Clin Invest. 1993;4:1753–1760. doi: 10.1172/JCI116385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Z, Fasco MJ, Kaminsky L. Optimization of Dnase I removal of contaminating DNA from RNA for use in quantitative RNA-PCR. Biotechniques. 1996;20:1012–1020. doi: 10.2144/96206st02. [DOI] [PubMed] [Google Scholar]
- 23.Adema GJ, de Boer AJ, Vogel AM, Loenen WAM, Figdor CG. Molecular characterization of the melanocyte lineage-specific antigen gp100. J Biol Chem. 1994;269:20126–20133. [PubMed] [Google Scholar]
- 24.Hecker KH, Roux KH. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. Biotechniques. 1996;20:478–485. doi: 10.2144/19962003478. [DOI] [PubMed] [Google Scholar]
- 25.Oh S, Fleischhauer K, Yang S. Isoelectric focusing subtypes of HLA-A can be defined by oligonucleotide typing. Tissue Antigens. 1993;41:135–142. doi: 10.1111/j.1399-0039.1993.tb01992.x. [DOI] [PubMed] [Google Scholar]
- 26.Sturm RA, O'Sullivan BJ, Box NF, Smith AG, Smit SE, Puttick ER, Parsons PG, Dunn IS. Chromosomal structure of the human TYRP1 and TYRP2 loci and comparison of the tyrosinase-related protein gene family. Genomics. 1995;29:24–34. doi: 10.1006/geno.1995.1211. [DOI] [PubMed] [Google Scholar]
- 27.Rammensee HG, Friede T, Stefanovic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 28.Sidney J, Grey HM, Southwood S, Celis E, Wentworth PA, del Guercio MF, Kubo RT, Chesnut RW, Sette A. Definition of an HLA-A3-like supermotif demonstrates the overlapping peptide-binding repertoires of common HLA molecules. Hum Immunol. 1996;45:79–93. doi: 10.1016/0198-8859(95)00173-5. [DOI] [PubMed] [Google Scholar]
- 29.Wang RF, Parkhurst MR, Kawakami Y, Robbins PF, Rosenberg SA. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J Exp Med. 1996;183:1131–1140. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guilloux Y, Lucas S, Brichard VG, Van Pel A, Viret C, De Plaen E, Brasseur F, Lethé B, Jotereau F, Boon T. A peptide recognized by human cytolytic T lymphocytes on HLA-A2 melanomas is encoded by an intron sequence of the N-acetylglucosaminyltransferase V gene. J Exp Med. 1996;183:1173–1183. doi: 10.1084/jem.183.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bullock TN, Eisenlohr LC. Ribosomal scanning past the primary initiation codon as a mechanism for expression of CTL epitopes encoded in alternative reading frames. J Exp Med. 1996;184:1319–1329. doi: 10.1084/jem.184.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Fur N, Kelsall SR, Silvers WK, Mintz B. Selective increase in specific alternative splice variants of tyrosinase in murine melanomas: a projected basis for immunotherapy. Proc Natl Acad Sci USA. 1997;94:5332–5337. doi: 10.1073/pnas.94.10.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eberle J, Garbe C, Wang N, Orfanos CE. Incomplete expression of the tyrosinase gene family (tyrosinase, TRP-1, and TRP-2) in human malignant melanoma cells in vitro. Pigm Cell Res. 1995;8:307–313. doi: 10.1111/j.1600-0749.1995.tb00679.x. [DOI] [PubMed] [Google Scholar]
- 34.Dirksen WP, Sun Q, Rottman FM. Multiple splicing signals control alternative intron retention of bovine growth hormone pre-mRNA. J Biol Chem. 1995;270:5346–5352. doi: 10.1074/jbc.270.10.5346. [DOI] [PubMed] [Google Scholar]
- 35.Lang KM, van Santen VL, Spritz RA. The two intervening sequences of human β- and γ-globin pre-mRNA are excised in a preferred temporal order in vitro. EMBO (Eur Mol Biol Organ) J. 1985;4:1991–1996. doi: 10.1002/j.1460-2075.1985.tb03882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukamoto K, Jackson I, Urabe K, Montague P, Hearing V. A second tyrosinase-related protein, TRP-2, is a melanogenic enzyme termed DOPAchrome tautomerase. EMBO (Eur Mol Biol Organ) J. 1992;11:519–526. doi: 10.1002/j.1460-2075.1992.tb05082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lethé, B., P. van der Bruggen, F. Brasseur, and T. Boon. 1997. MAGE-1 expression threshold for the lysis of melanoma cell lines by a specific cytotoxic T lymphocyte. Melanoma Res. 7(Suppl. 2):S83–S88. [PubMed]
- 38.Labarriere N, Diez E, Pandolfino MC, Viret C, Guilloux Y, Le Guiner S, Fonteneau JF, Dreno B, Jotereau F. Optimal T cell activation by melanoma cells depends on a minimal level of antigen transcription. J Immunol. 1997;158:1238–1245. [PubMed] [Google Scholar]
- 39.Schwartz R. Acquisition of immunologic self-tolerance. Cell. 1989;57:1073–1081. doi: 10.1016/0092-8674(89)90044-5. [DOI] [PubMed] [Google Scholar]
- 40.Slingluff C, Hunt D, Engelhard V. Direct analysis of tumor-associated peptide antigens. Curr Opin Immunol. 1994;6:733–740. doi: 10.1016/0952-7915(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 41.Sidney J, del Guercio MF, Southwood S, Engelhard V, Appella E, Rammensee H, Falk K, Rotzschke O, Takiguchi M, Kubo RT, et al. Several HLA alleles share overlapping peptide specificities. J Immunol. 1995;154:247–259. [PubMed] [Google Scholar]
- 42.Missale G, Redeker A, Person J, Fowler P, Guilhot S, Schlicht HJ, Ferrari C, Chisari FV. HLA-A31– and HLA-Aw68–restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J Exp Med. 1993;177:751–762. doi: 10.1084/jem.177.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang RF, Johnston SL, Southwood S, Sette A, Rosenberg SA. Recognition of an antigenic peptide derived from tyrosinase-related protein-2 by CTL in the context of HLA-A31 and -A33. J Immunol. 1998;160:890–897. [PubMed] [Google Scholar]