Abstract
Cyclic-AMP stimulation of GTP cyclohydrolase I (GCH1) gene transcription was investigated in PC12 cells, the protein kinase A-deficient PC12 cell line 126-1B2 and C6 cells using transient transfection assays of proximal promoter reporter constructs and wild type or dominant negative proteins, chromatin immunoprecipitation and real-time quantitative PCR. These studies show that protein kinase A is necessary and sufficient for cAMP-dependent transcription conferred by both the cAMP regulatory element and the adjacent CCAAT-box. In intact cells these cis-elements were shown to bind cAMP response element binding protein, CCAAT-enhancer binding protein beta and nuclear factor-Y, with each protein controlling a different aspect of the cAMP response. Cyclic-AMP acting through protein kinase A stimulated promoter recruitment of CCAAT-enhancer binding protein beta, nuclear factor-Y and RNA polymerase II while depleting the promoter of cyclic-AMP response element binding protein. Stimulation of transcription by cAMP was not associated with increased acetylation of histones H3 and H4 at proximal promoter nucleosomes, indicating that histone acetyltransferases are not involved in this response. Nonetheless, pharmacological inhibition of histone deacetylase activity did increase histone H4 acetylation and the recruitment of RNA polymerase II, indicating that histone acetyltransferases are normally associated with the proximal promoter. Only in C6 cells, however, did inhibition of histone deacetylases stimulate transcription and synergize with cAMP. These experiments provide the first glimpse of the GCH1 gene promoter functioning within intact cells and supply evidence for the involvement of histone acetyltransferase-containing complexes in GCH1 gene transcription.
Keywords: acetylation, C/EBPβ, cAMP, chromatin immunoprecipitation assay, CREB, gene transcription, GTP cyclohydrolase I, histone, NF-Y, nucleosome, Pol II, protein kinase A
GTP cyclohydrolase I (GCH1; EC 3.5.4.16) catalyzes the first, rate-limiting and regulated step in the biosynthesis of 5,6,7,8-tetrahydrobiopterin, the reduced pteridine cofactor essential for the synthesis of monoamine and nitric oxide neurotransmitters and the catabolism of phenylalanine (Thony et al. 2000). GCH1 is expressed by a limited number of cell types (Hirayama et al. 1993; Lentz and Kapatos 1996), where it can be controlled by a host of signal transduction pathways, including those using the second messenger cAMP. Cyclic-AMP induces GCH1 expression in some cells, such as adrenal medullary cells (Abou-Donia et al. 1986), midbrain and hypothalamic dopamine neurons (Zhu et al. 1994; Bauer et al. 2002), mesangial cells (Pluss et al. 1996) and pheochromocytoma-derived PC12 cells (Anastasiadis et al. 1998) but not in others, such as the pineal gland (Kapatos et al. 1981).
Cell type-specificity of the GCH1 gene response to cAMP suggests that novel cAMP signaling mechanisms may be at play, especially since the actions of cAMP are known to be propagated not only by cAMP-dependent protein kinase A (PKA) but also by mitogen-activated protein kinase-(MAPK) and phosphatidylinositol-3-kinase (PI3 K)-dependent signaling pathways (Grewal et al. 2000; Mei et al. 2002). Two recent reports that GCH1 transcription is enhanced by MAPK-dependent signaling pathways support this view (Ito et al. 2005; Sarraj et al. 2005). Alternatively, the selectivity of the cAMP response might be mediated exclusively at the level of GCH1 transcription. In this scenario, GCH1 gene regulatory elements would interact with a milieu of transcription factors and/or co-activators that is unique to cells that respond to cAMP with an increase in GCH1 transcription.
Our earlier work using rat gene GCH1 promoter reporter constructs and transient transfection assays showed that cAMP increases GCH1 transcription in PC12 cells but not in C6 cells (Kapatos et al. 2000), an astrocytoma cell line which expresses low levels of GCH1 (D’Sa et al. 1996). The selective response to cAMP was found to be conferred through protein–DNA interactions that occur within the first 142 bp upstream from the transcription cap site and to require two cis-regulatory elements that are separated by a single turn of the double helix; a non-canonical cAMP response element (CRE) and a high-affinity CCAAT-box (CAT-box) (see Fig. 1b). These studies also showed that the CRE contributes approximately 70% of the promoter response to cAMP while the CAT-box confers the remainder. This division of labor raises the possibility that cAMP, acting through different signal transduction pathways, might control GCH1 transcription by selectively affecting CRE and CAT-box cognate binding proteins.
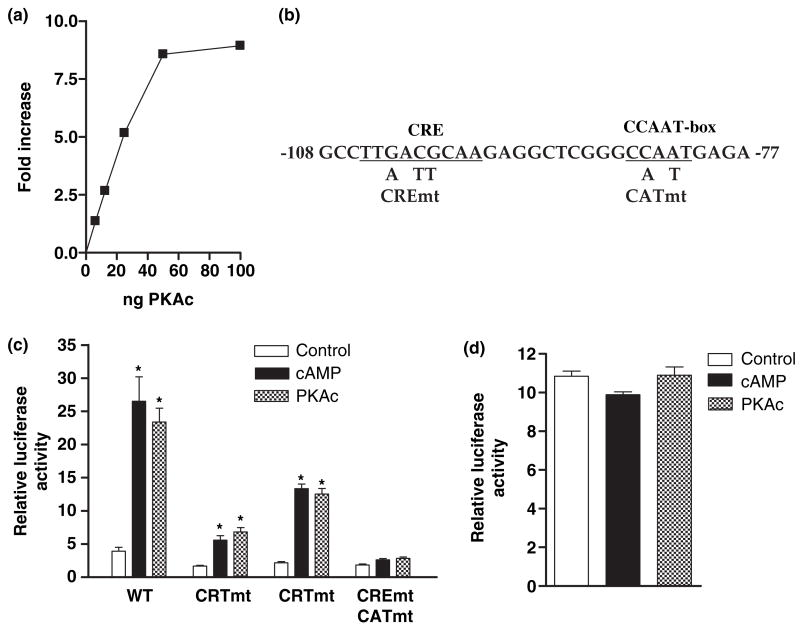
Fig. 1.
Over-expression of the catalytic subunit of protein kinase A (PKAc) mimics the cell type-specific effect of cAMP on GCH1 transcription. (a) PC12 cells were co-transfected with the WT GCH1 promoter construct p0.27rGCH-GL3, pRL-TK and amounts of pRC/RSV-PKAc DNA (PKAc) ranging from 5 to 100 ng. About 18 h later cultures were lysed and assayed for firefly and Renilla luciferase activities using the Dual Luciferase assay. Firefly activity was normalized to Renilla activity and expressed as Relative Luciferase Activity. (b) DNA sequence of the rat GCH1 CRE and CAT-box region from −108 to −77 and mutations introduced to produce CREmt, CATmt and CREmtCATmt. (c) PC12 cells were transfected with p0.27rGCH-GL3 WT, CREmt, CATmt or CREmtCATmt, pRL-TK and where noted 25 ng of pRC/RSV-PKAc. 18 h later conditioned media containing 5 mmol/L 8Br-cAMP (cAMP) was added to wells not co-transfected with PKAc and 4 h later all cells were harvested for dual-luciferase assay. (d) C6 cells were transfected with p0.27rGCH-GL3 WT, pRL-TK and where noted 25 ng of pRC/RSV-PKAc (PKAc). 18 h later conditioned media containing 5 mmol/L 8Br-cAMP (cAMP) was added to wells not co-transfected with PKAc and 4 h later all cells were harvested for dual-luciferase assay. *p ≤ 0.05 when compared with basal construct activity.
