Abstract
Many pathogenic bacteria can use heme compounds as a source of iron. Pathogenic Escherichia coli strains are capable of using hemoglobin as an iron source. However, the mechanism of heme acquisition from hemoglobin is not understood for this microorganism. We present the first molecular characterization of a hemoglobin protease (Hbp) from a human pathogenic E. coli strain. The enzyme also appeared to be a heme-binding protein. Affinity purification of this bifunctional protein enabled us to identify the extracellular gene product, and to clone and analyze its gene. A purification procedure developed for Hbp allowed us to perform functional studies. The protein interacted with hemoglobin, degraded it and subsequently bound the released heme. These results suggest that the protein is involved in heme acquisition by this human pathogen. Hbp belongs to the so-called IgA1 protease-like proteins, as indicated by the kinetics of its membrane transfer and DNA sequence similarity. The gene of this protein appears to be located on the large pColV-K30 episome, that only has been isolated from human and animal pathogens. All these characteristics indicate that Hbp may be an important virulence factor that may play a significant role in the pathogenesis of E. coli infections.
Keywords: hemoglobin protease, virulence factor, pColV-K30 episome, secretion, heme binding
The low concentration of free iron in the mucous membranes and in tissues is one of the first lines of host defense against bacterial infection. In humans, the majority of iron is located intracellularly complexed in ferritin, hemoglobin or heme proteins. Hemoglobin and heme, when released by lysis of erythrocytes, are bound by the plasma proteins haptoglobin and hemopexin, respectively. The small quantities of extracellular iron are complexed to carrier proteins like transferrin present in serum and lactoferrin which is present within mucosal surfaces (1, 2). Therefore, possession of specialized iron acquisition systems is a prerequisite for successful multiplication of pathogenic bacteria in their host.
The most common sites of human infections due to Escherichia coli are the gastrointestinal and urinary tract. Furthermore, this microorganism is also a well-recognized cause of septicemia and wound infections. Ferric iron, heme, and hemoglobin significantly enhance the susceptibility of animals and humans to E. coli infections (3, 4), and several high affinity iron uptake systems have been identified (1, 5). The molecular mechanisms of these iron uptake systems have been studied thoroughly. One way to sequester iron involves the synthesis of siderophores and specific transport systems for the uptake of the iron-loaded siderophores into the cell. Siderophores like enterobactin and aerobactin are able to remove iron from transferrin or lactoferrin. A large number of pathogenic bacterial species use heme compounds as a source of iron, and various outer membrane proteins have been isolated and characterized which proposed function is to bind heme to the bacterial cell surface (2, 6). In these systems the outer membrane receptor directly recognizes the heme compounds. A second more complex way to obtain heme involves an extracellular protein that binds heme and shuttles it back to a specific outer membrane receptor (7). This so-called hemophore-dependent heme acquisition system has been detected in Haemophilus influenzae (8) and Serratia marcescens (9). E. coli can also use heme and hemoglobin as iron sources, but little is known about the mechanisms of heme utilization involved (10). The ability to use heme compounds as a source of iron has been implicated as an important determinant of virulence in E. coli (11).
This study describes the molecular characterization of a hemoglobin protease from the human pathogenic E. coli strain EB1. This strain was isolated from an intra-abdominal wound infection (12). We present evidence that this protein is part of a hemophore-dependent heme acquisition system. The protein interacts with hemoglobin, degrades it, and subsequently binds the released heme. The protein belongs to the so-called IgA1 protease-like autotransporter proteins as indicated by the kinetics of its membrane transfer and DNA sequence similarity. Since the heme-binding protein is capable of degrading hemoglobin, its gene was named hbp for hemoglobin protease (Hbp).1
Materials and Methods
Bacterial Strains, Phages, Plasmids, and Media.
E. coli strain EB1 (O8-K43) is a clinical isolate from a patient with a wound infection (12). This strain was identified with the API 20E system (API S.A., Montalieu Vercieu, France) and used as a source of chromosomal and plasmid DNA for all cloning and PCR experiments. E. coli K-12 strain DH5α (GIBCO BRL, Gaithersburg, MD) and the plasmids pBluescript II SK+ (Stratagene, La Jolla, CA) and pBR322 were used in routine cloning procedures (13). Strain BL21 (F− ompTr− Bm− B) (DE3), and the plasmids pET11d and pLysS were used for subcloning and controlled expression of hbp (14). E. coli strain TG1 (13) and the phage vector M13mp19 were used for shotgun cloning and preparation of DNA templates for sequencing. The kanamycin resistance gene cartridge plasmid pUC4-K (Pharmacia Biotech Sverige, Uppsala, Sweden) and the vector pSG335 (15) were used in insertional mutagenesis experiments. E. coli strain AN299-23 (16) and plasmid pACYC184 (17) were used in a colicin V assay and in conjugation experiments. The plasmid pColV-K30 was isolated from strain LG1522 (18). Cells were routinely grown in YT medium (19). To prepare Hbp from the culture supernatant a chemically defined medium (MA-medium) described by Miller, 1992) was used. If required, antibiotics were added to the culture medium (13).
General Methods.
Recombinant DNA techniques were carried out as described (13). A digoxygenin labeling and detection kit (Boehringer Mannheim GmbH, Mannheim, Germany) was used to probe Southern blots. DNA sequencing was performed using the Taq Dye Primer Cycle Sequencing Kit and the 373A Automated DNA Sequencer of Applied Biosystems Inc. (Foster City, CA). Protein concentrations were determined according to Bradford (20) with bovine serum albumin as standard. SDS-PAGE and immunoblotting were carried out as described by (21, 22). Bound antibodies were visualized on immunoblots by enhanced chemiluminescence (Nycomed Amersham, Buckinghamshire, UK).
Amino Acid Sequencing of the NH2-terminus of Hbp.
1 ml of a 100× concentrated culture supernatant from EB1 (see below) was precipitated with TCA (10% vol/vol). Precipitated proteins were separated on a SDS–polyacrylamide gel (11%), and subsequently electroblotted onto an Immobilon-P transfer membrane filter (Milipore Corp., Bedford, MA). The fixed proteins were stained with Amidoblack. The Hbp band was cut out and subjected to automated Edman degradation using the model 473A protein sequencer of Applied Biosystems.
Cloning and Sequencing of DNA.
Primers were prepared based on the nucleotide sequence of tsh. The primers used were the probe-forward primer 5′-GTGTTAAGGGCGATAACC-3′ and the probe-reverse primer 5′-CTTCTTCAAGGGTAAAGG-3′. Chromosomal DNA of strain EB1 was used as a template for these primers in PCR experiments. A PCR product of 1 kb long was obtained (see Fig. 1) and used as a probe in Southern blot experiments with the chromosomal DNA of strain EB1. A HindIII–EcoRI DNA fragment of 12.6 kb, that positively reacted in this assay, was isolated and ligated into pBR322. Subsequently, the insert of this recombinant plasmid pHE12.6 was sequenced. A random sequencing strategy for sequencing of single stranded DNA was used (23). The DNA of the plasmids pHE12.6 and pET-Hbp (see below) was partially digested with Sau3A1, AluI, or PalI and DNA fragments between 750 and 2,000 bp in length were purified and cloned into M13mp19. Over 126 and 40 clones from pHE12.6 and pET-Hbp, respectively, were sequenced. At least 10 clones for each kilobase of DNA. Synthesized oligonucleotide primers and the cycle sequencing protocol were used on the original double stranded DNA fragments of pHE12.6 and pET-Hbp to complete the sequence. Each strand was sequenced at least twice. Regions of uncertainty were resequenced in both directions. The overall redundancy was between 6–8.
Figure 1.
