Abstract
Hematopoietic stem cells (HSCs) in adult marrow are believed to be derived from fetal liver precursors. To study cell kinetics involved in long-term hematopoiesis, we studied single-sorted candidate HSCs from fetal liver that were cultured in the presence of a mixture of stimulatory cytokines. After 8–10 d, the number of cells in primary cultures varied from <100 to >10,000 cells. Single cells in slow growing colonies were recloned upon reaching a 100–200 cell stage. Strikingly, the number of cells in subclones varied widely again. These results are indicative of asymmetric divisions in primitive hematopoietic cells in which proliferative potential and cell cycle properties are unevenly distributed among daughter cells. The continuous generation of functional heterogeneity among the clonal progeny of HSCs is in support of intrinsic control of stem cell fate and provides a model for the long-term maintenance of hematopoiesis in vitro and in vivo.
Keywords: asymmetric cell divisions, stem cells, hematopoiesis, cell cycle, long-term hematopoietic cell cultures
Blood formation originates in a small population of hematopoietic stem cells (HSCs)1 that have been defined as pluripotent cells with self-renewal capacity (1, 2). The mechanisms underlying the proliferation and differentiation of HSCs are incompletely understood (3, 4). Both extrinsic (e.g., growth factors and cell–matrix interactions) and intrinsic factors (e.g., developmentally controlled transcription factors) are involved in the regulation of HSCs. Although hematopoietic growth factors and an adequate microenvironment are crucial for the survival and proliferation of HSCs, self-renewal/differentiation decisions in HSCs seem to be derived largely independently of cytokines and are postulated to be determined intrinsically (4–8).
Studies aimed at dissecting the molecular mechanism involved in stem cell regulation have been hampered by difficulties in obtaining populations of HSCs devoid of more differentiated progenitor cells. Apart from a paucity of distinguishing phenotypic features, these difficulties are also related to available HSCs assays. Thus, despite intense efforts to establish determinants by which primitive HSCs can be defined prospectively, available in vivo and in vitro stem cell assays only allow retrospective identification of HSCs. In mice, candidate HSCs are believed to be among the cells expressing a Thy-1.1loSca1hiLin−/lo phenotype (9, 10). This cell population represents <0.1% of murine fetal liver and is highly enriched for multipotent progenitors (11). However, cells with this phenotype display considerable heterogeneity with respect to “stem cell” properties such as CFU-S formation (12), and only a minority has the ability to reconstitute lympho-myelopoiesis in lethally irradiated recipients (10, 13). In the last two decades, a large amount of effort has also been directed towards the development of in vitro assays for human HSCs (for review see reference 14). One prominent example of such an assay is the long-term culture–initiating cell (LTC-IC) assay (15). LTC-ICs are hematopoietic cells that are capable of generating myeloid colony-forming cells after at least 5 wk of culture in the presence of irradiated feeder cells. LTC-ICs are highly enriched among cells with a CD34+CD38− phenotype, and the yield of LTC-ICs in such cells from adult human marrow and umbilical cord blood is ∼20 (16) and ∼50% (17), respectively. Based on these considerations, human CD34+CD38− cells are expected to be highly enriched for HSCs. However, the frequency of CD34+CD38− cord blood cells capable of initiating hematopoiesis in immune deficient mice is only 0.1% (18–20). The heterogeneity within the CD34+CD38− compartment of human cells is reminiscent of the functional heterogeneity as outlined above for murine cells with a Thy-1.1loSca1hiLin−/lo phenotype.
To study the functional heterogeneity of candidate HSCs, we followed the fate of single-sorted fetal liver CD34+CD38− cells that were cultured in cytokine-supplemented serum-free medium. By combining observations on in vitro growth with a detailed characterization of individual cells produced in culture, we observed that functional heterogeneity is continuously generated among the clonal progeny of HSCs. Furthermore, the growth characteristics of cell clones allowed us to more closely and, to a certain degree, semiprospectively define cells with the highest proliferative capacity. Our observations provide an explanation for the extreme functional heterogeneity among highly purified candidate HSCs. Several possible mechanisms for the asymmetric cell divisions that best explain our findings are discussed.
Materials and Methods
Purification of Fetal Liver Stem Cell Candidates
Cells with CD34+CD38−CD71−CD45RA− phenotype were isolated from previously frozen samples of fetal liver obtained from elective, therapeutic abortions in wk 10–16 of gestation. The use of human material was approved by local Institutional Review Boards as well as the Ethical Screening Committee of the University of British Columbia. Cells were separated using Ficoll-Hypaque and processed for flow cytometry cell sorting as previously described (6, 21). In brief, cells were labeled with OKT-9–FITC (anti-CD71), 8G12-Cy-5 (anti-CD34), 8d2-PE (anti-CD45RA), and Leu-17–PE (anti-CD38; Becton Dickinson, San Jose, CA) and sorted using a dual laser FACStar® Plus (Becton Dickinson) equipped with an argon and helium-neon laser (Fig. 1). Cells either were collected in serum-free medium or were individually sorted directly into round-bottomed tissue culture plates (Nunc, Roskilde, Denmark) containing serum-free medium supplemented with hematopoietic growth factors (as indicated below) using an automatic cell deposition unit (Becton Dickinson). The experimental design of the culture experiments is shown in Fig. 2.