In vitro assays indicate that the rat and human GCH1 CRE binds members of the basic leucine zipper (bZIP) family of transcription factors, including CREB and C/EBPβ (Kapatos et al. 2000; Hirayama et al. 2001; but see Sarraj et al. 2005). Although the GCH1 CRE appears to be promiscuous, at least in vitro, evidence suggests that the GCH1 CAT-box exclusively recruits the transcription factor Nuclear Factor Y (NF-Y) (Kapatos et al. 2000; Hirayama et al. 2001) in close association with RNF4 (Wu et al. 2004). NF-Y is a heterotrimer composed of A, B and C subunits which must be fully assembled to bind DNA (Sinha et al. 1995; Kim et al. 1996; Mantovani 1999). RNF4 is a small RING protein which acts as both a co-activator and a regulator of nuclear trafficking (Moilanen et al. 1998; Poukka et al. 2000).
Like CREB and C/EBPβ (Gonzalez and Montminy 1989; Metz and Ziff 1991; Wilson and Roesler 2002), NF-Y is involved in cAMP responsiveness of a number of genes (Muro et al. 1992; Boularand et al. 1995; Rangan et al. 1996; Baler et al. 1997; Cote et al. 2002). Unlike CREB and C/EBPβ, however, NF-Y has never been shown to be a substrate for PKA (but see Faniello et al. 1999). In contrast, CREB (Xing et al. 1998), C/EBPβ (Nakajima et al. 1993) and NF-Y (Alabert et al., 2006) are each regulated either directly or indirectly by MAPK-dependent phosphorylation, again raising the possibility that non-traditional signaling pathways can be involved in the activation of GCH1 transcription by cAMP.
Signaling molecules such as cAMP ultimately influence the activity or recruitment of transcription factors at gene promoters and enhancesomes which then serve to attract co-activators that directly recruit RNA polymerase II (Pol II) and/or modify chromatin structure (Carey 1998; Naar et al., 2001). Promoter-associated chromatin can be remodeled by protein complexes that covalently modify histone tails in nucleosomes wrapping promoter DNA, producing a ‘histone code’ that is recognized by other proteins capable of further chromatin modifications (Jenuwein and Allis 2001; Turner 2002). This broadly accepted model proposes that acetylation of core histones H3 and H4 at N-terminal lysines by histone acetyltransferases (HAT) is associated with enhanced gene expression while deacetylation by histone deacetylases (HDAC) is associated with gene repression. Supporting this model are studies which have repeatedly shown that actively transcribed genes are characterized by promoter-poised nucleosomes acetylated at H3K9 and H4K8 (Santos-Rosa et al. 2002; Liang et al. 2004; Bernstein et al. 2005; Kim et al. 2005). Although the control of GCH1 gene expression continues to be an active area of research, nothing is currently known regarding the involvement of HAT proteins and histone acetylation in GCH1 gene transcription.
We report here that PKA is necessary and sufficient for cAMP-dependent GCH1 transcription conferred by both the proximal promoter CRE and the CAT-box. We also present evidence that CREB, C/EBPβ and NF-Y bind to these cis-elements and are involved in the cAMP response. We show that cAMP stimulates Pol II, C/EBPβ and NF-Y recruitment to the GCH1 proximal promoter in a PKA-dependent fashion but that this recruitment is not characterized by increased acetylation of histones H3 and H4 at GCH1 proximal promoter nucleosomes. We also find, however, that GCH1 transcription can be stimulated in a cell type-specific manner by pharmacological inhibition of HDAC, suggesting that HAT proteins have important roles in controlling GCH1 transcription.
Materials and Methods
Plasmids
The construction and characterization of the wild type pGL3-based luciferase reporter construct p0.27rGCH-GL3 containing 142 bp of the rat GCHI proximal promoter spanning from the transcription start site to the end of the GC-box as well as the CRE mutated, CAT-box mutated and combined CRE and CAT-box mutations has been reported previously (Kapatos et al. 2000 and see Fig. 1b). Renilla reporter plasmids driven by the human herpes virus thymidine kinase minimal promoter (pRL-TK) or no promoter (pRL-Null) were purchased (Promega, Madison WI, USA). The plasmid Rc/RSV-PKAc containing the constitutively active catalytic subunit of PKA (PKAc) was obtained from Dr R.A. Maurer, Oregon Health Sciences University. The A-ZIP family of dominant negative A-CREB, 4H-ATF4 and 4H-C/EBP were obtained from Dr C. Vinson, NCI, NIH and were subcloned into pRC/RVS. C/EBPβ was obtained from Dr P.F. Johnson, NCI, Frederick and was subcloned into pRC/RSV. pRC/RSV-CREB and CREB M1 were obtained from Dr M.R. Montminy, Joslin Diabetes Center, Harvard Medical School. ATF4 was obtained from Dr T. Hai of Ohio State University and was subcloned into pRC/RSV. Wild type NF-YA and dominant negative Δ4YA13m29 (NF-YAm29) were obtained from Dr R. Mantovani, Dipartmento di Scienze Biomoleculari e Biotecnologie, Universita’ di Milano, Milano, Italy. All plasmid DNA was purified by ion-exchange chromatography as recommended by the supplier (Qiagen, Valencia CA, USA).
Cell cultures and transfection
Wild type PC12 cells and the PKA-deficient PC12 cell line 126-1B2 (Dr J.A. Wagner, Cornell University) were passaged in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal calf serum, 10% horse serum, 100 μg/mL penicillin and 100 μg/mL streptomycin. The rat C6 astrocytoma cell line was grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and antibiotics. All cultures were maintained in a humidified atmosphere of 10% CO2 at 37°C. The day before transfection cells were plated onto poly-D-lysine coated 24-well plates. Cells were transfected using either LipofectaminePlus or Lipofectamine2000 (Invitrogen, Carlsbad, CA, USA) with a total of 0.5–0.8 μg of DNA per well. Cells were harvested 22 h after co-transfection with PKAc. In other experiments, 18 h after transfection cultures were treated for 4 h with cell conditioned media containing 5 mmol/L 8Br-cAMP. In some experiments, protein kinase inhibitors dissolved in DMSO (0.1% final) were added 30 min prior to application of 8Br-cAMP. Cells were lysed and firefly and Renilla luciferease were assayed using the Dual Luciferase system (Promega). Firefly luciferase activity was normalized to Renilla activity and is reported as relative luciferase activity.
Chromatin immunoprecipitation assay
ChIP was performed as described previously but with modifications designed to increase assay sensitivity and decrease between sample variability (West et al. 2004). 20 million PC12 or 126-1B2 cells or 10 million C6 cells were plated on poly-D-lysine coated 100 mm dishes and treated the next day with 8Br-cAMP (5 mmol/L), trichostatin A (TSA; 100 ng/mL) or sodium butyrate (NaBut; 10 mmol/L) dissolved in growth-conditioned medium. In some experiments, cells were pre-treated for 30 min with H89 or TSA prior to application of 8Br-cAMP. Treatments were terminated by rapid medium aspiration and addition of serum-free media containing 1% fresh formaldehyde. After rocking incubation for 10 min at 25°C, cells were washed with PBS and protein-DNA cross-linking terminated at 25°C by the addition of 0.125 mol/L glycine for 10 min. All further procedures were performed at 4°C in the presence of protease inhibitors.