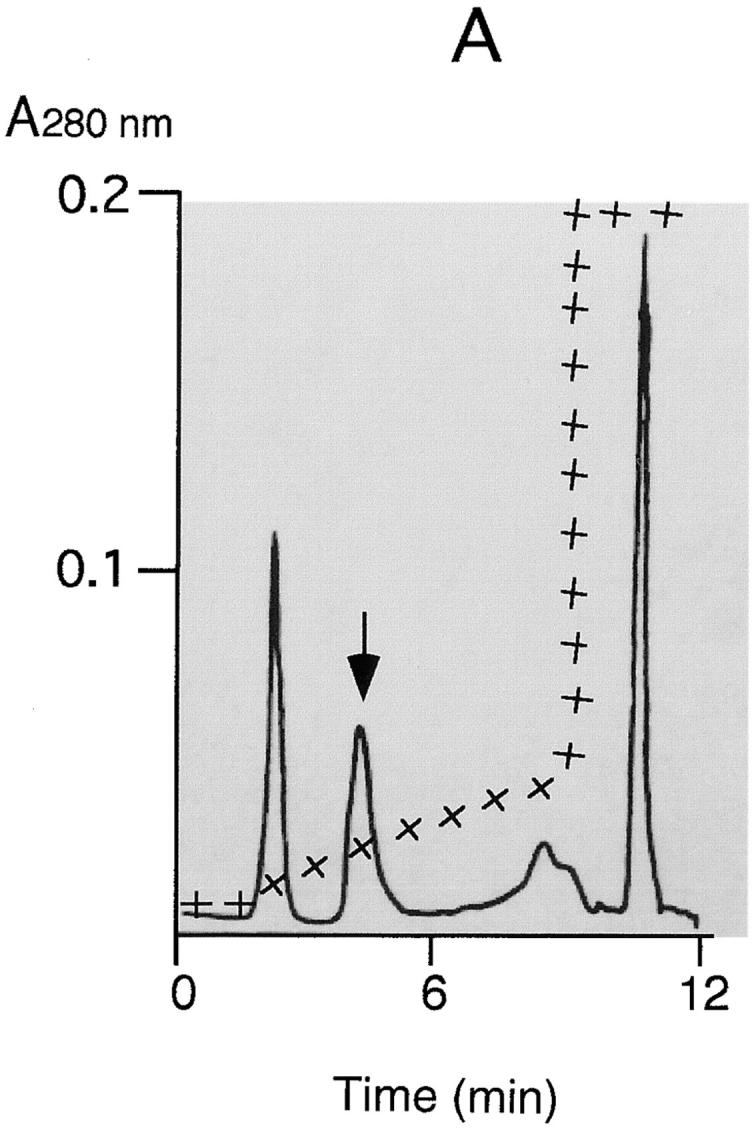
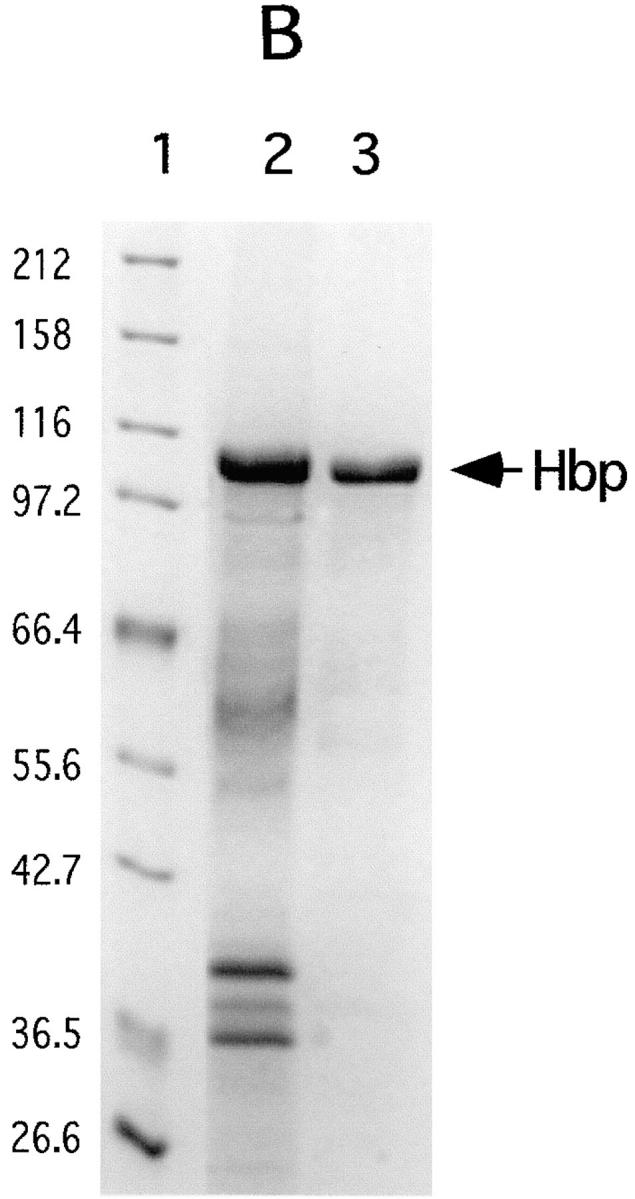
Purification of Hbp. Strain EB1 was cultivated in 5 l MA-medium by gently shaking at 37°C. At an OD660 of 0.6 the culture was cooled to 4°C and the bacteria were separated from the culture supernatant by microfiltration (CFP-2-E-4A membrane cartridge [0.2 μm]; A/G Technology Corporation). The obtained culture supernatant was concentrated a hundred times by using an ultrafiltration column (UFP-10-C-4A membrane cartridge with a NMWC of 10,000; A/G Technology Corporation). A buffer exchange with 50 mM Hepes, pH 7, was done during the concentration step. Six ml of concentrated culture supernatant was then applied to a 0.8-ml POROS 20QE column (PerSeptive Biosystems GmbH, Freiburg, Germany) equilibrated in 50 mM Hepes, pH 7. The column was washed with 8 ml Hepes and developed by a linear salt gradient of 50–200 mM KCl in Hepes. The flow rate during the runs was 4 ml/min. 1-ml fractions were collected and analyzed by SDS-PAGE. (A) POROS 20QE chromatography: elution profile of Hbp. The arrow indicates the Hbp peak. (+) indicates the concentration of KCl during the run (50–1,000 mM). (B) SDS-PAGE profile (11% wt/vol) of samples taken at the two steps of the purification process: the samples were TCA precipitated and analyzed by SDS-PAGE and Coomassie R-250 staining. Lane 1, molecular mass markers (kD); lane 2, the hundred times concentrated culture supernatant (3.5 μg); lane 3, purified 110-kD protein (2.5 μg).
Cloning of the Structural hbp Gene.
The T7 expression system was used for the cloning and IPTG-inducible expression of hbp (14). The coding sequence of hbp was amplified by PCR using the plasmid pHE12.6 as template DNA. Pfu DNA polymerase and Taq Extender PCR additive were used in these experiments as indicated by the manufacturer (Stratagene). Primers were designed to introduce NcoI and BamHI restriction sites at the 5′ and 3′ ends, respectively. The primers used were the 110-forward primer 5′-CAATTCCATGGACAGAATTTATTCTCTTCGCTA-3′, introducing a NcoI site (boldface letters), and the 110-reverse primer 5′-ATATTGGATCCAGAGATGTGTTCAGGAGTTAG-3′, introducing a BamHI site (boldface letters). The complete hbp gene was cloned into the expression vector pET11d (14) using the created NcoI and BamHI sites. However, a point mutation (boldface letters) was created at the 5′ end of the gene, 5′-ATGGACAG-3′ instead of ATGAACAG-3′, by the introduction of hbp into the NcoI–BamHI site of pET11d. This point mutation was reverted by using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). The constructed gene was sequenced to check for second site mutations. Plasmid pET-Hbp was introduced into E. coli BL21(DE3) carrying pLysS. Expression of hbp was induced by the addition of IPTG to a growing culture as described previously (14). Furthermore, the structural hbp gene was also cloned into the vector pBR322. The same procedure was followed as described above, only a different set of primers was used. Instead of the 110-forward primer, primer HindIII-forward in combination with the 110-reverse primer was used. Primer HindIII-forward (5′-GGTTGAAGCTTTATGTGCAGGCATAACGG-3′) introduces a HindIII site (boldface letters) 50 bp upstream of the putative promoter of hbp. The complete gene, including its own promoter, was cloned into pBR322 using the created HindIII and BamHI sites. The plasmid pBRHbp was introduced into E. coli DH5α.
Site-specific Mutagenesis of hbp.
Knockout mutants of hbp were created by the use of the counterselectable vector pSG335 (15). The assembly of the suicide vectors used in this assay required a number of intermediate constructions. The structural hbp gene and the flanking regions of the gene were amplified by PCR using the plasmid pET-Hbp as template DNA. The primers used were the 110-reverse primer (see above), introducing a BamHI site and the pET-primer 5′-CCACGTACGTTCCTCT-3′. The obtained DNA fragment was cloned into the XbaI–HindII site of pBluescript. The recombinant plasmid was designated pBS-Hbp. The hbp gene in pBS-Hbp was disrupted by cloning a kanamycin resistance cassette into the EcoRV site of the gene, obtaining the plasmid pBS-Hbp::kmr. The disrupted hbp gene was isolated from pBS-Hbp::kmr by digesting the plasmid with the enzymes XbaI and MluI. As a consequence of the digestion with the enzyme MluI the disrupted hbp gene is also lacking the last 84 bp at the 3′ end of the gene. The suicide vector pSG-Hbp4.8 was constructed by cloning the 4.8-kb XbaI–MluI DNA fragment into the counterselectable vector pSG335. The main characteristics of pSG335 are the presence of a sacB gene, which converts sucrose to a product that is lethal to E. coli, and the presence of a chloramphenicol resistance gene (15). Another suicide vector, pSG-Hbp2.2, was assembled by cloning a 2.2-kb XbaI–MluI DNA fragment into pSG335. This 2.2-kb DNA fragment, containing the kmr cassette flanked on both sides by ∼500 bp of the hbp gene, was obtained by PCR. The plasmid pBS-Hbp::kmr was used as template DNA. The primers used were the XbaI-forward primer 5′-CGATTT CTAGACAATGATGCCCCGGTC-3′, introducing a XbaI site (boldface letters), and the MluI-reverse primer 5′-CGAAAA CGCGTTGAAGACACTTTTATCTGC-3′, introducing a MluI site (boldface letters). The suicide vectors pSG-Hbp2.2 and pSG-Hbp4.8 were used to transform the wild-type strain E. coli EB1 selecting for resistance to kanamycin. Single colonies were subcultured to allow the necessary recombination and plasmid curing events to occur. Then cells were plated on YT containing kanamycin and 3% sucrose, but lacking NaCl (15, 24). The plates were incubated at 30°C. Colonies grown on this selective medium were screened for their inability to synthesize Hbp by means of immunoblotting.
Subcellular Localization of Hbp.
Subcellular fractions of strain BL21(DE3)(pLysS, pET-Hbp) and the wild-type EB1 strain were prepared essentially as previously described (25, 26). Cells were collected by centrifugation, washed, and stored as frozen cells at −30°C. To obtain a total cell lysate, cells were lysed by freezing and thawing combined with a short ultrasonic treatment (27). Cell debris was removed from the cell lysate by centrifugation at 14,000 g for 10 min at 4°C. Subsequently, soluble proteins (cytoplasmic and periplasmic proteins) and membranes were separated by ultracentrifugation. The cytoplasmic membranes were separated from outer membranes by selective solubilization of cytoplasmic membranes using 0.5% sodium lauryl sarcosinate (Sarkosyl). For detection of Hbp in the different subcellular fractions, immunoblotting was carried out with a specific anti-Hbp antiserum.
Heme-binding Assays.