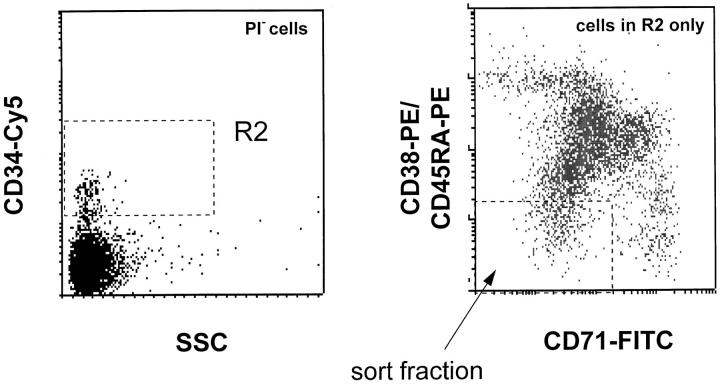
Figure 1.
FACS® profile of the CD34+CD38− candidate HSCs sorted from fetal liver to initiate single cell cultures in 96-well plates. Viable, propidium iodide negative (PI−) CD34+CD38− cells with a low side scatter (SSC) and low levels of CD45RA and CD71 expression were recovered from the indicated sort windows.
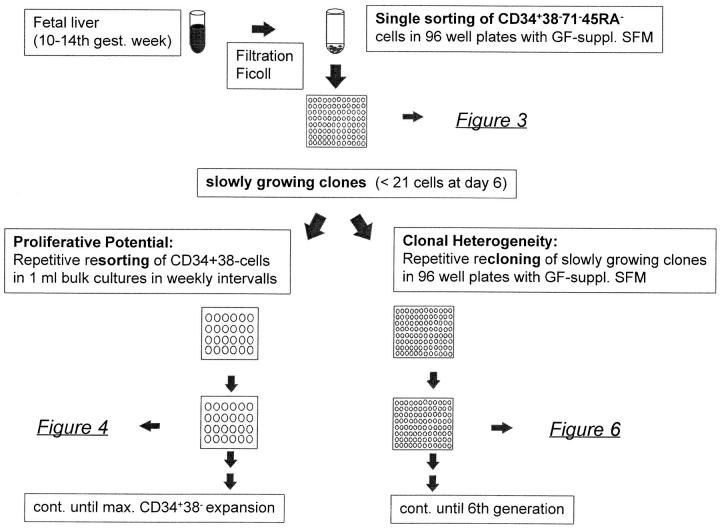
Figure 2.
Experimental design of study.
Culture of Cells
Single-cell Cultures.
Single-sorted cells were cultured in 96-well round-bottomed plates (60 cells/plate) with each well containing 100 μl of serum-free medium (21) containing 100 ng/ml each of Steel factor (or stem cell factor) and Flt-3 ligand and 20 ng/ml each of IL-3, IL-6, and G-CSF. After 5–7 d of culture, another 100 μl of growth factor–containing medium was added. After 6–9 d, the number of cells per well varied over a wide range from wells with >10,000 cells to wells with <100 cells. Slowly growing colonies with 100–250 cells after 8–13 d in culture were recloned and the number and morphology (shape, size, and refractive index) of cells in each subclone were scored at various time intervals. Slowly growing colonies were recloned again between days 8 and 19. Such recloning of subclones was repeated until the sixth generation.
Expansion Culture.
For the evaluation of the proliferative potential slow growing colonies that had reached a level of 5 × 104 cells were transferred in 1-ml cultures of 24-well plates. CD34+CD38− cells were resorted every 5–10 d as described above if cultures became confluent (>106 cells/well) to initiate subcultures until the percentage of CD34+CD38− cells per well dropped below 0.1% of viable cells.
Results
Characterization of Primary Cultures Derived from Single CD34+CD38− Fetal Liver Cells.
Single CD34+CD38− fetal liver stem cell candidates were sorted in individual wells of microtiter plates containing serum-free medium supplemented with cytokines (see Materials and Methods). The plating efficiency (number of wells containing visible single cells after sorting) was >90%, of which >80% appeared viable after 24 h. The cell number, morphology, and the percentage of CD34+CD38− cells in primary clones was analyzed at different time intervals. After 6–9 d, the number of cells in the cultures varied over a wide range, indicating extensive heterogeneity among fetal liver CD34+CD38− cells. Rapidly growing clones of >10,000 cells as well as slow growing clones of <50 cells were observed. All wells with growing cells (n = 121) were transferred to 1-ml cultures after 5 × 104 or more cells were present between 10 and 21 d of culture. Cells with a CD34+CD38− phenotype were resorted from expanded 1-ml cultures when these cultures became confluent (0.5-2.0 × 106 cells) and used for continuation of cultures using identical culture conditions. This procedure was repeated until no more CD34+ cells were produced. Three different categories (A–C) were defined based on the ability to produce CD34+CD38− cells in culture. Clones producing CD34+CD38− cells up to day 16 were categorized as A, whereas colonies producing CD34+CD38− cells for up to day 59 or more were categorized as B and C, respectively.