Cells were scraped into PBS and collected by centrifugation at 400 g. Cell pellets were suspended in 1 mL of lysis buffer (1% SDS, 10 mmol/L EDTA, 50 mmol/L Tris–HCl, pH 8.0) and incubated for 10 min at 4°C. DNA was fragmented to an average size of 400 bp by sonication using an Autotune Series High Intensity Ultrasonic Processor. Cell debris was pelleted, supernatant harvested and 5% set aside as input DNA. Samples were diluted 10-fold with dilution buffer (0.01% SDS, 1.0% Triton X-100, 1.0 mmol/L EDTA, 150 mmol/L NaCl, 15 mmol/L Tris–HCl, pH 8.0) and pre-cleared by the addition of 20 μg of rabbit IgG followed by end-over-end rocking for 1 h. 200 μl of salmon sperm DNA/BSA blocked Protein A-agarose (50% slurry) was added for 1 h. Beads were collected by centrifugation and supernatants transferred to fresh non-stick tubes.
At this stage fresh protease inhibitors were added and samples were divided into 4–5 parts, each receiving 5 μg of antibody directed against either RNA polymerase II (Santa Cruz Biotechnology, Santa Cruz, CA, USA, H-224), CREB (Santa Cruz, H-74), C/EBPβ (Santa Cruz, C-19), NF-YB (Santa Cruz, FL-207), K9,14 di-acetylated H3 or K5,8,12,16 tetra-acetylated H4 (both from Upstate Biotechnology, Lake Placid, NY, USA). Anti-Green Fluorescent Protein (GFP; Santa Cruz, SC8334) or normal rabbit immunoglobulin G (IgG; Santa Cruz, SC-2027) served as negative controls. Samples were incubated overnight and immune complexes were collected with 40 μL of Protein A-agarose followed by incubation for 2 h. Beads were pelleted and washed for 10 min sequentially with low-salt wash (0.1% SDS, 1% TritonX-100, 2 mmol/L EDTA, 150 mmol/L NaCl, 20 mmol/L Tris–HCl, pH 8.0), high-salt wash (0.1% SDS, 1% Triton X-100, 2 mmol/L EDTA, 500 mmol/L NaCl, 20 mmol/L Tris–HCl, pH 8.0), LiCl wash (0.25 mol/L LiCl, 1% NP40, 1% deoxycholate, 1 mmol/L EDTA, 10 mmol/L Tris–HCl, pH 8.0) and finally TE. To the beads was added 175 μL of 10 mmol/L Tris–Cl, pH 8.0 containing 1% SDS, 0.1 mol/L NaHCO3, 0.2 mol/L NaCl and 1 μg/mL RNaseA. Input DNA was treated identically. In order to reverse cross-links and digest protein, beads and input DNA were incubated overnight at 65°C and were then made 10 mmol/L in EDTA and 40 μg in Proteinase K followed by incubation at 50°C for 1 h. DNA was purified using the Qiaquick PCR Purification system (Qiagen) and eluted with 100 μL of 10 mmol/L Tris–HCl, pH 8.0.
Samples were analyzed by real time quantitative PCR (QPCR) (Roche LightCycler, Mannheim, Germany) using QuantiTect SYBR-Green PCR reagent (Qiagen). PCR primers were designed to amplify either the proximal (−46 GCCGCGCCTCTCTTTTTATG −27 and −146 TGTGCAACTGCGGGGTTTAG −167) or distal (−5396 TCACTGGCTCATGTACT-GAATG −5374 and −5439 GACCTGCTTCCTCTCAATACAG −5417) sequence of the rat GCH1 promoter or the proximal c-fos proximal promoter (−122 TCCTACATGCGGAGGGTCC-AGGAGAC-97 and −304 GAG-TAGTAGGCGCCTCAGCTGGCCG −328). Cycling parameters included a hot start at 95°C for 900 s and then 40 cycles of 94°C for 15 s at 20°C/s, 55°C for 25 s at 20°C/s and 72°C for 20 s at 2°C/s with single acquisition mode. Melting curve analysis was always performed to determine amplicon homogeneity: 95°C for 5 s at 20°C/s, 65°C for 15 s at 20°C/s and 95°C for 0 s at 0.1°C/s with continuous acquisition. All samples were run in triplicate. Standard curves for each PCR product were generated by plotting Ct versus serial dilutions of pooled input DNA. The enrichment for each sample was calculated by subtracting non-specific immunoprecipitation and dividing by input DNA. The resulting values were then normalized to control samples immunoprecipitated with the identical antibody. Normalization enabled data to be pooled across different experiments and be expressed as fold increase.
Quantitative real time RT-PCR analysis
Total RNA was isolated from cultures of PC12 or C6 cells and treated with DNase I as recommended by the supplier (Qiagen, RNeasy). 25–150 ng of RNA was reverse transcribed (Qiagen, Omniscript) in a reaction primed with random hexamers. 10–20% of this reaction served as template for quantitative real-time RT-PCR (QRTPCR) analysis using QuantiTect SYBR-Green PCR reagents to amplify either rat GCH1 (forward, caagggataccaggagacca; reverse, tctcgtcatggtcctcatca), c-fos (forward gaagggaaaggaataagatggc; reverse, ttctcgtcttcaagttgatctg) or β-actin (forward, gtcgtaccactggcattgtg; reverse, ctctcagctgtggtggtgaa) cDNA. Control reactions minus reverse transcriptase were included for each primer pair and quantity of RNA. Standard curves for each transcript were generated using serial dilutions of pooled RNA from control samples and distinct reverse transcription reactions. GCH1 and c-fos mRNA abundance were expressed relative to that of β-actin and normalized across experiments.
Western blot analysis
Approximately 10 μg of a PC12 cell soluble acid protein extract was analyzed using 12% SDS-PAGE and antibodies directed against K9,14 di-acetylated H3 or K5,8,12,16 tetra-acetylated H4 (Bradford 1976). A chemiluminescence-based secondary antibody conjugated peroxidase reaction was performed and detected by X-ray film.
Data analysis
All experiments were performed at least three times and assayed in triplicate. Data were pooled and analyzed by one-way or two-way ANOVA with individual post hoc tests performed using Bonferroni’s Multiple Comparison Test (PRISM, GraphPad). Differences were accepted as significant at p < 0.05.
Results
Over-expression of the catalytic subunit of PKA mimics cell type-specific cAMP-dependent GCH1 transcription
We first sought to determine whether eliminating cAMP from the cAMP signaling cascade by over-expressing the catalytic subunit of PKA (PKAc) could mimic the effects of cAMP on GCH1 transcription in PC12 cells. Transfection with varying amounts of PKAc DNA stimulated transcription from the WT GCH1 proximal promoter construct in a concentration-dependent manner, reaching a plateau of approximately ninefold at 100 ng of DNA (Fig. 1a). About 25 ng of PKAc DNA produced a four-to six-fold stimulation of GCH1 transcription, which is roughly equivalent to that of a maximally effective concentration of 8Br-cAMP (Fig. 1c). This amount of PKAc DNA was used in all subsequent experiments.