Heme–protein complexes in a polyacrylamide gel can be detected by chemiluminescence as described (28). The supernatant of a mid-logarithmic phase culture of EB1 was concentrated as described above. Hemin or human hemoglobin (Sigma), to a final concentration of 0.30 mM or 1.5 μM respectively, was added to 50 μl of concentrated culture supernatant. The concentration of Hbp in these mixtures was 2 μg. The samples in 50 mM Hepes, pH 7.0 were incubated at 37°C for 1 h. Subsequently, 25 μl of the samples were run on a 11% nondenaturing polyacrylamide gel. Potential heme-protein complexes in the gel were detected by chemiluminescence.
Heme affinity chromatography was used to investigate the binding specificity of Hbp for heme. Hbp produced by strain DH5α(pHE12.6) was obtained from 1 liter of a mid-logarithmic phase culture. 50 ml of this culture supernatant was mixed with 100 μl of hemin agarose (Sigma Chemical Co., St. Louis, MO), as described (29). The mixtures were incubated at 4°C overnight. The gel beads were collected and washed six times with 0.5 M NaCl and 10 mM sodium phosphate, pH 7.0. The proteins still retained to the gelmatrix were eluted from the beads with SDS sample buffer as described (9). The eluted proteins were analyzed by SDS-PAGE and Coomassie R-250 staining.
Hemoglobin Protease Activity.
Denatured hemoglobin as substrate was used to detect an endopeptidase activity by the purified Hbp protein and culture supernatants of the different E. coli strains (30). The determination of the catalytic type of the peptidase was performed using 3,4-dichloroisocoumarin (3,4-DCI), a specific serine peptidase inhibitor. A new assay was developed to determine the specificity of Hbp for hemoglobin. Purified Hbp (0.5 μg / 50 μl) was mixed with 0.1 μg native hemoglobin in 100 mM phosphate buffer, pH 6.0. The mixture, Hbp and hemoglobin alone were incubated at 37°C for 4 h. Subsequently, 25 μl samples were run on an 8% nondenaturing polyacrylamide gel. Hemoglobin in the gel was detected by incubating the gel first in a chemiluminescence substrate solution and then sprinkling the gel with a H2O2 solution to initiate heme-associated peroxidase activity. The gel was immediately exposed to a X-Omat AR film (Eastman Kodak Company, Rochester, NY) to visualize hemoglobin. Subsequently, hemoglobin in this native gel was also visualized by Coomassie R-250.
Antibodies.
The polyclonal anti-Hbp antiserum was raised in rabbit against a peptide which consists of 15 amino acid residues of Hbp located in the NH2-terminal part of the protein (NELGYQLFRDFAENK). Cross-reactivity was verified by immunoblotting in the presence or absence of the peptide against which the antiserum was raised.
Conjugational Transfer of the EB1 pColV Plasmid.
Strain AN299-23 was first transformed with pACyC184. The resulting strain AN299-23(pACyC184) was used as recipient strain. The EB1 donor cells and recipient cells mixed in a ratio of 8:2 were allowed to conjugate at 37°C in a static liquid culture for 2 h. Thereafter, cells were plated on MA-agar plates containing chloramphenicol resulting in the loss of the EB1 donor cells. The residual colonies were tested on their capacity to produce colicin V. Colicin V activity of possible transconjugants was determined by using a soft agar overlay technique as described previously (31). Colicin-producing colonies gave a clear killing zone surrounding the cell colony. E. coli AN299-23(pACYC184) was used as a colicin V susceptible indicator strain.
Hemagglutination Assay.
Hemagglutination activity of EB1 cells was assayed by their ability to agglutinate rabbit, chicken or human erythrocytes on glass slides as described elsewhere (32). Cells from liquid cultures as well from YT-agar plates both grown at 26 or 37°C were tested in this assay. In addition, purified Hbp (1 μg), purified outer membrane proteins (50 μg) obtained from cells grown at 26 or 37°C, and concentrated culture supernatants were also tested on their ability to agglutinate the different erythrocytes. The outer membrane vesicles were prepared as described elsewhere (33).
Computer Analysis.
For analysis and alignment of DNA sequences the following programs and packages have been used: Gene Works 2.3, DNA Strider 1.0, Sequencher 2.1 and the GCG-package. Putative gene products were searched for similarity to sequences reported previously in nonredundant protein data banks (NCBI BLASTX Search).
Results
Detection of an Extracellular Heme-binding Protein.
E. coli can use heme and hemoglobin as iron sources (10). In Serratia marcescens (9), the extracellular protein HasA is involved in the uptake of heme from hemoglobin, whilst in Hemophilus influenzae the extracellular protein HxuA recognizes heme bound to hemopexin (8). E. coli may also secrete proteins that are able to scavenge heme from the environment. To test this hypothesis, supernatant of a culture in the mid-exponential phase of growth from the human pathogenic E. coli strain EB1 was examined for the presence of extracellular heme-binding proteins. EB1 was cultured in a chemically defined medium to avoid contamination of the extracellular proteins by constituents of the culture medium. The culture supernatant was concentrated by means of ultrafiltration and then analyzed by SDS-PAGE. At least four proteins with estimated molecular masses of 110, 40, 38, and 37 kD were present (Fig. 1 B, lane 2). These proteins were assayed for heme-binding properties. The proteins were incubated with hemin and subsequently subjected to nondenaturing PAGE. Heme–protein complexes in the gel were visualized by chemiluminescence. One prominent heme-associated protein band was detected. That band was cut out from the gel and reanalyzed by means of SDS-PAGE revealing its relative molecular mass of 110 kD (data not shown). This heme-binding protein also proved to possess proteolytic activity against hemoglobin and it was designated Hbp for hemoglobin protease (see below).
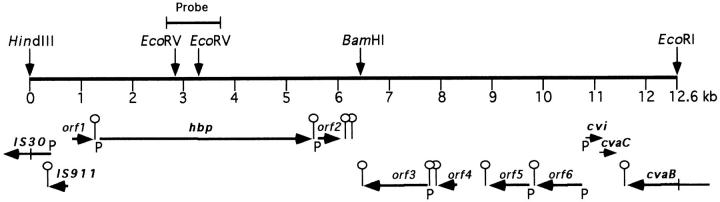
DNA Sequence of the HindIII–EcoRI Fragment of 12.6 kb.
Analysis of Hbp revealed the NH2-terminal amino acid sequence GTVNNELGYQLFRD. This amino acid sequence was found to be 100% identical to that of the deduced amino acid sequence of Tsh (temperature sensitive hemagglutination; reference 34). The Tsh protein has a deduced molecular weight of 118,000 daltons and was found to be significantly homologous to IgA1 proteases, a family of proteins that are secreted by some pathogenic bacteria. The resemblance in characteristics between Tsh detected in the avian pathogenic E. coli strain ×7122 and Hbp isolated from a human pathogen suggests that the proteins are identical. A DNA probe was prepared based on the nucleotide sequence of tsh and a 12.6-kb HindIII–EcoRI DNA fragment was isolated by means of Southern blot experiments. The nucleotide sequence of this fragment was determined and analyzed for the location of open reading frames (ORFs; Fig. 2). In total, 12 ORFs were detected (Table 1).
Figure 2.
Organization of ORFs of the 12,606-bp contig. The scale is given in kb. The positions of the unique HindIII, BamHI, and EcoRI sites and two EcoRV sites are indicated. ORFs are indicated by thick black arrows and the arrowheads show the direction of transcription/ translation. The names of the genes are indicated above the arrows; bold characters indicate genes of which the nucleotide sequence has been published before, plain characters indicate newly discovered ORFs. Possible terminator structures and promoter regions found by computer analysis are indicated by the symbol (o) or a P, respectively. The position where the DNA-probe hybridized to the 12.6-kb fragment in Southern blot experiments is indicated by a straight line. The sequence data of this contig have been deposited in the EMBL Nucleotide Sequence Database under accession number AJ223631.
Table 1.
Putative ORFs in the 12.6-kb DNA Fragment
| ORF | Endpoints | Size of product | Ribsosome binding site consensus sequence and initiation codon (bold) | |||
|---|---|---|---|---|---|---|
| nt | aa | |||||
| IS30 | 1 < 402* | 383 | TGCGGAGGTTTTTTGAATG | |||
| IS911 | 362 < 727 | 122 | CAACACAGGTGCTCCAATG | |||
| orf1 | 823 > 1242 | 164 | GCCTGAAGGAGATGAAATG | |||
| hbp | 1368 > 5498 | 1377 | TCAGGAGTAATTAAAAATG | |||
| orf2 | 5610 > 6056 | 143 | ACATCTCTGGAGATTTATG | |||
| orf3 | 6486 < 7742 | 442 | GGATAAACTCCGCCGGATG | |||
| orf4 | 7943 < 8287 | 115 | TACAGGAACTGAAGCGATG | |||
| orf5 | 8926 < 9723 | 266 | GCAGGAAAAAACACGTATG | |||
| orf6 | 9860 < 10738 | 293 | CAAACAGCTCCCGTTTGTG | |||
| cvi | 10929 > 11162 | 78 | AATGGGATAAAAAGTAATG | |||
| cvaC | 11143 >11451 | 103 | ATAAAAAGGAGATCATATG | |||
| cvaB | 11627 < 12606‡ | 698 |
ORF has been cut at the HindIII site. Total size of the original IS30 ORF is 1,151 nucleotides.
ORF has been cut at the EcoRI site. Total size of the original cvaB ORF is 2,096 nucleotides.
The amino acid sequences of the putative gene products were searched for similarity to sequences reported previously in nonredundant protein data banks. The results are summarized in Table 2. Hbp appears to be nearly identical to Tsh (34). The minor differences found between the nucleotide sequences of these two genes resulted in amino acid substitutions Q209K and A842T. Other known genes contained on the 12.6-kb DNA fragment are IS911, IS30 (35, 36), and genes that are part of the colicin V operon (37). The enzyme IS30 is required for excising and inserting mobile genetic elements (35). IS911 probably plays a role in the translational frame shifting in the control of transposition by IS30 (38). Whether these genes were involved in rearrangements leading to the integration of the hbp gene into its current position remains to be determined. Orf2 showed a high homology with fms, a gene that encodes a deformylase (39). A high homology score was also observed for orf3 and orf4 with two plasmid encoded genes y4hp and y4ho of Rhizobium sp. NGR234 (40). The function of these genes is unknown. Orf5 and Orf6 showed reasonable homology to gene products of Mycobacterium tuberculosis (41) and Bacillus subtilus (these data are available from GenBank/EMBL/DDBJ under accession number Y14081), respectively. These two hypothetical proteins show significant similarity to oxidoreductases. Whether the putative genes located on the 12.6-kb DNA fragment are involved in Hbp-mediated heme uptake or in the secretion of Hbp remains to be determined.