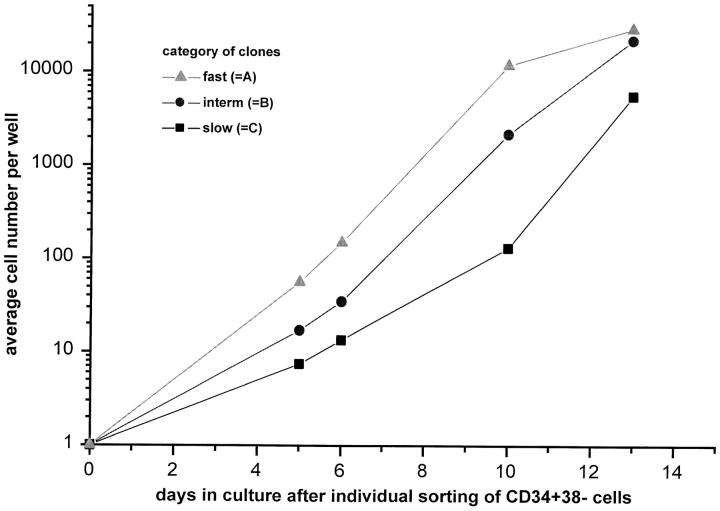
Retrospective analysis revealed that colonies that gave rise to the highest number of CD34+CD38− cells for the longest time period corresponded to primary clones with slow growth properties in the first 9 d of culture (Table 1). This fraction represented 16% of the clones analyzed, whereas the majority of clones (60%) were fast growing (>103 cells at day 9) with a relatively low expansion potential. Furthermore, the number of cells in fast growing colonies already started to plateau at day 10, whereas the cell number in slow growing clones was still expanding at day 12 (Fig. 3). These observations underscore the functional heterogeneity of the CD34+CD38− cell compartment in human fetal liver and suggest that categories A and B represent the property of more differentiated progenitor cells.
Table 1.
Production of CD34+CD38− Cells by Single Cells in Serum-free Medium Supplemented with Steel Factor, Flt-3, IL-3, IL-6, and G-CSF
| Category | Production of CD34+CD38− cells up to day | No. of clones (n = 121) | Proportion (%) | Maximum CD34+CD38− expansion* | Average cell-cycle time (h) up to day 9‡ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | 16 | 73 | 60 | 4.4 × 103 | 20.8 ± 0.2 | |||||
| B | 59 | 29 | 24 | 6.6 × 107 | 24.8 ± 0.1 | |||||
| C | 129 | 19 | 16 | 2.7 × 1012 | 32.0 ± 0.1 |
Obtained by repeated sorting and reculture of CD34+CD38− cells in 1-ml cultures until no more CD34+CD38− cells were produced.
Obtained by dividing the culture period in hours by the log2 of the estimated number of cells (n) at day 9.
Figure 3.
Clonal heterogeneity in early proliferative response of single-sorted CD34+CD38− fetal liver cells cultured in growth factor–supplemented SFM. Single CD34+CD38− fetal liver cells were sorted into the wells of round-bottomed microtiter plates containing serum-free medium supplemented with Steel factor, Flt-3 ligand, IL-3, IL-6, and G-CSF at 37°C. At the indicated time intervals, the number of cells in each well was scored.
Expansion Potential of Single-sorted CD34+CD38− Fetal Liver Cells.
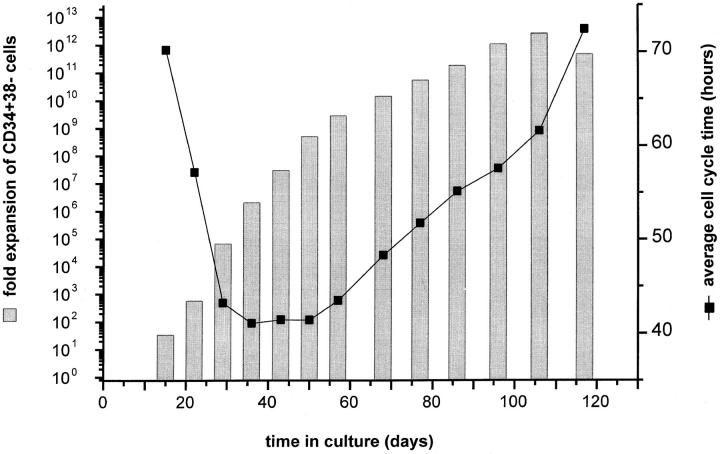
To further characterize the expansion potential of slow growing clones, the experiment described above was repeated with CD34+CD38− cells from a different fetal liver. The CD34+CD38− expansion potential per clone varied over a wide range, from 1.43 × 104 to 2.7 × 1012, with an average expansion of 8.65 × 108 CD34+CD38− cells in the 27 clones that were analyzed. The maximum continued production of CD34+CD38− cells was 129 d. Strikingly, a single clone produced 2.7 × 1012 CD34+CD38− cells over 106 d in culture (corresponding to at least 41 “self-renewal” population doublings). Interestingly, in this clone (Fig. 4), the calculated average cell cycle time of CD34+CD38− cells in culture was very long up to day 15 (70.2 h), decreased from day 29 to 57 (to 43.3 h), and finally gradually increased again up until day 117 (72.4 h). These findings indicate that after an initial period of slow growth, the CD34+CD38− cells divided more rapidly before decreased turnover and eventual loss of CD34+CD38− cell production. Similar growth kinetics were observed for the other clones with lower proliferative potential examined in these experiments.