We next determined whether co-transfection with PKAc enhances transcription through both the GCH1 proximal promoter CRE and CAT-box elements (Fig. 1b). These experiments showed quite clearly that mutation of the CRE (CREmt) or the CAT-box (CATmt) reduced PKAc-dependent transcription by 70–80% and 20–30%, respectively, while mutation of both elements (CREmtCATmt) decreased promoter activity to control levels (Fig. 1c). The same pattern of transcription from these reporter constructs was detected following incubation with 8Br-cAMP (Fig. 1c). Like the responsiveness of the CRE and CAT-box to cAMP, cell type specificity is also a defining characteristic of cAMP-dependent GCH1 transcription (Kapatos et al. 2000). Figure 1d shows that transcription in C6 cells from the WT GCH1 promoter was not enhanced by co-transfection with PKAc or by incubation with 8Br-cAMP. The cellular specificity of the cAMP response therefore appears to arise downstream from PKA signaling.
PKA is necessary for cAMP-dependent GCH1 transcription
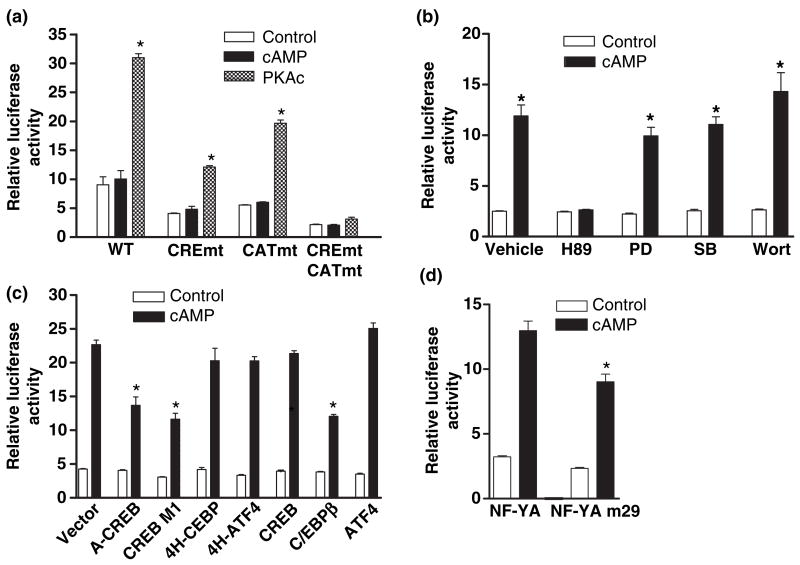
In order to confirm that PKA is necessary for cAMP-dependent transcription, we next examined GCHI proximal promoter function in the PKA-deficient cell line 126-1B2 (Van Buskirk et al. 1985). GCH1 transcription in 126-1B2 cells from the WT or mutated promoter constructs was found to be completely refractory to incubation with 8Br-cAMP (Fig. 2a). In contrast, complementation of 126-1B2 cells by co-transfection with PKAc stimulated GCHI transcription (Fig. 2a). Moreover, the pattern of stimulation from the CREmt and CATmt reporter constructs was virtually identical to that observed for PC12 cells treated with 8Br-cAMP or co-transfected with PKAc (see Fig. 1c). These results demonstrate that with the exception of PKA, 126-1B2 cells contain all the machineries necessary for cAMP-dependent GCH1 transcription conferred by both the CRE and the CAT-box elements. This conclusion is supported by the complete lack of GCH1 mRNA induction in 126-1B2 cells following 4 h incubation with 8Br-cAMP, a treatment which increased PC12 cell GCH1 transcript levels by four-fold (Fig. 5a). The capacity of PKAc over-expression to stimulate transcription in 126-1B2 cells is also in stark contrast to the negative response of C6 cells to this same treatment, and supports our contention that cell type-specific cAMP-dependent GCH1 transcription involves signaling mechanisms downstream of PKA.
Fig. 2.
Protein kinase A (PKA) is necessary and sufficient for cAMP-dependent GCH1 transcription which also involves CREB, C/EBPβ and NF-Y. (a) PKA-deficient 126-1B2 cells were co-transfected with p0.27rGCH-GL3 WT, CREmt, CATmt or CREmtCATmt, pRL-TK and where noted 25 ng of pRC/RSV-PKAc. 18 h later conditioned media containing 5 mmol/L 8Br-cAMP (cAMP) was added to wells not co-transfected with PKAc and 4 h later all cells were harvested for dual-luciferase assay. (b) PC12 cells transfected 18 h earlier with p0.27rGCH-GL3 WT and pRL-TK were pre-incubated for 30 min with either 20 μmol/L of H89, an inhibitor of PKA, 30 μmol/L of PD98059 (PD), an inhibitor of MAPKK, 10 μmol/L of SB203580 (SB), an inhibitor of p38 MAPK, 100 nm of wortmannin (Wort), an inhibitor of PI3 K or DMSO (Vehicle) at 0.1% and then treated with 5 mmol/L 8Br-cAMP (cAMP) for 4 h before harvesting for assay. (c) PC12 cells were transfected with p0.27rGCH-GL3 WT, pRL-TK and 100 ng of pRC/RSV-based expression vectors encoding for wild type or dominant negative forms of CREB (A-CREB, CREB M1 and CREB), C/EBP (4H-CEBP and C/EBPβ) or ATF4 (4H-ATF4 and ATF4) and then treated with 5 mmol/L 8Br-cAMP (cAMP) for 4 h before harvesting for assay. (d) PC12 cells were transfected with p0.27rGCH-GL3 WT, pRL-TK and expression vectors encoding for wild type (NF-YA) or dominant negative (NF-YA m29) NF-YA and then treated with 5 mmol/L 8Br-cAMP (cAMP) for 4 h before harvesting for assay. *p ≤ 0.05 when compared with respective control activity.
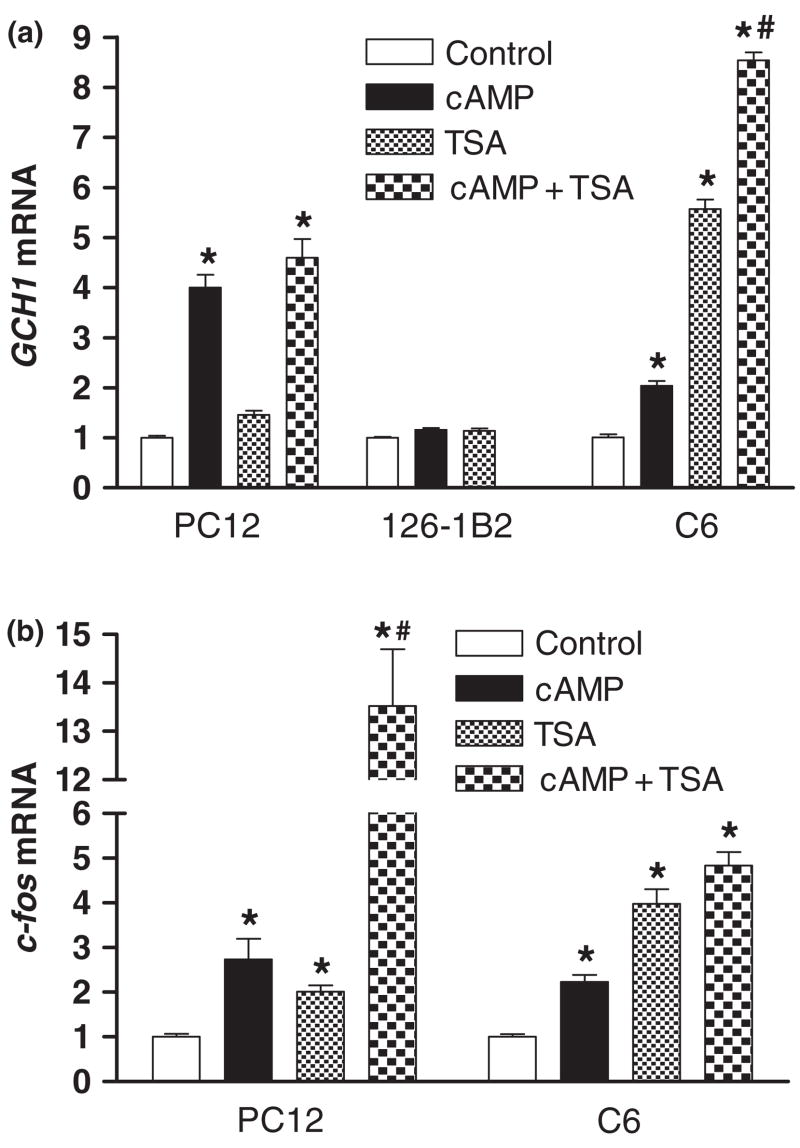
Fig. 5.