Table 2.
Similarity of Predicted ORF Products to Proteins of Other Organisms
| Product | Similar protein in database | Identity observed | BLAST score | |||
|---|---|---|---|---|---|---|
| IS30 | Transposase of E. coli | 100%, from startcodon to HindIII restriction site | 705 | |||
| IS911 | Hypothetical insertion element IS911 of E. coli | 99%, entire length | 621 | |||
| Orf1 | No significant similarities | — | — | |||
| Hbp | Tsh gene product of E. coli | 99%, entire length | 6768 | |||
| Orf 2 | Fms gene product of E. coli | 98%, entire length | 436 | |||
| Orf 3 | Y4hP gene product of Rhizobium sp. NGR234 | 39–58%, entire length | 299 | |||
| Orf 4 | Y4hO gene product of Rhizobium sp. | 53%, entire length | 325 | |||
| Orf 5 | MTCY31.20 gene product of Mycobacterium | 32-42%, NH2-terminal part | 116 | |||
| tuberculosis, probably a mono-oxygenase | ||||||
| Orf 6 | yhjI gene product of Bacillus subtilis, probably a | 29-45%, entire length | 172 | |||
| dehydrogenase | ||||||
| Cvi | Immunity protein for colicin V of E. coli | 100%, entire length | 404 | |||
| CvaC | Colicin V protein of E. coli | 100%, entire length | 449 | |||
| CvaB | Export protein of colicin V of E. coli | 100%, from EcoRI restriction site to stopcodon | 1441 |
Hbp Is Plasmid Encoded.
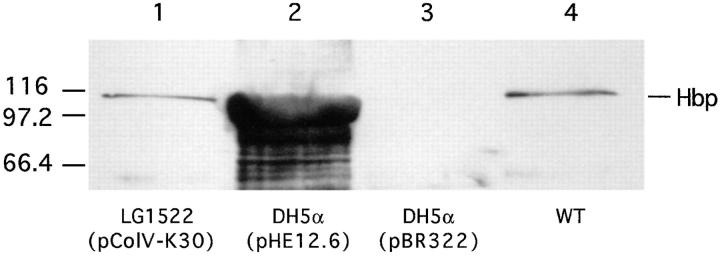
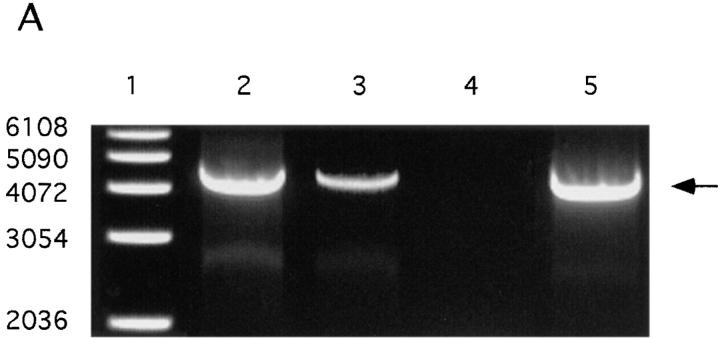
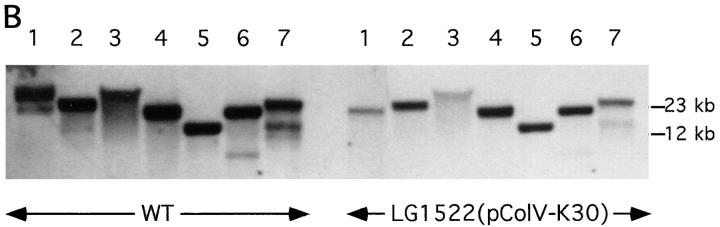
DNA sequence analysis of the HindIII–EcoRI DNA fragment of 12.6 kb showed that besides hbp also a part of the colicin V operon is included in this fragment (37). It is known that the structural colV genes are contained on so-called ColV plasmids (18, 31). Therefore, we examined whether or not Hbp is plasmid encoded. An in vitro assay for colicin V production was used to screen strain EB1. The strain was found to be positive for colicin V production (data not shown). Strain LG1522 (pColV-K30) was used as a positive control strain. This K-12 strain obtained the pColV-K30 plasmid by conjugational transfer from a wild-type strain (18). Immunoblot analysis of the culture supernatant of LG1522 (pColV-K30) with anti-Hbp antiserum showed that this strain expressed Hbp on the same level as EB1 (Fig. 3). These results strongly suggest that hbp in strain LG1522 is contained on the pColV-K30 plasmid. Conjugational transfer of the EB1 pColV plasmid to the recipient strain AN299-23(pACyC184) was performed to find evidence that in EB1 hbp is also plasmid encoded. Based on the colocalization of the hbp gene and the ColV operon, one should expect to find the presence of hbp in ColV+ transconjugants. A ColV+ transconjugant, strain TC003, was isolated. Analysis of the restriction endonuclease profile of the plasmid DNA from TC003 showed that this strain contains the pColV plasmid from EB1 (data not shown). The pColV plasmids of the wild-type strain EB1, LG1522 (pColV-K30) and TC003 were used as templates in PCR experiments. As shown in Fig. 4 A, lane 5, the transconjugant TC003 contains the hbp gene, whereas the recipient strain AN299-23 is lacking this gene (lane 4). These results demonstrated that in strain EB1 hbp is plasmid encoded and in strain LG1522 hbp is located on pColV-K30 (Fig. 4 A, lane 3). The pColV plasmids of strain EB1 and LG1522 were also used as templates in Southern blot hybridization experiments. The Southern blot hybridization profiles of the plasmids from both strains appeared to be identical (Fig. 4 B). Taken together, this strongly suggests that in strain EB1 hbp is also located on a pColV-K30 plasmid, and that the gene is located at ∼190° of the genetic map of pColV-K30 (42).
Figure 3.
Analysis of the secretion of Hbp by the wild-type strain EB1, DH5α carrying pHE12.6 or pBR322 and strain LG1522 carrying pColV-K30. Supernatants of mid logarithmic phase cultures grown in MA-medium were collected. The proteins in the supernatants were TCA precipitated, separated by SDS-PAGE and immunodetected with anti-Hbp antiserum. The amounts loaded are expressed as OD660 equivalents. Lanes 1 and 4, 1.0 OD660 units; lanes 2 and 3, 2.0 OD660 units.
Figure 4.
Screening of plasmid DNA on the presence of hbp by PCR and Southern blot hybridization experiments. The pColV plasmids of the wild-type strain EB1, the K-12 strain LG1522 (pColV-K30) and the transconjugant TC003 were isolated, purified on an agarose gel and used as templates. Strain TC003 obtained the pColV plasmid from EB1 by conjugational transfer. (A) Amplification of hbp by PCR. Primers used in these experiments were the 110-forward and 110-reverse (see Materials and Methods). Lane 1, 1 kb DNA ladder; lane 2, wild-type strain EB1; lane 3, K-12 strain LG1522 (pColV-K30); lane 4, recipient strain AN299-23(pACyC184); and lane 5, transconjugant TC003. (B) Southern blot hybridization with a 1,022-bp long DNA fragment from hbp as probe. Arrow points to 4,130-bp PCR product. Plasmid DNA of both strains was digested with various restriction enzyme combinations and hybridized with the digoxygenin labeled 1,022-bp DNA fragment as probe. Single lanes represent plasmid digests with HindIII, EcoRI, SacI, or SalI only (lanes 1–4, respectively) or EcoRI in addition to HindIII (lane 5), SalI (lane 6), SacI (lane 7).
Purification of Hbp from Culture Supernatants.
Hbp was purified from concentrated culture supernatant of strain EB1 by ion exchange chromatography. The protein eluted from the column between 80 and 120 mM KCl was free of contamination by other proteins as shown by SDS-PAGE (see Fig. 1). The fractions containing the purified protein were pooled, quickly frozen and stored at −80°C. The amount of 110-kD protein purified from one liter of culture supernatant was ∼100 μg. The total quantity of 110-kD protein secreted by EB1 in MA-medium was ∼300 μg/liter. The culture supernatant of strain EB1 contained large amounts of polysaccharide slime. Under anaerobic growth conditions the production of these macromolecules was enhanced, as was the production of Hbp. It was not possible to scale up the purification procedure, because the POROS 20QE column was obstructed by these macromolecules during scaling up runs. To overcome this problem, strain E. coli DH5α(pHE12.6) was used to oversecrete Hbp without the coproduction of a polysaccharide slime (see Fig. 3). The quantity of 110-kD protein detected in the supernatant of a culture from DH5α(pHE12.6) was ∼2–4 mg/liter. The amount of purified protein obtained from one liter culture supernatant of this strain was ∼1 mg.
Heme-binding Properties of Hbp.
The heme-binding properties of Hbp were tested by incubating concentrated supernatant proteins of strain EB1 with hemoglobin or hemin, which is the oxidized form of heme. The mixtures were subjected to nondenaturing PAGE and heme-associated protein bands were detected by chemiluminescence. As shown in Fig. 5 B, Hbp was visualized by this assay indicating that this protein can bind heme (lane 2). Furthermore, the protein was able to acquire heme from hemoglobin (Fig. 5 B, lane 1). This implies that Hbp had to interact with hemoglobin in such a way that heme will be released from hemoglobin. A mechanism for the release of heme could be a proteolytic degradation of hemoglobin by Hbp.