Figure 4.
Expansion potential and average cell cycle time of CD34+CD38− cells resorted from expanded cultures initiated with a single-sorted CD34+38− fetal liver cell. Initial culture and subculture of the CD34+CD38− cells was in serum-free media supplemented with cytokines. The average cell cycle time of CD34+CD38− was calculated as described in Table 1.
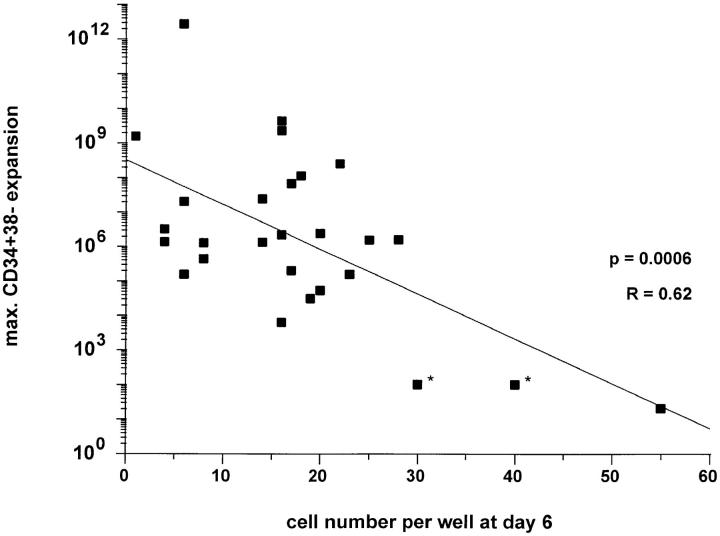
In general, these results document that extensive heterogeneity exists in the ability to produce CD34+CD38− cells among slow growing CD34+CD38− fetal liver cells. To examine whether the early proliferative response of single slow growing clones is a good predictor of maximum expansion potential, the cell numbers in clones at different time intervals were plotted against the total number of CD34+CD38− cells produced by that clone. Linear regression analysis revealed a significant negative correlation (P = 0.0006) between the cell number at day 6 and the maximum CD34+38− expansion potential (Fig. 5). This result suggests that the proliferative potential and cell cycle characteristics in primitive hematopoietic cells are linked.
Figure 5.
Correlation between early proliferative response at day 6 of slow growing clones derived from single-sorted CD34+CD38− fetal liver cells and maximum CD34+38− expansion potential of different clones. The maximum expansion potential was determined by repeated subculture of CD34+CD38− cells sorted from confluent 1-ml cultures initially seeded with the content of a single well. *Estimated cell numbers: amount of CD34+38− was not determined due to low cell numbers; expansion was calculated on total cells for these clones.
Heterogeneity in Subclones Derived from Slow Growing CD34+CD38− Fetal Liver Clones.
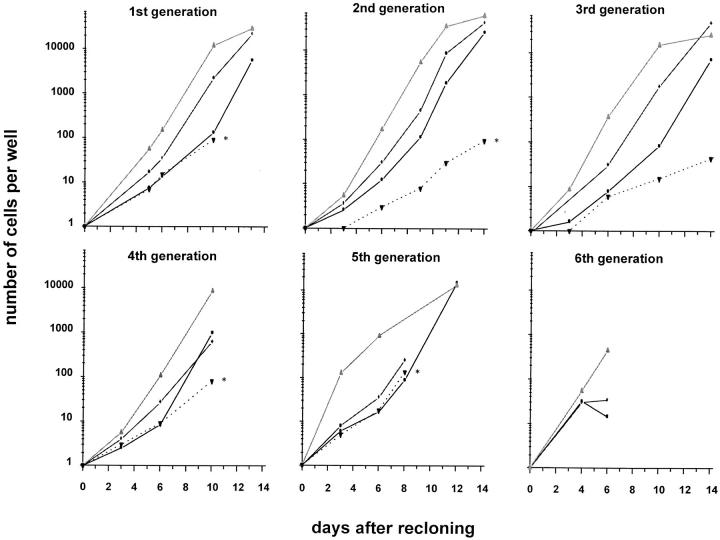
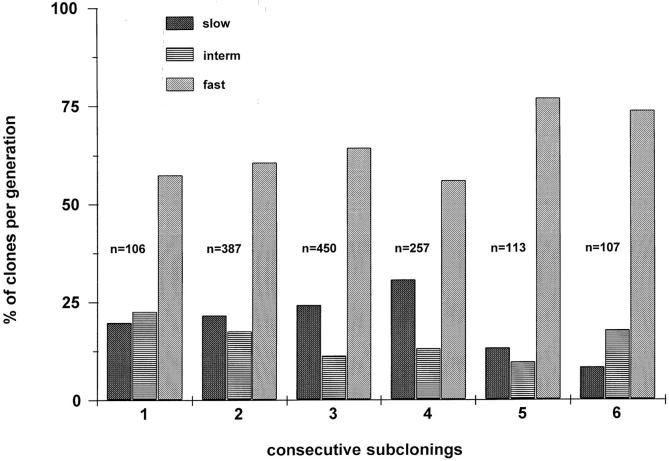
To further analyze the functional heterogeneity within the most primitive, slow growing CD34+CD38− fetal liver cells, we recloned cells recovered from category C at days 8–19, when cell numbers had reached between 100 and 250 cells. Individual subclones were analyzed with respect to cell number and morphology at different time intervals and, in some cases, also analyzed for total CD34+CD38− cell production as described above for primary clones. Surprisingly, the clonal heterogeneity observed in the primary single-sorted CD34+CD38− fetal liver cells was preserved. This is shown for five consecutive generations of subclones of a single slow growing colony of CD34+CD38− cells in Fig. 6. Furthermore, as shown in Fig. 7, the distribution pattern of the three categories remained more or less constant through multiple generations of recloning, with fast growing clones representing the majority. Slowly growing clones that did not show morphological features of terminal (macrophage) differentiation represented 20% of primary clones and between 20 and 30% of second to fourth generation subclones, and decreased to <10% by the sixth generation (Fig. 7). Our results indicate that the number of CD34+CD38− cells with extensive replating potential in slow growing clones decreases upon multiple rounds of recloning.
Figure 6.