Inhibition of histone deacetylase activity increases GCH1 mRNA levels in C6 but not PC12 cells. (a) Real-time reverse transcription quantitative PCR analysis of GCH1 mRNA in PC12 cells, 126-1B2 or C6 cells incubated for 4 h with 5 mmol/L 8Br-cAMP (cAMP), 100 ng/mL of trichostatin A (TSA) or cAMP + TSA.(b) Real-time reverse transcription quantitative PCR analysis of c-fos mRNA in PC12 or C6 cells incubated for 4 h with 5 mmol/L 8Br-cAMP (cAMP), 100 ng/mL of trichostatin A (TSA) or cAMP + TSA. *p ≤ 0.05 when compared with respective control activity. #p ≤ 0.05 when compared with cAMP or TSA alone.
Protein kinase A is sufficient for cAMP-dependent GCH1 transcription
Although these experiments show that PKA is necessary for cAMP responsiveness from both the CRE and CAT-box elements, PKA-dependent effects in PC12 cells might occur either early or late in a cAMP signaling cascade. Indeed, cAMP signaling that is dependent upon PKA but does not utilize PKA as the ultimate downstream kinase is known to involve MAPKK (Kawasaki et al. 1998; Grewal et al. 2000), p38 MAPK (Cao et al. 2001), MAPK (Grewal et al. 2000) or PI3 K (Mei et al. 2002). Many of the effects of cAMP that are independent of PKA also involve cAMP-binding proteins and nucleotide exchange factors (Epac/cAMP-GEF) that stimulate MAPKKK activity (De Rooij et al. 1998; Kawasaki et al. 1998).
To discriminate between these possibilities, PC12 cells transfected with the WT GCH1 reporter were pre-incubated for 30 min with either 20 μmol/L of H89, an inhibitor of PKA, 30 μmol/L of PD98059, an inhibitor of MAPKK, 10 μmol/L of SB203580, an inhibitor of p38 MAPK, 100 nm of wortmannin, an inhibitor of PI3 K or the DMSO vehicle at 0.1% and then treated with 8Br-cAMP for 4 h before harvesting. These concentrations of inhibitors were chosen based upon published studies showing maximal effectiveness and specificity in PC-12 cells (Pugazhenthi et al. 1999; Grewal et al. 2000; Liu et al. 2001). Figure 2b shows that incubation with the PKA inhibitor H89 completely blocked the WT promoter response to cAMP, whereas inhibitors of MAPKK, p38 MAPK or PI3 K were without effect. We conclude that PKA is necessary and sufficient for cAMP-dependent enhancement of GCH1 transcription confered by both the CRE and the CAT-box.
CREB, C/EBPβ and NF-Y are involved in cAMP-dependent GCH1 transcription
In vitro assays using PC12 nuclear extracts have shown that the rat GCH1 CRE binds CREB, ATF4 and C/EBPβ while the CAT-box recruits NF-Y (Kapatos et al. 2000). To investigate the functional contributions of these proteins to cAMP-dependent GCH1 transcription, wild type and dominant negative forms were transfected into PC12 cells along with the WT GCH1 promoter construct. The dominant negative constructs A-CREB, 4H-ATF4 and 4H-C/EBPβ do not bind DNA but instead target the basic leucine zipper region of their respective target proteins to prevent DNA binding. (Olive et al., 1997; Ahn et al., 1998). In contrast, CREB M1 competes with endogenous CREB for binding to the CRE but contains an alanine substitution at the critical regulatory site serine 133 which prevents phosphorylation by PKA (Gonzalez and Montminy 1989) and recruitment of the co-activator CBP (Chrivia et al. 1993). The NF-Y dominant negative Δ4YA13m29 (NF-YA m29) does not interfere with NF-Y heterotrimer assembly but does prevent NF-Y binding to the CAT-box (Mantovani et al. 1994). Concentration-effect curves were generated for each of these constructs and all effects on transcription were found to be maximum at 100 ng DNA per well.
These co-transfection experiments showed that the dominant negative proteins A-CREB and CREB M1 but not wild type CREB decreased cAMP-dependent GCH1 transcription by 50–60% without affecting basal activity (Fig. 2c). Binding of CREB to the CRE and the phosphorylation of CREB at serine 133 by PKA thus appear to be essential for cAMP stimulation but not for basal levels of activity. Moreover, the partial inhibition observed for either CREB dominant negative is comparable to the fraction of the cAMP response that is lost following mutation of the CRE. Over-expression of wild type C/EBPβ but not dominant negative 4H-C/EBP also inhibited the promoter response to cAMP by 50–60% without altering basal activity. When compared with over-expression of wild type NF-YA, the dominant negative NF-YA m29 decreased cAMP-dependent GCH1 transcription by 20–30% without affecting basal activity (Fig. 2d). This level of inhibition is also comparable to the fraction of the cAMP response lost following mutation of the CAT-box. Neither ATF4 nor dominant negative 4H-ATF4 altered GCH1 transcription. Overall, these results provide convincing evidence that CREB, C/EBPβ and NF-Y each have important roles in the regulation of GCH1 transcription by cAMP.
CREB, C/EBPβ, NF-Y and Pol II are associated with the GCH1 proximal promoter
We next used PC12 cells and chromatin immunoprecipitation (ChIP) assays to determine whether endogenous CREB, C/EBPβ, NF-Y and Pol II are associated with the endogenous GCH1 promoter. Sonication parameters were first developed to efficiently and reproducibly reduce formaldehyde cross-linked genomic DNA to an average size of 400 bp which was shown in preliminary studies to be optimal for ChIP (Fig. 3a). Immunoprecipitated DNA from ChIP assays was detected by PCR using gene specific primers to amplify a 141 bp product from the GCH1 proximal promoter (−167 to −27) or a 65 bp product from the distal GCH1 promoter (−5439 to −5375) (Fig. 3b). Proximal promoter primers amplify a region of the gene which contains the GC-box, CRE and the CAT-box (Kapatos et al. 2000) while distal promoter primers amplify a region located approximately 5.4 kb upstream from the transcription start site (Fig. 3b). Distal promoter amplifications serve as a gene-specific control because deletion analysis has shown that this region of the gene is not involved in cAMP-dependent transcription (Kapatos et al. 2000; Hirayama et al. 2001).