Figure 5.
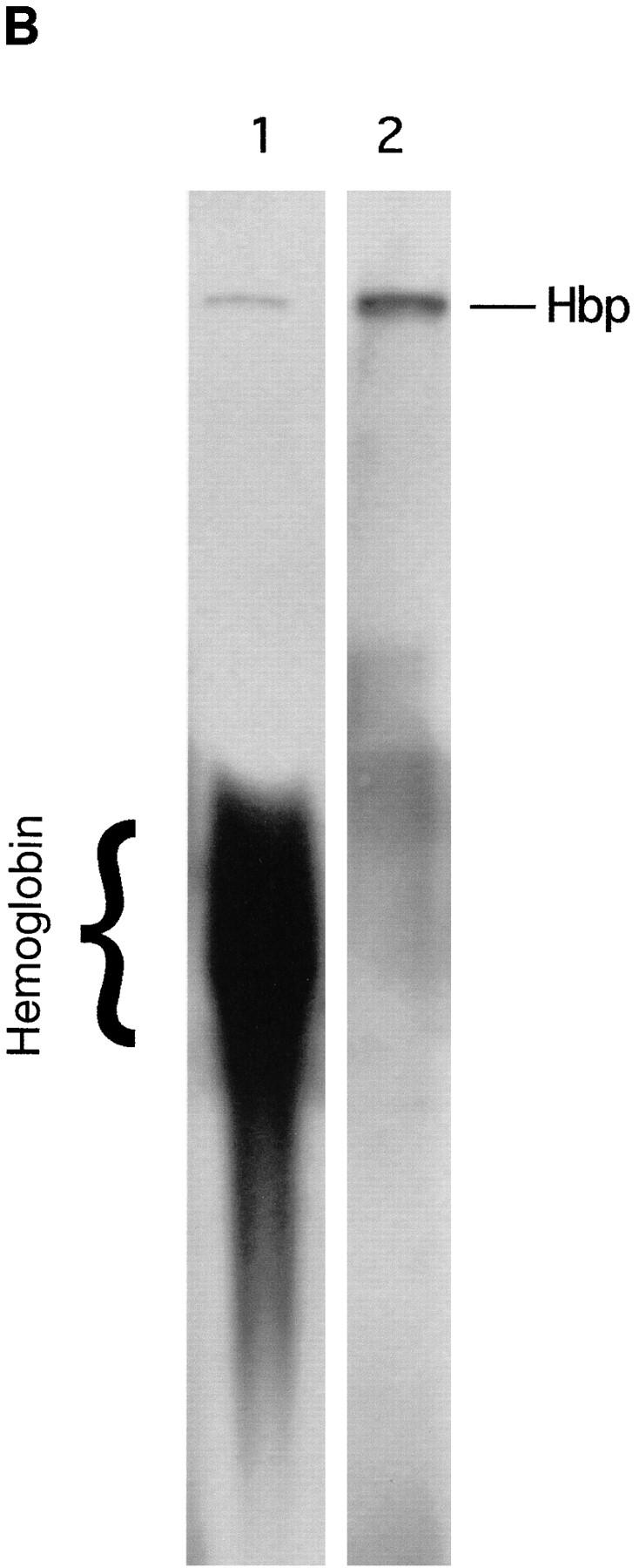
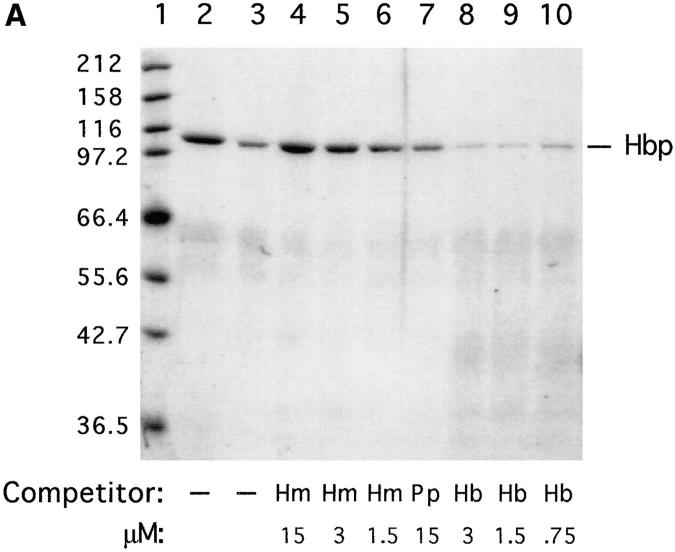
Heme-binding properties of Hbp. (A) Isolation of Hbp by hemin affinity chromatography and binding specificity as measured by competition experiments. 50 ml of culture supernatant of strain DH5α- (pHE12.6) was incubated with hemin-agarose, as described in Materials and Methods. Lane 1, marker proteins (in kD); lane 2, Hbp not retained on hemin-agarose; lane 3, Hbp retained on hemin-agarose alone; lanes 4–6, Hbp retained in the presence of 15, 3 and 1.5 μM hemin (Hm); lane 7, Hbp retained in the presence of 15 μM protoporphyrin IX (Pp); lanes 8–10, Hbp retained in the presence of 3.0, 1.5, and 0.75 μM hemoglobin. (B) Detection of heme-protein complexes in a nondenaturing polyacrylamide gel system. Concentrated supernatant proteins of strain EB1 were incubated with hemin or hemoglobin for 1 h. Samples of these mixtures were run on a 11% polyacrylamide gel. Heme-associated protein bands in the gel were visualized by means of chemiluminescence as described in Materials and Methods. Lane 1, supernatant proteins incubated with hemoglobin; lane 2, supernatant proteins incubated with hemin. The positions of heme-saturated Hbp and hemoglobin in the gel are indicated.
The heme-binding properties of Hbp were further investigated by heme affinity chromatography. The retention of Hbp on hemin-agarose was examined by using a nonconcentrated culture supernatant of strain DH5α(pHE12.6), containing 40 nM Hbp. As shown in Fig. 5 A, lane 3, retention of Hbp by this resin was observed. Pre- or coincubation of the culture supernatant with 3–0.75 μM of human hemoglobin could adequately inhibit the interaction of Hbp with hemin-agarose (8 μM of ligand; Fig. 5 A, lane 8–10). No inhibition of binding was observed after the incubation of Hbp with protoporphyrin IX, the precursor molecule of heme (Fig. 5 A, lane 7). The binding of Hbp to hemin-agarose was not due to nonspecific ionic interactions, because a high ionic strength washing buffer was used in these experiments. Unexpectedly, a more profound interaction of Hbp with hemin-agarose was detected by the pre- or coincubation of the culture supernatant with low concentrations of hemin (Fig. 5 A, lanes 4–6). At higher concentrations of hemin (≥ 50 μM) the interaction of Hbp with hemin-agarose was prevented (data not shown). The protein bound to hemin-agarose could be completely eluted under nondenaturing conditions by using 3 μM human hemoglobin, 50 μM hemin or an 1 M NaCl and 20% ethanol solution (data not shown). Taken together, the results demonstrate that Hbp is a heme-binding protein. In addition to heme, hemoglobin was also capable to compete very efficiently with hemin-agarose. Furthermore, these data indicate that Hbp has a higher affinity for hemoglobin than for hemin.
Hemoglobin Protease Activity.
Hbp belongs to a group of related proteins named the Tsh family (43). All members of this family contain a consensus motif for the catalytic site of serine proteases, suggesting that all these proteins are proteolytic enzymes. Therefore, the recruitment of heme from hemoglobin could be the result of a proteolytic breakdown of hemoglobin by Hbp. A general protease assay with denatured hemoglobin as substrate was used to detect an endopeptidase activity by Hbp. As shown in Table 3, a proteolytic activity of Hbp against denatured hemoglobin was detected. To prove that this activity was caused by a serine protease, the specific inhibitor 3,4-DCI, was used. Indeed, preincubation of Hbp with this irreversible inhibitor completely abolished the observed protease activities (Table 3). A knockout mutant of Hbp (mutant mHbp18) was obtained by site-directed mutagenesis experiments with the suicide vector pSG-Hbp2.2 (see below). The culture supernatant of mutant mHbp18 showed almost no proteolytic activity (Table 3). The observed background activity by this strain is probably due to leakage of intracellular enzymes into the growth medium. The insertion of the kanamycin cassette into hbp in mHbp18 could also result in inactivation of the orf2 downstream of hbp by polarity, since both genes are transcribed in the same orientation (as shown in Fig. 2). Although a putative terminator structure was found between hbp and orf2, the possibility exists that orf2 contributes to the proteolytic activity of Hbp. To exclude this possibility strain DH5α(pBRHbp) was used in the protease assay. This strain contains only the structural hbp gene under control of its own promoter. Hbp is secreted by this strain (data not shown). A proteolytic activity by DH5α(pBRHbp) comparable to that of EB1 was detected (Table 3). This indicates that orf 2 is not responsible for the observed proteolytic activity. Taken together, these results indicate that Hbp is a serine protease.
Table 3.
Detection of a Proteolytic Activity by Hbp
| Samples | Activity | Activity with 3,4 DCI | Specific activity | Activity | ||||
|---|---|---|---|---|---|---|---|---|
| units | units | units/mg | % | |||||
| Supernatant of strain EB1 | 0.40 | 0.18 | 220 | 100 | ||||
| Purified Hbp from nonconcentrated supernatant | 0.42 | 0.00 | 168 | 76 | ||||
| Purified Hbp from concentrated supernatant | 0.69 | 0.00 | 69 | 31 | ||||
| Supernatant of mHbp18 | 0.01 | ND | — | — | ||||
| Supernatant of DH5α(pBRHbp) | 0.30 | ND | 200 | 91 |
Culture supernatants of the different strains were collected at an OD660 of 0.6. The obtained supernatant of the wild-type strain EB1 was concentrated a hundred times and then applied to a POROS 20QE column, or fresh supernatant was directly run on this column (Fig. 1). The knockout mutant mHbp18 lacks the expression of Hbp (as shown in Fig. 8 A). Strain DH5α(pBRHbp) contains only the structural hbp gene under control of its own promoter. The samples of the different supernatants and the POROS 20QE eluates were incubated with denatured hemoglobin (pH 6.5) at 37°C for 10 min. The serine protease inhibitor 3,4-dichloroisocoumarin (3,4-DCI, 3 mM) was preincubated with Hbp at 30°C for 30 min.