Clonal heterogeneity of subclones is preserved through at least five consecutive generations of recloning of slow growing clones derived from single-sorted CD34+CD38− fetal liver cells. The average number of cells in multiple clones of categories A, B, and C (see text) is plotted. A single “category C” clone (dotted line and *) was recloned when 100–200 cells were present and this was repeated for subsequent category C subclones that were produced from a single initial clone.
Figure 7.
Percentage of slow, intermediate, and fast growing clones in the progeny of single-sorted CD34+CD38− fetal liver cells shown in Fig. 6. n = number of clones/generation. Figure represents pooled data from two experiments.
Compared with the findings in slow growing clones, recloning of intermediate or fast growing clones revealed a more homogeneous growth pattern in subclones. In such subclones, the vast majority (>90%) either developed into mature macrophages or produced up to a few thousand CD34+CD38− cells over 2–3 wk before terminal myeloid differentiation.
Discussion
The data presented in this paper provide insight into three major aspects of stem cell biology. First, the data show that CD34+CD38− candidate HSCs from fetal liver display extensive functional heterogeneity when cultured as single cells in defined culture conditions. Secondly, this heterogeneity is generated in the slow growing progeny of single-sorted fetal liver cells. Thirdly, cells with the highest overall proliferative potential could be recognized by their slow growth kinetics, allowing early identification of colonies containing cells with a very high proliferative potential.
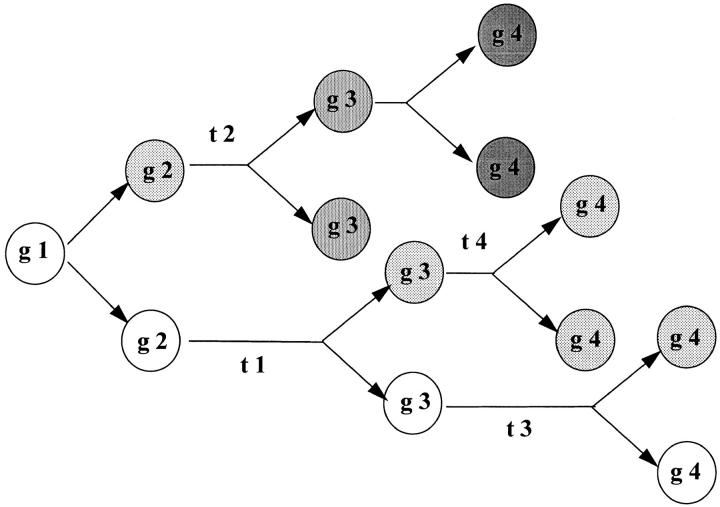
The majority of subclones derived from slow growing CD34+CD38− cells were fast growing clones with a low proliferative potential. However, a minority of subclones showed similar growth kinetics to the parental clones, and this heterogeneity was preserved through four to six generations of recloning. Because culture conditions were kept constant in all these experiments, our findings support the conclusion that differences in the fate of individual stem cells are continuously and intrinsically generated. What could be the mechanisms involved in the heritable functional heterogeneity within the clonal progeny of slow growing candidate HSCs? The most simple hypothesis is that daughter cells are endowed with different cell fates via asymmetric cell divisions (Fig. 8). Such asymmetric cell divisions would result in one daughter cell being similar to the mother cell and the other daughter cell more committed to terminal differentiation. Alternatively, differences in cell fate could be acquired after mitosis by unknown mechanisms. Direct evidence for asymmetric divisions in early hematopoiesis was previously reported using time lapse video recordings of CD34+CD71loCD45RAlo bone marrow progenitor cells in culture (22). In our study, we also observed a distinct polarity among slow growing CD34+CD38− cells: such cells were small, motile, and appeared as “commas” with most of the cellular volume preceding a cytoplasmic tail of one to two cell diameter (5–10 μm). In previous studies, it was reported that ∼20% of the cell divisions in early hematopoiesis would qualify as asymmetric (5, 6), whereas extensive amplification of cell numbers during proliferation and expansion of more committed hematopoietic progenitors would involve primarily symmetric divisions (3).