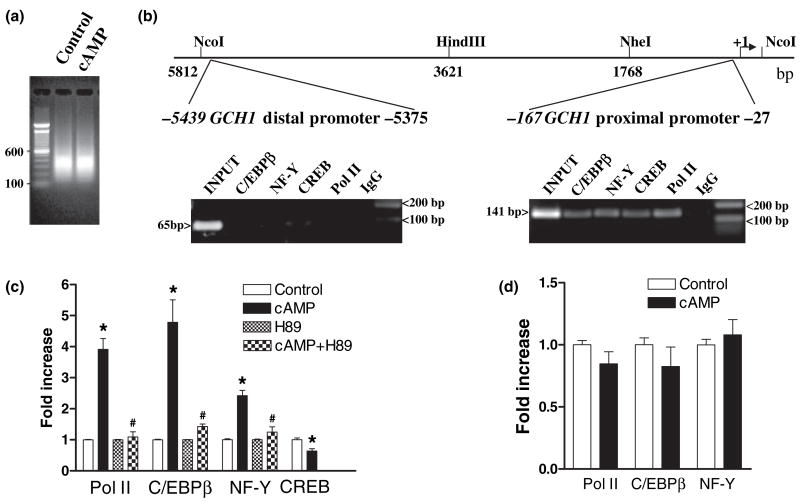
Fig. 3.
Protein kinase A-dependent recruitment of PolII, C/EBPβ and NF-Y but not CREB to the GCH1 proximal promoter. (a) Sonication of cross-linked DNA from control and 8Br-cAMP-treated PC12 cells reproducibly generates average size fragments of 400 bp. (b) A schematic of the rat GCH1 5′ flanking region showing the locations of proximal and distal promoter regions amplified by PCR relative to the transcription start site ( + 1). Electrophoresis of PC12 ChIP PCR reactions using GCH1 proximal and distal promoter primer pairs identified amplicons of the correct size and shows that under basal conditions Pol II, CREB, C/EBPβ and NF-Y are associated with the proximal but not distal promoter. (c) Real-time quantitative PCR analysis of PC12 ChIP samples using GCH1 proximal promoter primers shows that Pol II, C/EBPβ and NF-Y but not CREB are recruited to the proximal promoter in response to 1 h incubation with 5 mmol/L 8Br-cAMP (cAMP) and that recruitment is blocked by the PKA inhibitor H89 (cAMP + H89) which has no effect on its own (H89). (d) Real-time quantitative PCR analysis of 126-1B2 cell ChIP samples using GCH1 proximal promoter primers shows that Pol II, C/EBPβ and NF-Y are not recruited to the proximal promoter in response to 1 h incubation with 5 mmol/L 8Br-cAMP (cAMP). *p ≤ 0.05 when compared with respective control activity. #p ≤ 0.05 when compared with cAMP.
Our initial ChIP analysis used extracts from untreated PC12 cells and precipitating antibodies directed against the large subunit of Pol II, CREB, C/EBPβ or the B subunit of the NF-Y heterotrimer. Abundant PCR products of the correct size were detected in proximal promoter amplifications of ChIP DNA precipitated by each of these antibodies (Fig. 3b). In no case were PCR signals above control precipitations observed for distal promoter amplifications (Fig. 3b). These results indicate that under basal conditions Pol II, CREB, C/EBPβ and NF-YB are associated with the GCH1 proximal but not distal promoter. Based upon the results of transient transfection assays (Figs 2c and d), we tentatively conclude that CREB and NF-Y bound to the promoter enhance transcription whereas binding of C/EBPβ represses transcription.
PKA-dependent recruitment of Pol II, C/EBPβ and NF-Y to the GCH1 proximal promoter
Further studies investigated whether the interaction of these proteins with the promoter is modified by cAMP and requires PKA. QPCR of immunoprecipitated DNA derived from PC12 cells incubated under control conditions or with 8Br-cAMP for 1 h revealed that Pol II, C/EBPβ and NF-Y were each recruited to the GCH1 proximal promoter in response to cAMP, resulting in a two- to three-fold increase in Pol II, a four- to five-fold increase in C/EBPβ and a two- to three-fold increase in NF-Y (Fig. 3c). In contrast, the abundance of CREB at the proximal promoter declined by 40%. ChIP signals above background were never detected in distal promoter amplifications (data not shown). Cyclic-AMP-dependent recruitment of Pol II, C/EBPβ and NF-Y is therefore specific to the proximal promoter.
Proximal promoter levels of Pol II, C/EBPβ or NF-Y remained unaltered following incubation with the PKA inhibitor H89. But, H89 treatment did prevent cAMP-dependent recruitment of these proteins (Fig. 3c). Similarly, treatment of PKA-deficient 126-1B2 cells with 8Br-cAMP did not alter proximal promoter loading by Pol II, C/EBPβ, or NF-Y (Fig. 3d). These results agree with data showing that GCH1 transcription in 126-1B2 cells is refractory to 8Br-cAMP. Moreover, it would appear that PKA activity per se is not necessary for assembly of the basal transcriptional machinery because incubation of PC12 cells with H89 did not alter the abundance of these proteins at the promoter and untreated PC12 and 126-1B2 cells express essentially equal levels of GCH1 mRNA (data not shown).
Recruitment of Pol II, C/EBPβ and NF-Y to the GCH1 proximal promoter does not involve acetylation of histones H3 and H4
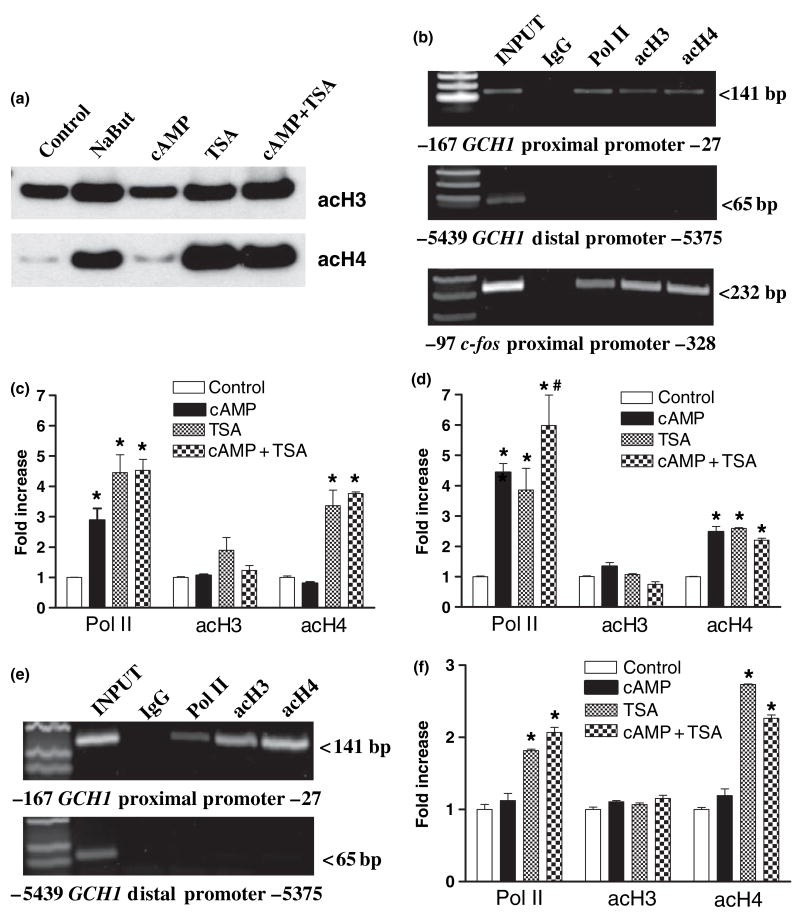
CREB, C/EBPβ and NF-Y each interact with HAT proteins, including CBP, p300, PCAF and Gcn5 (Chrivia et al. 1993; Mink et al. 1997; Currie 1998). Based upon this knowledge the next series of experiments set out to determine whether acetylation of nucleosomes poised at the GCH1 proximal promoter correlates with cAMP-dependent transcription. Initial studies using western blotting of PC12 cell acid soluble protein extracts showed that whole cell levels of K9,14 di-acetylated histone H3 (acH3) and K5,8,12,16 tetra-acetylated histone H4 (acH4) were unaffected by 1 hour incubation with 8Br-cAMP (Fig. 4a). Gel analysis of ChIP assays from untreated PC12 cells then localized GCH1 nucleosomes marked at acH3 and acH4 to the proximal but not distal promoter (Fig. 4b). Subsequent ChIP with QPCR showed that levels of acH3 and acH4 at these nucleosomes remained unchanged 1 h after incubation with 8Br-cAMP, even though Pol II was actively recruited in these experiments (Fig. 4c).