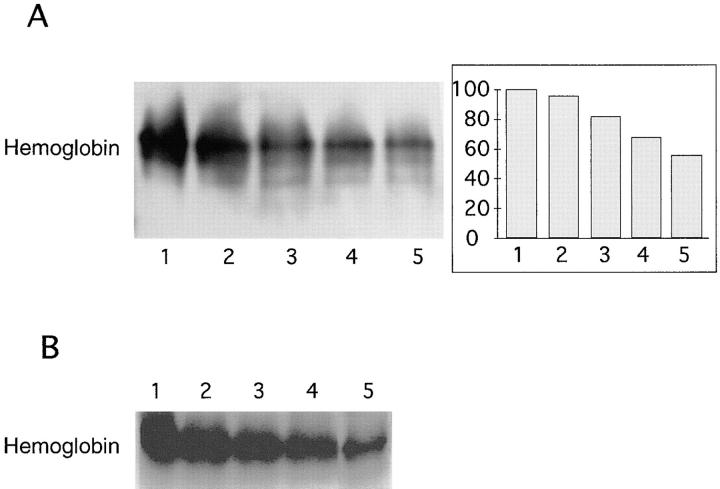
An assay was developed to determine the specificity of Hbp for hemoglobin. Native hemoglobin was incubated with purified Hbp in a molar ratio of 1:3 at 37°C for 4 h. Subsequently, the protein samples were subjected to nondenaturing gel electrophoresis and hemoglobin in the gel was visualized by chemiluminescence. The intrinsic heme-associated peroxidase activity of hemoglobin was initiated by H2O2. Hydrolysis of hemoglobin by Hbp will give a release of heme and consequently a lower signal of hemoglobin in this assay should be expected. The gel was also Coomassie R-250 stained to verify that the release of heme was indeed due to a proteolytic activity of Hbp. As shown in Fig. 6, both the chemiluminescence assay (A) and the Coomassie blue staining of the gel (B) revealed a decrease in hemoglobin after the incubation with Hbp. The enzyme degraded hemoglobin in a pH range between pH 8.0 and 5.0, but is optimally active on hemoglobin at pH 6.0 (lane 5). Albumin, human lactoferrin and human immunoglobulin A were not degraded by Hbp (data not shown). These results indicated that Hbp released heme from hemoglobin by specifically degrading this protein.
Figure 6.
Hemoglobin protease activity by Hbp at different pH. Hemoglobin was incubated with purified Hbp, as described in Materials and Methods. Samples were analyzed by nondenaturing PAGE and PhotoImaging. (A) Hemoglobin visualized by means of chemiluminescence. Lane 1, hemoglobin not incubated with Hbp, pH 6.0; lanes 2–5, hemoglobin incubated with Hbp at pH 8.0, 7.0, 6.5, and 6.0, respectively. A quantitation of the PhotoImage data is presented at the right hand of the image. The initial amount of hemoglobin present in the reaction mixture was set at 100%. (B) Hemoglobin visualized by means of Coomassie R-250 staining. Lanes 1–5, same samples as in A.
Subcellular Localization of Hbp.
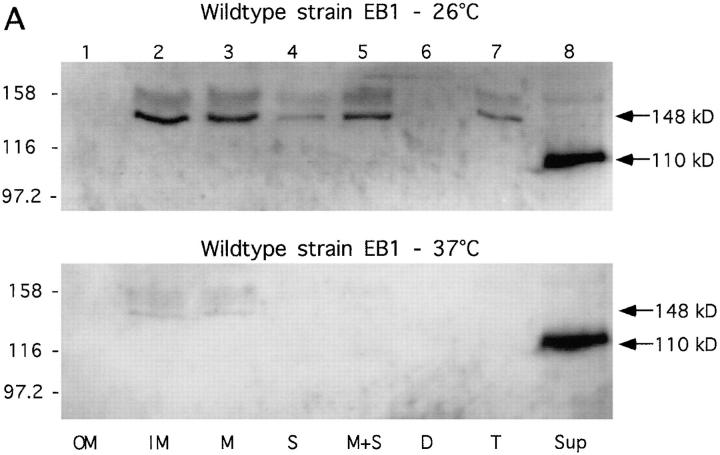
Based on amino acid sequence homology between Tsh and immunoglobulin A1 proteases it was suggested that tsh encodes an IgA protease-type exoprotein (34). Since Tsh and Hbp are quite similar, this assumption also counts for Hbp. A common feature of these so-called autotransporter proteins is that all necessary translocation functions are carried by the polyprotein precursor itself and that these proteins are processed during their translocation over the cell envelope (44). Based on sequence analysis of Hbp it should be expected that Hbp, with a size of 110 kD, will be present in the cytoplasm and cytoplasmic membrane as a 148-kD precursor, and in the periplasm and outer membrane as a 142-kD intermediate form. Subcellular localization experiments with the wild-type strain EB1 and strain BL21(pET-Hbp) were performed to find these putative intermediary products of Hbp in the cell. Strain BL21(pET-Hbp), that contains only the structural hbp gene, was used to check if the gene product indeed carries all the necessary translocation functions. In addition, the effect of different growth temperatures on the secretion of the protein by these two strains was examined. As shown in Fig. 7 A, the wild-type strain EB1 was very efficient in translocating Hbp across its cell envelope. At a growth temperature of 37°C only the mature protein was found in the culture supernatant fraction (Fig. 7 A, lane 8, bottom). The 148-kD precursor of Hbp was detected in the cytoplasmic membrane fraction at 26°C (Fig. 7 A, lane 2), probably due to a hampered transport of the protein across this membrane. The 142-kD intermediate of Hbp was not found in the cellular fractions by this assay. The kinetics of processing and transport of the protein through the cell envelope were probably too fast. In Neisseria gonorrhoeae, comparable results in these kinetics have been found (44, 45). The kinetics of these processes were not influenced by the lower growth temperature. Moreover, no difference was found in the total amount of protein secreted at the different growth temperatures (Fig. 7 A, lane 8).
Figure 7.
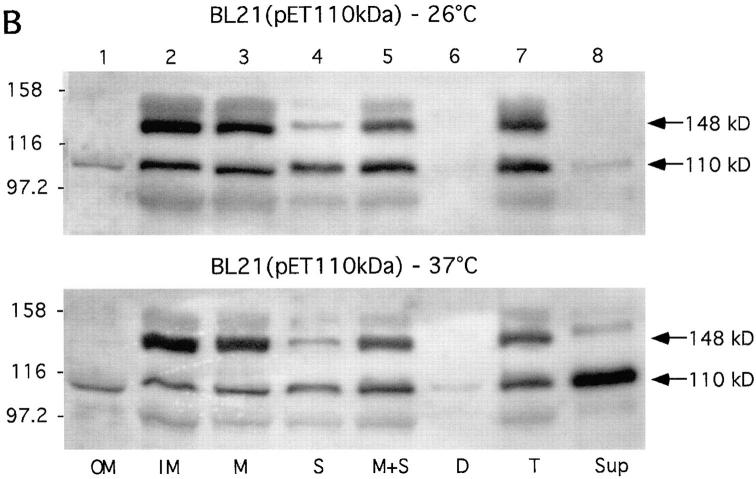
Subcellular localization of Hbp. Cells of the wild-type strain EB1 (A) and IPTG-induced cells of strain BL21(pET-Hbp) (B) were grown in LB medium at 26°C and 37°C. Outer membrane (OM), inner membrane (IM) and crude membrane (M) fractions were derived from 1.3 OD660 units. Soluble (S), membrane and soluble (M+S), cellular debris (D), and total lysate (T) fractions were derived from 0.33 OD660 units. Culture supernatant (Sup) fractions were derived from 3.0 OD660 units. The fractions were analyzed by immunoblotting with the anti-Hbp antiserum. The secretory intermediates of Hbp and molecular mass markers (kD) are indicated at the right and left side of the panels, respectively.
The secretion of Hbp by strain BL21(pET-Hbp) at 37°C was similar to that of the wild-type strain, but in this strain a significant accumulation of the 148-kD precursor was detected in the cytoplasmic membrane at 37°C (Fig. 7 B, lane 2). Furthermore, traces of the mature protein were detected in the soluble and cytoplasmic membrane fractions suggesting that part of the precursor of Hbp has been converted into the mature form. At 26°C these translocation defects were even more pronounced (Fig. 7 B, top): almost no secretion of Hbp occurred at this growth temperature (lane 8).