Figure 8.
The intrinsic timetable model of stem cell biology. In this model, the functional properties of stem cells are (i) linked to the number of preceding cell divisions or generations (g) and (ii) unevenly distributed among daughter cells upon each cell division. It is postulated that each stem cell division is asymmetric and results in daughter cells that differ in cell cycle properties. As a result, the time interval between successive generations (t 1, t 2, etc.) is variable between clones of the same generation and the interval between divisions is subject to both intrinsic (developmental) and extrinsic (microenvironment and growth factors) control. Clonal variations in turn-over time result in an extreme hierarchy of stem cells varying in replicative history and related functional properties that is difficult to reconcile with the concept of stem cells as a homogeneous population of cells.
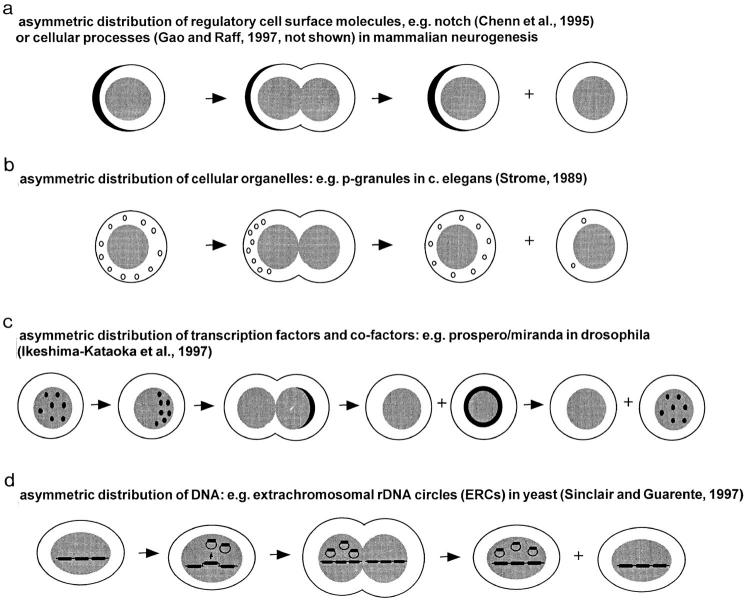
To date, limited data on the mechanism involved in asymmetric division of HSCs are available. Investigations of asymmetric division in plants (for review see reference 23), Caenorhabditis elegans (24), Drosophila and other species (for review see reference 25), as well as in mammalian neural stem cells (26–28) have elucidated several distinct possibilities, and several working models based on these better defined systems are shown in Fig. 9. Similar mechanisms are implicated in asymmetric cell divisions in Drosophila and mammalian neural progenitor cells (for review see references 29, 30). Many different proteins involved in these processes have been identified, some of which, i.e., the notch- (27) and numb- (28) families of proteins, may also be involved in early hemopoiesis (Fig. 9 a). Indeed, Drosophila neuroblast stem cells have been shown to undergo self-renewing asymmetric divisions in culture (31), very similar to the data on fetal liver HSCs reported here. Recently, Milner et al. showed that constitutive expression of the intracellular domain of notch-1 in the 32D myeloid progenitor cell line leads to inhibition of differentiation and to expansion of undifferentiated cells in response to G-CSF (32). These effects have recently been shown to be transmitted by the Notch-1-ligand Jagged-1 (33). The pros gene, which encodes for the homeodomain-containing nuclear protein prospero, is involved in asymmetric divisions of Drosophila neuroblasts (Fig. 9 c and references 34, 35). This might be of special interest in light of recent data on the role of homeobox transcription factors HOXA10 (36), HOXB4 (37), and others in hematopoiesis (38, 39). Studies on oligodendrocyte precursor cells by Gao and Raff (26) also resemble the observations reported here. Gao and Raff concluded that the differentiation and maturation of oligodendrocyte precursor cells is an intrinsic property of the cell and that asymmetric divisions of precursors contribute to differences in cell fate. Interestingly, the cell cycle time of different precursors was also found to be related to cell fate in their studies. Finally, it also seems important to exclude asymmetric inheritance of extrachromosomal rDNA circles (40) as an explanation for the divergence in the fate of HSCs (Fig. 9 d).
Figure 9.
Possible mechanisms of asymmetric cell division in HSCs.