Fig. 4.
Recruitment of Pol II, C/EBPβ and NF-Y does not involve acetylation of histones H3 and H4 at GCH1 proximal promoter poised nucleosomes. (a) Western blots of equal amounts of acid-soluble proteins from PC12 cells treated for 1 h with 10 mmol/L sodium butyrate (NaBut), 5 mmol/L 8Br-cAMP (cAMP), 100 ng/mL of trichostatin A (TSA) or cAMP + TSA and probed with antibodies directed against K9,14 di-acetylated H3 (acH3) or K5,8,12,16 tetra-acetylated H4 (acH4). (b) Electrophoresis of PC12 ChIP PCR reactions using GCH1 proximal and distal promoter primer pairs shows that under basal conditions Pol II, acH3 and acH4 are associated with the proximal but not distal promoter. Similar analysis using c-fos proximal promoter primers identifies amplicons of the correct size associated with Pol II, acH3 and acH4 ChIP samples. (c) Real-time quantitative PCR analysis of PC12 ChIP samples using GCH1 proximal promoter primers. Pol II is equally recruited in response to cAMP, TSA and cAMP + TSA. None of the treatments alter levels of acH3. In contrast, TSA and cAMP + TSA equally elevate acH4 levels. (d) Real-time quantitative PCR analysis of PC12 ChIP samples using c-fos proximal promoter primers. Pol II is recruited in response to cAMP and TSA but Pol II recruitment following cAMP + TSA is greater than either treatment alone. None of the treatments alter levels of acH3. In contrast, cAMP, TSA and cAMP + TSA equally elevate levels of acH4. (e) Electrophoresis of C6 ChIP PCR reactions using GCH1 proximal and distal promoter primer pairs shows that under basal conditions Pol II, acH3 and acH4 are associated with the proximal but not distal promoter. (f) Real-time quantitative PCR analysis of C6 ChIP samples using GCH1 proximal promoter primers. Pol II was not recruited in response to cAMP but was recruited following TSA and cAMP + TSA. None of the treatments alter levels of acH3. In contrast, TSA and cAMP + TSA both elevate acH4 levels. *p ≤ 0.05 when compared with respective control activity. #p ≤ 0.05 when compared with cAMP or TSA alone.
Stimulation of transcription from the c-fos promoter by cAMP requires HAT activity (Fass et al. 2003). The c-fos promoter was therefore analyzed as a positive control using the identical ChIP samples used to study the GCH1 promoter. Gel analysis of ChIP samples from PC12 cells using PCR primers that amplify a 232 bp product of the c-fos proximal promoter (−328 to −97) detected Pol II, acH3 and acH4 (Fig. 4b). Pol II at the inactive c-fos promoter has been interpreted to indicate poising of the pre-initiation complex (Pinaud and Mirkovitch 1988; Fass et al. 2003). It would seem that acH3 and acH4 are also characteristic of this poised promoter. QPCR analysis of the c-fos promoter showed that cAMP increased Pol II recruitment by 4-fold and the acetylation of histone H4 by two- three-fold but had no effect on acetylation of histone H3 (Fig. 4d). These data clearly demonstrate that cAMP stimulation of the c-fos promoter recruits a HAT complex which then acetylates histone H4 but that this sequence of events does not occur at the GCH1 proximal promoter.
Basal differences in GCH1 transcription do not involve acetylation of histones H3 and H4
The C6 cell line expresses very low basal levels of GCH1 mRNA (D’Sa et al. 1996; Vann et al. 2000, 2002). In fact, QRTPCR indicates that C6 cells contain approximately 10% of the GCH1 mRNA found in PC12 cells (data not shown). A comparison between PC12 and C6 cells of Pol II loading and histone acetylation might therefore be informative about the relationship of GCH1 gene transcription and chromatin structure. Gel analysis of ChIP samples showed, however, that like PC12 cells C6 cells have significant amounts of acH3 and acH4 located at the GCH1 proximal but not distal promoter (Fig. 4e). This observation makes it unlikely that differences in acetylation of nucleosomes at the proximal promoter are responsible for differences in basal levels of transcription. Finding Pol II as well as acH3 and acH4 at the C6 cell promoter suggests that like c-fos, GCH1 transcription in these cells is poised for activation. Although GCH1 transcription in C6 cells may be poised, treatment with 8Br-cAMP did not alter proximal promoter levels of Pol II or the acetylation of histones H3 and H4 (Fig. 4f).
Inhibitors of histone deacetylase increase GCH1 Pol II recruitment and histone H4 acetylation
The next series of experiments set out to determine whether evidence for HAT proteins at the GCH1 promoter can be obtained by pharmacologically inhibiting HDAC and monitoring levels of acH3 and acH4 in associated nucleosomes. Initial studies using western blotting showed that levels of acetylated histones H3 and H4 in PC12 cell were increased 3- to 20-fold, respectively, following 1 h incubation with the HDAC inhibitors sodium butyrate (NaBut; 10 mmol/L) or trichostatin A (TSA; 100 ng/mL) (Fig. 4a). The robust increase in acetylation of histone H4 but not H3 following HDAC inhibition suggests that the acetyl groups on lysines 9 and 14 of histone H3 turn over slowly, which may account for the lack of effect of cAMP and HDAC inhibition on levels of acH3 at individual genes. Western blotting also showed that incubation with 8Br-cAMP did not potentiate or inhibit TSA-dependent acetylation of histone H3 or H4 proteins.
ChIP and QPCR were used next to assess Pol II recruitment and the acetylation of GCH1 proximal promoter nucleosomes in PC12 cells following incubation with TSA alone or in combination with 8Br-cAMP. As observed following 8Br-cAMP treatment, TSA stimulated Pol II recruitment by two- to four-fold without affecting histone H3 acetylation (Fig. 4c). Unlike cAMP, however, TSA increased the acetylation of histone H4 by three- to fourfold, a strong indication that proteins with HAT activity are associated with the GCH1 proximal promoter even though these proteins do not appear to be either activated or recruited during cAMP stimulation. Neither Pol II loading nor histone acetylation was enhanced further by combining 8Br-cAMP and TSA treatments. TSA treatment alone or in combination with 8Br-cAMP did not induce Pol II loading or histone H4 acetylation at the GCH1 distal promoter (data not shown). Moreover, an identical pattern of Pol II recruitment and histone H4 acetylation was observed following treatment of PC12 cells with NaBut (data not shown). ChIP analysis of C6 cells treated with TSA alone or in combination with 8Br-cAMP generated a pattern of Pol II recruitment and histone acetylation similar to what was observed in PC12 cells (Fig. 4f).