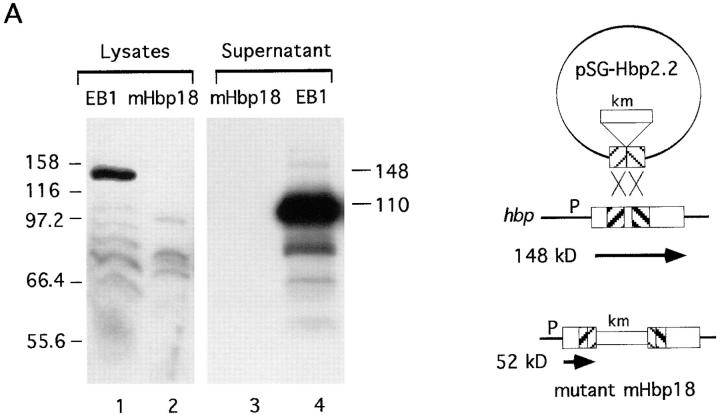
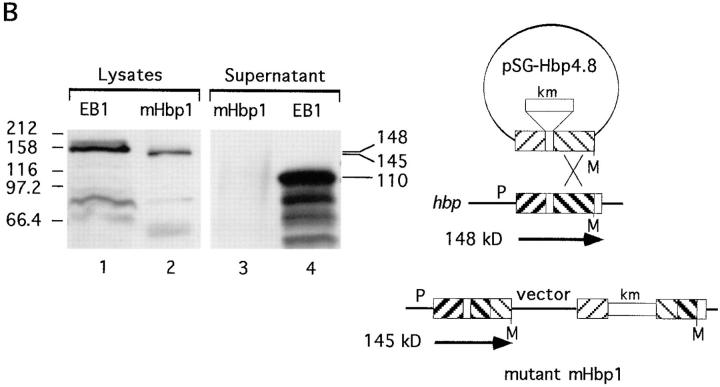
Two independent hbp-knockout mutants were made by site-directed mutagenesis (Fig. 8). Mutant mHbp18 was the result of a double crossover event (Fig. 8 A). This mutant lacks the expression of Hbp and loosed its proteolytic activity against hemoglobin (as shown in Table 3). The expected truncated Hbp of 52 kD was only detectable in total cell lysates of mHbp18 at OD660 units of >2.0 in these immunoblot experiments (data not shown). The other mutant (mHbp1) was mutated in the COOH-terminal part of Hbp, due to a single crossover event (Fig. 8 B). The COOH-terminal domain of IgA protease precursors provides the essential transport function for the proteases across the outer membrane. Mutations in this so-called β-domain blocked the translocation of the passenger proteins across the membrane (44). Mutant mHbp1, lacks the last 84 bp at the 3′-end of the gene and expresses a truncated precursor protein of 144.783 kD (Fig. 8 B, lane 2). Due to this mutation secretion of Hbp in the environment was inhibited (Fig. 8 B, lane 3). These results strongly suggested that Hbp is an autotransporter, but the efficiency of the translocation of this protein over the cell envelope depends on the host that is used.
Figure 8.
Analysis of the expression of Hbp by Hbp-mutants. The mutants mHbp18 and mHbp1 were obtained by site-directed mutagenesis experiments with the suicide vectors pSG-Hbp2.2 and pSG-Hbp4.8, respectively. See Materials and Methods for a detailed description of the construction of these suicide vectors. Culture supernatant fractions and total lysate fractions were derived from 4.0 and 0.5 OD660 units, respectively. The fractions were analyzed by immunoblotting with the anti-Hbp antiserum. (A) Expression of Hbp by mHbp18. Lanes 1 and 4, lysate and supernatant of the wild-type strain EB1; lanes 2 and 3, lysate and supernatant of mHbp18. (B) Expression of Hbp by mHbp1. Lanes 1 and 4, same samples as in A; lanes 2 and 3, lysate and supernatant of mHbp1. The secretory intermediates of Hbp and molecular mass markers (kD) are indicated at the right and left side of the panels, respectively. A schematic representation of the recombination events of the suicide vectors is depicted at the right side of this figure. Mutant mHbp18 was the result of a double crossover and mutant mHbp1 of a single crossover event. Symbols in this diagram are: M, MluI restriction site; P, promoter; km, kanamycin cassette.
Discussion
E. coli possesses four iron uptake systems that use siderophores such as enterobactin and aerobactin, produced by E. coli, or the fungal siderophores ferrichrome and coprogen. Iron acquisition by this bacterium can also occur in a process mediated by citrate (1, 5). Pathogenic E. coli strains are able to use heme compounds as iron sources, but so far little is known about the mechanisms involved in this kind of iron uptake (10). The results of this study suggest that the human pathogenic E. coli strain EB1 contains a hemophore-dependent heme acquisition system. The bacterium secretes a heme-binding protein (Hbp) with an estimated size of 110 kD, that degrades hemoglobin. It is likely that Hbp is the shuttle protein of this heme-scavenging system in E. coli.
Recently, an exported protease (PssA) from a Shiga toxin-producing E. coli has been characterized (43). Sequence comparison showed that PssA is related to the family of autotransporter proteins, especially to SepA of S. flexneri (46), Tsh of E. coli (34) and EspC from EPEC (47). These proteins belong to a family of proteins that is only distantly related to the IgA1 proteases of Neisseria and Haemophilus (43). It was proposed to designate this group of related proteins the Tsh family. The protein PssA showed a moderate serine protease activity in a casein-based assay. The conservation of the putative catalytic center of serine proteases in all members of the Tsh family suggests similar proteolytic activities for these proteins. However, the natural substrate for these proteins remained unclear because casein is unlikely to be the natural substrate. We found that Hbp has a high affinity to hemoglobin and acquires its heme from hemoglobin by degradation of this protein. It is tempting to speculate that hemoglobin is also the natural substrate for the other members of the Tsh family. An activity of ∼300 pmol/h/mg of Hbp was found and the enzyme showed a pH optimum of 6.0–6.5, which is consistent with the physiological pH during an infection, since at the site of infection the pH will fall to 6.0 in response to inflammation.
This study shows that elution of Hbp from hemin-agarose is possible with hemin or hemoglobin. Otherwise, very rigid conditions, like boiling the beads in denaturing buffers or high salt buffers, are needed to elute Hbp from hemin-agarose, indicating high affinity binding of Hbp to hemin and hemoglobin. Hbp shares these characteristics with HasA of S. marcescens, a heme-binding shuttle protein responsible for the uptake of heme from hemoglobin by an unknown mechanism (9). We demonstrated that Hbp can obtain heme from hemoglobin by a proteolytic interaction. A remarkable observation in the heme affinity chromatography experiments was that low concentrations of free heme molecules favor the binding of Hbp to hemin-agarose. Presumably, heme induced a conformational change of Hbp in such a way that the protein gets a higher affinity to hemin-agarose. This suggests the presence of two heme-binding sites in Hbp, one with a moderate and the other with a high affinity for heme.
Hbp has to be shuttled back to the cell surface if it is involved in a heme-acquisition system. Hence, E. coli must possess a specific outer membrane receptor that will recognize the heme-saturated Hbp protein. Results of preliminary studies suggested that E. coli indeed has a receptor that is able to discriminate between the apo- and holo-form of Hbp (unpublished results). Recently, a putative 69-kD heme receptor (ChuA) in E. coli has been described (48). Utilization of heme and hemoglobin by E. coli was ChuA dependent. It would be of interest to find out whether this receptor is also involved in the Hbp-mediated heme uptake. Work is in progress to further characterize the receptor for Hbp in E. coli EB1.
This study also demonstrates that hbp is located on pColV-K30. The pColV-K30 plasmid (144 Kbp) belongs to a heterogeneous group of virulence plasmids (42). The plasmids encode the production of a colicin known as colicin V and are commonly found in invasive strains of E. coli from human and domestic animals (18). ColV plasmids encode several virulence-related properties in addition to colicin V (42). One of the properties that clearly contributes to the virulence of pColV-K30 is the aerobactin iron assimilation system. This shuttle-mediated iron uptake system has been extensively reviewed (5, 11, 42). It seems that pColV-K30 encodes two shuttle-mediated iron uptake systems; one for the acquisition of iron and another for heme. It would be of great interest to find out if these two iron uptake systems are in some way related. In pColV-K30 hbp is located on a HindII–BamHI DNA fragment of 6,453 bp. We examined HindIII–BamHI restriction enzyme profiles of different pColV plasmids (42) for the presence of similar sized HindII–BamHI DNA fragments. The profiles reveal DNA fragments of ∼6,500 bp in pColV-292, pColV-F70 and pColV-F54, suggesting that hbp is present in these plasmids. We propose that Hbp contributes to the virulence properties of pColV plasmids.
We show that Hbp is synthesized as a polyprotein precursor which is processed following export to the cell surface. The kinetics of protein translocation across the outer membrane and subsequent cleavage are very rapid, whilst the transfer through the cytoplasmic membrane appears to be a slower process. Hbp is also efficiently secreted by a K12-strain in which the structural hbp gene was introduced. Furthermore, secretion of Hbp is inhibited by a mutation in the β-domain of its precursor. Comparable results were found for the IgA protease of N. gonorrhoeae (44, 45). These results strongly suggest that Hbp is an autotransporter protein. Autotransporters are large polyproteins that are organized in functional domains and are eventually processed at the cell surface. The necessary translocation functions are carried by the polyprotein precursor itself. The β-domain of the precursor is responsible for the transport of the protein across the outer membrane (44). However, in strain BL21(pET-Hbp) we found that the secretion of the protein was less efficient. Accumulation of the 148-kD precursor in the cytoplasmic membrane and traces of the mature protein were detected in the periplasmic and cytoplasmic membrane fractions of this strain. Accumulation of the 148-kD precursor in the membrane might cause changes in the conformation of the precursor in such a way that the proteolytic site of the β-domain becomes accessible to periplasmic proteases. It seems that the efficiency of transport across this membrane and or cleavage of the signal peptide is dependent on the host type used. Thus, it cannot be excluded that other, yet unknown factors are involved in this pathway of secretion.
It has been shown that tsh is responsible for a mannose-resistant hemagglutination activity in E. coli (34). This activity was best expressed at 26°C. However, it remains unclear which role an extracellular Tsh protein might play in hemagglutination, since microbial adhesins are normally membrane-associated. E. coli strain EB1 was also tested for its ability to agglutinate erythrocytes from human and different animals. The EB1 cells were cultured under different conditions and subsequently tested as described in Materials and Methods. Purified Hbp, outer membrane proteins and concentrated culture supernatants were also tested for hemagglutination activity. None of these experiments yielded a positive reaction (data not shown). The reason for the apparent difference between Hbp and Tsh in hemagglutination activity remains unclear. It seems that only under very specific conditions hemagglutination activity by Tsh can be detected.