It is tempting to speculate that our observations provide an explanation for the recent observation that committed progenitor cells do not mediate early hematopoietic reconstitution after blood cell transplantation in mice (41). According to this scenario, slow growing clones, capable of engrafting lethally irradiated recipients, would produce rapidly growing and differentiating subclones while simultaneously producing more primitive precursors. Data on ontogeny-related changes in the functional properties and telomere length of fetal liver compared with cord blood and adult bone marrow cells (42, 43) have led to the speculation that the replicative life span of phenotypically identical HSCs may decrease with age and could be limited to <100 cell divisions (4). A model of early stem cell biology that is compatible with both asymmetric divisions of the HSCs and ontogeny-related changes in HSC function is shown in Fig. 8.
In conclusion, the data shown here provide further evidence that the fate of the most primitive HSC is primarily determined intrinsically and regulated only in a permissive way by extrinsic factors in agreement with previous studies in model systems (8, for review see reference 44). The mechanisms underlying the asymmetric divisions of HSCs documented here appear to be a fruitful area for further studies.
Acknowledgments
We thank Colleen MacKinnon for typing the manuscript.
This work was supported by National Institutes of Health grant AI-29524 and by a grant from the National Cancer Institute of Canada with funds from the Terry Fox Run as well as a grant from the Deutsche Forschungsgemeinschaft (to T.H. Brummendorf ).
Abbreviations used in this paper
- HSC
hematopoietic stem cells
- LTC-IC
long-term culture–initiating cell
References
- 1.Till JE, McCulloch EA. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res. 1961;14:213–222. [PubMed] [Google Scholar]
- 2.Metcalf, D. 1977. Hemopoietic Colonies. In Vitro Cloning of Normal and Leukemic Cells. Springer-Verlag, Berlin/ Heidelberg, Germany. 227 pp. [PubMed]
- 3.Morrison SJ, Shah NM, Anderson DJ. Regulatory mechanisms in stem cell biology. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- 4.Lansdorp PM. Self-renewal of stem cells. Biol Blood Marrow Transplant. 1997;3:171–178. [PubMed] [Google Scholar]
- 5.Suda T, Suda J, Ogawa M. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc Natl Acad Sci USA. 1984;81:2520–2524. doi: 10.1073/pnas.81.8.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayani H, Dragowska W, Lansdorp PM. Lineage commitment in human hemopoiesis involves asymmetric cell division of multipotent progenitors and does not appear to be influenced by cytokines. J Cell Physiol. 1993;157:579–586. doi: 10.1002/jcp.1041570318. [DOI] [PubMed] [Google Scholar]
- 7.Stoffel, R., B. Ledermann, F.J. de Sauvage, and R.C. Skoda. 1997. Evidence for a selective-permissive role of cytokine receptors in hematopoietic cell fate decisions. Blood. 90:123a(abstr.). [DOI] [PMC free article] [PubMed]
- 8.Fairbairn LJ, Cowling GJ, Reipert BM, Dexter TM. Suppression of apoptosis allows differentiation and development of a multipotent hemopoietic cell line in the absence of added growth factors. Cell. 1993;74:823–832. doi: 10.1016/0092-8674(93)90462-y. [DOI] [PubMed] [Google Scholar]
- 9.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 10.Uchida N, Weissman IL. Searching for hemopoietic stem cells: evidence that Thy-1.1lo Lin− Sca-1+cells are the only stem cells in C57BL/Ka-Thy-1.1 bone marrow. J Exp Med. 1992;175:175–184. doi: 10.1084/jem.175.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci USA. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spangrude GJ, Johnson GR. Resting and activated subsets of mouse multipotent hematopoietic stem cells. Proc Natl Acad Sci USA. 1990;87:7433–7437. doi: 10.1073/pnas.87.19.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith LG, Weissman IL, Heimfeld S. Clonal analysis of hematopoietic stem-cell differentiation in vivo. Proc Natl Acad Sci USA. 1991;88:2788–2792. doi: 10.1073/pnas.88.7.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bock TA. Assay systems for hematopoietic stem and progenitor cells. Stem Cells (Basel) 1997;15:185–195. doi: 10.1002/stem.5530150824. [DOI] [PubMed] [Google Scholar]
- 15.Sutherland HJ, Lansdorp PM, Henkelman DH, Eaves AC, Eaves CJ. Functional characterization of individual human hematopoietic stem cells cultured at limiting dilution on supportive marrow stromal layers. Proc Natl Acad Sci USA. 1990;87:3584–3588. doi: 10.1073/pnas.87.9.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petzer AL, Hogge DE, Lansdorp PM, Reid DS, Eaves CJ. Self-renewal of primitive human hematopoietic cells (long-term culture–initiating cells) in vitroand their expansion in defined medium. Proc Natl Acad Sci USA. 1996;93:1470–1474. doi: 10.1073/pnas.93.4.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conneally E, Cashman J, Petzer A, Eaves C. Expansion in vitro of transplantable human cord blood stem cells demonstrated using a quantitative assay of their lympho-myeloid repopulating activity in nonobese diabetic–scid/scidmice. Proc Natl Acad Sci USA. 1997;94:9836–9841. doi: 10.1073/pnas.94.18.9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larochelle A, Vormoor J, Hanenberg H, Wang JCY, Bhatia M, Lapidot T, Moritz T, Murdoch B, Xiao XL, Kato I, et al. Identification of primitive human hematopoietic cells capable of repopulating NOD/SCID mouse bone marrow: implications for gene therapy. Nat Med. 1996;2:1329–1337. doi: 10.1038/nm1296-1329. [DOI] [PubMed] [Google Scholar]
- 19.Bhatia M, Wang JCY, Kapp U, Bonnet D, Dick JE. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc Natl Acad Sci USA. 1997;94:5320–5325. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eaves C, Miller C, Cashman J, Conneally E, Petzer A, Zandstra P, Eaves A. Hematopoietic stem cells: inferences from in vivo assays. Stem Cells (Basel) 1997;15:1–5. doi: 10.1002/stem.5530150802. [DOI] [PubMed] [Google Scholar]
- 21.Lansdorp PM, Dragowska W. Long-term erythropoiesis from constant numbers of CD34+cells in serum-free cultures initiated with highly purified progenitor cells from human bone marrow. J Exp Med. 1992;175:1501–1509. doi: 10.1084/jem.175.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denkers IAM, Dragowska W, Jaggi B, Palcic B, Lansdorp PM. Time lapse video recordings of highly purified human hematopoietic progenitor cells in culture. Stem Cells (Basel) 1993;11:243–248. doi: 10.1002/stem.5530110312. [DOI] [PubMed] [Google Scholar]
- 23.Gallagher K, Smith LG. Asymmetric cell division and cell fate in plants. Curr Opin Cell Biol. 1997;9:842–848. doi: 10.1016/s0955-0674(97)80086-5. [DOI] [PubMed] [Google Scholar]
- 24.Strome S. Generation of cell diversity during early embryogenesis in the nematode Caenorhabditis elegans. . Int Rev Cytol. 1989;114:81–123. doi: 10.1016/s0074-7696(08)60859-1. [DOI] [PubMed] [Google Scholar]
- 25.Knoblich JA. Mechanisms of asymmetric cell division during animal development. Curr Opin Cell Biol. 1997;9:833–841. doi: 10.1016/s0955-0674(97)80085-3. [DOI] [PubMed] [Google Scholar]
- 26.Gao F-B, Raff M. Cell size control and a cell-intrinsic maturation program in proliferating oligodendrocyte precursor cells. J Cell Biol. 1997;138:1367–1377. doi: 10.1083/jcb.138.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 28.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 29.Rhyu MS, Knoblich JA. Spindle orientation and asymmetric cell fate. Cell. 1995;82:523–526. doi: 10.1016/0092-8674(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 30.Lin H, Schagat T. Neuroblasts: a model for the asymmetric division of stem cells. Trends Genet. 1997;13:33–39. doi: 10.1016/s0168-9525(96)10050-0. [DOI] [PubMed] [Google Scholar]
- 31.Furst A, Mahowald AP. Cell division cycle of cultured neural precursor cells from Drosophila. . Dev Biol. 1985;112:467–476. doi: 10.1016/0012-1606(85)90419-1. [DOI] [PubMed] [Google Scholar]
- 32.Milner LA, Bigas A, Kopan R, Brashem-Stein C, Bernstein ID, Martin DIK. Inhibition of granulocytic differentiation by mNotch1. . Proc Natl Acad Sci USA. 1996;93:13014–13019. doi: 10.1073/pnas.93.23.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Milner LA, Deng Y, Iwata M, Banta A, Graf A, Marcovina S, Friedman C, Trask BJ, Hood L, Torok-Storb B. The human homolog of rat jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with notch1. Immunity. 1998;8:43–55. doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- 34.Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophiladevelopment. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- 35.Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs prospero to a daughter cell during Drosophilaasymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence HJ, Sauvageau G, Ahmadi N, Lopez AR, Lebeau MM, Link M, Humphries K, Largman C. Stage- and lineage-specific expression of the HOXA10 homeobox gene in normal and leukemic hematopoietic cells. Exp Hematol. 1995;23:1160–1166. [PubMed] [Google Scholar]
- 37.Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, Humphries RK. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. . Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 38.Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, Largman C, Lawrence HJ, Humphries RK. Differential expression of homeobox genes in functionally distinct CD34+subpopulations of human bone marrow cells. Proc Natl Acad Sci USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence HJ, Sauvageau G, Humphries RK, Largman C. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells (Basel) 1996;14:281–291. doi: 10.1002/stem.140281. [DOI] [PubMed] [Google Scholar]
- 40.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 41.Zijlmans JMJM, Visser JWM, Later L, Veer, Kleiverda K, Heemskerk DPM, Kluin PM, Willemze R, Fibbe WE. The early phase of engraftment after murine blood cell transplantation is mediated by hematopoietic stem cells. Proc Natl Acad Sci USA. 1998;95:725–729. doi: 10.1073/pnas.95.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lansdorp PM, Dragowska W, Mayani H. Ontogeny-related changes in proliferative potential of human hematopoietic cells. J Exp Med. 1993;178:787–791. doi: 10.1084/jem.178.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cross MA, Enver T. The lineage commitment of haemopoietic progenitor cells. Curr Opin Genet Dev. 1997;7:609–613. doi: 10.1016/s0959-437x(97)80007-x. [DOI] [PubMed] [Google Scholar]