Recently, the complete nucleotide sequence was determined of the plasmid from Rhizobium sp. NGR234 that endows the bacterium with the ability to associate symbiotically with leguminous plants (40). The results of this study showed that pNGR234a has functioned as a transposon trap. Potential genes detected on this plasmid appear to be all involved in the symbiosis between Rhizobium and legumes. No genes essential for transcription, translation or primary metabolism were found (40). Presumably, colicin V virulence plasmids also function as ‘transposon traps' and consequently these plasmids may only contain virulence genes. Therefore, it will be interesting to clarify the functions of the orfs located on the 12.6-kb DNA fragment.
In conclusion, the results of this study demonstrate that Hbp is a heme-binding protein that obtains its heme from hemoglobin by degradation of this protein. The protein is likely to be the shuttle protein of a hemophore-dependent heme acquisition system in this human pathogenic E. coli strain. Hbp is a member of the Tsh family and its structural gene is located on a ColV virulence plasmid. All these characteristics strongly suggest that Hbp is an important virulence factor that may play a significant role in the pathogenesis of E. coli infections. However, the role of this protease as a virulence factor remains to be tested in vivo. Further studies are needed to prove if the other IgA1 protease-like autotransporters are also heme-binding proteins with a proteolytic activity specific for hemoglobin. We are currently investigating the nature of binding of Hbp at the cell surface of E. coli and the proteolytic properties of the protein in more detail.
Acknowledgments
This work was supported by the Netherlands Organization for Technology (STW).
Abbreviations used in this paper
- Hbp
hemoglobin protease
- ORF
open reading frame
References
- 1.Mietzner TA, Morse SA. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 2.Otto BR, Verweij-van Vught AMJJ, MacLaren DM. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 3.Bornside GH, Bouis PJ, Cohn I. Hemoglobin and Escherichia coli, a lethal intraperitoneal combination. J Bacteriol. 1968;95:1567–1571. doi: 10.1128/jb.95.5.1567-1571.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullen JJ, Rogers HJ. Bacterial iron metabolism and immunity to Pasteurella septica and Escherichia coli. . Nature. 1969;224:380–382. doi: 10.1038/224380a0. [DOI] [PubMed] [Google Scholar]
- 5.Crosa JG. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee BC. Quelling the red menace: haem capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- 7.Ghigo JM, Letoffe S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. . J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cope LD, Thomas SE, Latimer JT, Slaughter CA, Mullereberhard U, Hansen EJ. The 100 kDa haem: haemopexin-binding protein of Haemophilus influenzae: Structure and localization. Mol Microbiol. 1994;13:863–873. doi: 10.1111/j.1365-2958.1994.tb00478.x. [DOI] [PubMed] [Google Scholar]
- 9.Letoffe S, Ghigo JM, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescensextracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law D, Wilkie KM, Freeman R, Gould FK. The iron uptake mechanisms of enteropathogenic Escherichia coli: the use of haem and haemoglobin during growth in an iron-limited environment. J Med Microbiol. 1992;37:15–21. doi: 10.1099/00222615-37-1-15. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths, E. 1987. The iron uptake systems of pathogenic bacteria. In Iron and Infection: Molecular, Physiological and Clinical Aspects. J.J. Bullen and E. Griffiths, editor. John Wiley & Sons, Chichester, UK. 69–137.
- 12.Verweij-van Vught AMJJ, Namavar F, Sparrius M, Vel WAC, MacLaren DM. Pathogenic synergy between Escherichia coli and Bacteroides fragilis: studies in an experimental mouse model. J Med Microbiol. 1985;19:325–331. doi: 10.1099/00222615-19-3-325. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 14.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 15.Geissler S, Drummond M. A counterselectable pACYC184-based lacZα-complementing plasmid vector with novel multiple cloning sites; construction of chromosomal deletions in Klebsiella pneumoniae. Gene (Amst) 1993;136:253–255. doi: 10.1016/0378-1119(93)90474-h. [DOI] [PubMed] [Google Scholar]
- 16.Wagegg W, Braun V. Ferric citrate transport in Escherichia colirequires outer membrane receptor protein FecA. J Bacteriol. 1981;145:156–163. doi: 10.1128/jb.145.1.156-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang ACY, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams PH. Novel iron uptake system specified by ColV plasmids: an important component in the virulence of invasive strains of Escherichia coli. . Infect Immunol. 1979;26:925–932. doi: 10.1128/iai.26.3.925-932.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J.H. 1992. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 456 pp.
- 20.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oudega B, Koningstein G, Rodrigues L, Ramon M, Hilbert H, Dusterhoft A, Pohl TM, Weitzenegger T. Analysis of the Bacillus subtilisgenome: cloning and nucleotide sequence of a 62 kb region between 275 degrees (rrnB) and 284 degrees (pai) Microbiol. 1997;143:2769–2774. doi: 10.1099/00221287-143-8-2769. [DOI] [PubMed] [Google Scholar]
- 24.Blomfield IC, Vaughn V, Rest RF, Eisenstein BI. Allelic exchange in Escherichia coli using the Bacillus subtilis sacBgene and a temperature-sensitive pSC101 replicon. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 25.Filip C, Fletcher G, Wulff JL, Earhart CF. Solubilization of the cytoplasmatic membrane of Escherichia coliby the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindberg F, Tennent JM, Hultgren SJ, Lund B, Normark S. PapD, a periplasmic transport protein in P-pilus biogenesis. J Bacteriol. 1989;171:6052–6058. doi: 10.1128/jb.171.11.6052-6058.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luirink, J., C.M. tenHagen-Jongman, C.C. Van der Weijden, B. Oudega, S. High, B. Dobberstein, and R. Kusters. 1994. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO (Eur. Mol. Biol. Organ.) J. 13:2289–2296. [DOI] [PMC free article] [PubMed]
- 28.Otto BR, Kusters JG, Luirink J, deGraaf FK, Oudega B. Molecular characterization of a heme-binding protein of Bacteroides fragilisBE1. Infect Immun. 1996;64:4345–4350. doi: 10.1128/iai.64.10.4345-4350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsutsui K, Mueller GC. Affinity chromatography of heme-binding proteins: an improved method for the synthesis of hemin-agarose. Anal Biochem. 1982;121:244–250. doi: 10.1016/0003-2697(82)90475-4. [DOI] [PubMed] [Google Scholar]
- 30.Sarath, G., R.S. delaMotte, and F.W. Wagner. 1989. Protease assay methods. In Proteolytic Enzymes: A Practical Approach. R.J. Beynon and J.S. Bond, editors. IRL Press, Oxford. 25–55.
- 31.Frick KK, Quackenbush RL, Konisky J. Cloning of immunity and structural genes for Colicin V. J Bacteriol. 1981;148:498–507. doi: 10.1128/jb.148.2.498-507.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klemm P, Jorgensen BJ, Kreft B, Christiansen G. The export systems of type 1 and F1C fimbriae are interchangeable but work in parental pairs. J Bacteriol. 1995;177:621–627. doi: 10.1128/jb.177.3.621-627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Vrije T, Tommassen J, De Kruijff B. Optimal posttranslational translocation of the precursor of PhoE protein across Escherichia colimembrane vesicles requires both ATP and the protonmotive force. Biochem Biophys Acta. 1987;900:63–72. doi: 10.1016/0005-2736(87)90278-1. [DOI] [PubMed] [Google Scholar]
- 34.Provence DL, Curtiss R., III Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia colistrain. Infect Immunol. 1994;62:1369–1380. doi: 10.1128/iai.62.4.1369-1380.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalrymple B, Caspers P, Arber W. Nucleotide sequence of the prokaryotic mobile genetic element IS30. EMBO (Eur Mol Biol Organ) J. 1984;3:2145–2149. doi: 10.1002/j.1460-2075.1984.tb02104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burland V, Plunkett G, Sofia HJ, Daniels DL, Blattner FR. Analysis of the Escherichia coligenome VI: DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilson L, Mahanty HK, Kolter R. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO (Eur Mol Biol Organ) J. 1990;9:3875–3884. doi: 10.1002/j.1460-2075.1990.tb07606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 39.Guillon JM, Mechulam Y, Schmitter JM, Blanquet S, Fayat G. Disruption of the gene for Met-tRNAf met formyltransferase severely impairs growth of Escherichia coli. . J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freiberg C, Fellay R, Bairoch A, Broughton WJ, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobiumand legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 41.Philipp WJ, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom BR, Jacobs WR, Cole ST. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. . Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waters VL, Crosa JH. Colicin V virulence plasmids. Microbiol Rev. 1991;55:437–450. doi: 10.1128/mr.55.3.437-450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Djafari S, Ebel F, Deibel C, Kramer S, Hudel M, Chakraborty T. Characterization of an exported protease from Shiga toxin-producing Escherichia coli. . Mol Microbiol. 1997;25:771–784. doi: 10.1046/j.1365-2958.1997.5141874.x. [DOI] [PubMed] [Google Scholar]
- 44.Klauser T, Pohlner J, Meyer TF. The secretion pathway of IgA protease-type proteins in gram-negative bacteria. Bioessays. 1993;15:799–805. doi: 10.1002/bies.950151205. [DOI] [PubMed] [Google Scholar]
- 45.Pohlner J, Halter R, Beyreuther K, Myer TF. Gene structure and extracellular secretion of Neisseria gonorrhoeaeIgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 46.Benjellountouimi Z, Sansonetti PJ, Parsot C. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol Microbiol. 1995;17:123–135. doi: 10.1111/j.1365-2958.1995.mmi_17010123.x. [DOI] [PubMed] [Google Scholar]
- 47.Stein M, Kenny B, Stein MA, Finlay BB. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coliwhich is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J Bacteriol. 1996;178:6546–6554. doi: 10.1128/jb.178.22.6546-6554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres AG, Payne SM. Haem iron-transport system in enterohaemorrhagic Escherichia coliO157:H7. Mol Microbiol. 1997;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]