QPCR analysis of these same PC12 cell ChIP samples using c-fos primers showed that Pol II is also recruited to the this promoter in response to TSA treatment, as has been reported previously (Fass et al. 2003) (Fig. 4d). In contrast to GCH1, however, Pol II loading following the combination of 8Br-cAMP and TSA was greater than with either stimulus alone (Fig. 4d). Treatment with TSA did not modify c-fos histone H3 acetylation but did increase by two- to three-fold the acetylation of histone H4. Unlike the effect of TSA on Pol II recruitment, combined treatment did not enhance histone H4 acetylation beyond what was observed following either treatment alone.
Inhibition of histone deacetylase activity increases GCH1 mRNA levels in C6 but not in PC12 cells
The next series of experiments questioned whether Pol II recruitment induced by HDAC inhibition is sufficient to increase GCH1 transcription. As determined by QRTPCR, incubation of PC12 cells for 4 h with TSA did not increase GCH1 transcript levels or potentiate or inhibit the response of the GCH1 gene to 8Br-cAMP (Fig. 5a). Continued analysis of these same samples revealed that c-fos transcript levels in PC12 cells were increased two- three-fold by 8Br-cAMP or TSA treatment alone but that combined treatment synergistically increased c-fos mRNA by more than 13-fold (Fig. 5b). Surprisingly, a two-fold induction of GCH1 expression by cAMP was observed in C6 cells (Fig. 5a) even though this treatment did not alter Pol II loading or transcription monitored by transient transfection assays. It should be noted, however, that even after cAMP stimulation GCH1 mRNA levels in C6 cells are approximately 20% of untreated PC12 cells. Nevertheless, treatment of C6 cells with TSA increased GCH1 mRNA levels by more than five-fold and the combination of cAMP and TSA synergistically increased transcript levels by almost nine-fold (Fig. 5a). The induction of c-fos transcript levels in C6 cells was generally more subdued, increasing two-fold by 8Br-cAMP, four-fold by TSA and five-fold by combined treatments. Overall, these results indicate that TSA-induced Pol II recruitment is sufficient to increase GCH1 transcription in C6 cells but not in PC12 cells.
Discussion
A number of conclusions about the regulation of GCH1 transcription by cAMP can be reached from the present work. First, cAMP enhances transcription by acting entirely through PKA. Proteins involved in the transcriptional response to cAMP must therefore be substrates for PKA, as is already known for CREB and C/EBPβ (Gonzalez and Montminy 1989; Metz and Ziff 1991) and has been suggested by studies of NF-Y (Boularand et al. 1995; Rangan et al. 1996; Faniello et al. 1999). Second, the effect of PKA on transcription is conferred by both the CRE and the CAT-box. This makes GCH1 one of a small group of genes which utilize this combination of cis-elements to respond to cAMP with an increase in transcription (Muro et al. 1992; Osawa et al. 1996; Baler et al. 1997). The advantage offered by this tandem array of regulatory elements is unknown but, presumably involves a greater degree of transcriptional control. Third, CREB is constitutively bound by the proximal promoter and initiates cAMP-dependent transcription following phosphorylation of the critical regulatory site at serine 133 by PKAc. Fourth, cAMP acting through PKA stimulates recruitment of NF-Y and C/EBPβ to the proximal promoter. Loading of the promoter by NF-Y is required for maximal cAMP responsiveness. To our knowledge, this is the first time NF-Y has been shown to be recruited to a promoter in response to cAMP. In contrast, based upon the results of transient transfection assays, the role of C/EBPβ in the cAMP response appears to be one of inhibiting transcription. CREB and C/EBPβ compete in vitro for binding to the proximal promoter CRE (Kapatos et al. 2000), making it likely that by displacing CREB, C/EBPβ can either prevent or reverse cAMP stimulation. Indeed, one interpretation of the simultaneous localization of these proteins at the promoter by ChIP is that CREB and C/EBPβ are bound by different GCH1 alleles, with the C/EBPβ-bound allele able to support basal transcription but unable to respond to cAMP. In general, the non-canonical GCH1 CRE might provide a low-affinity site from which CREB can function yet be displaced by other bZIP proteins, such as C/EBPβ, ATF2 or c-Jun, which have different transcriptional properties (Kapatos et al. 2000; Hirayama et al. 2001; Sarraj et al. 2005).
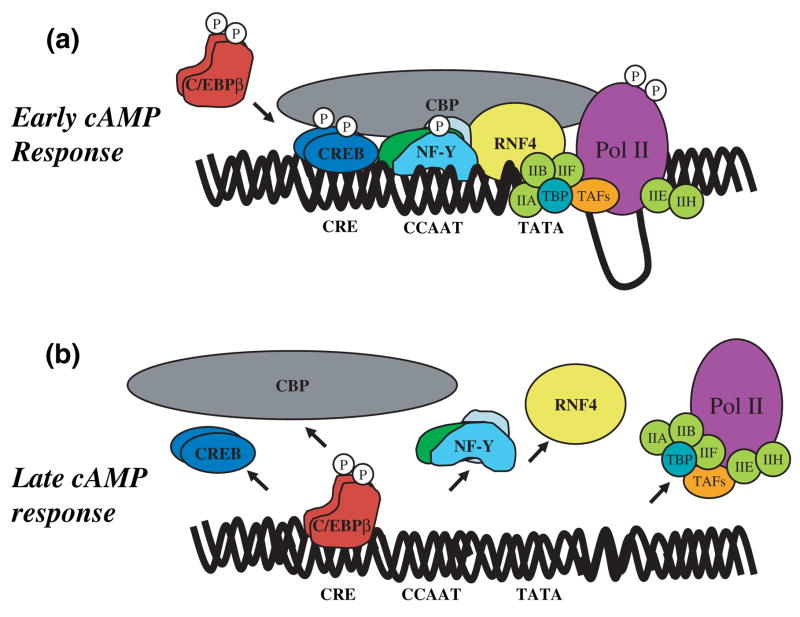
A model of protein-DNA interactions at the GCH1 proximal promoter during cAMP-dependent transcription is presented in Fig. 6. This model assumes that NF-Y and Pol II are recruited to the promoter before the arrival of C/EBPβ, an assumption based upon the observation that C/EBPβ in PC12 cells must be phosphorylated by PKA before entering the nucleus (Metz and Ziff 1991). Our model of the cAMP response is therefore divided into two phases. In the Early cAMP Response, cAMP activates PKA in the cytosol and PKAc is translocated to the nucleus. CREB bound to the CRE is phosphorylated by PKAc at serine 133 (Gonzalez and Montminy 1989) and subsequently recruits the co-activator CBP (Chrivia et al. 1993). CBP then interacts with the holoenzyme complex, bringing Pol II and the other general transcriptional machinery into contact with the promoter. In response, the CTD of Pol II is phosphorylated at serine 5 by Cdk7, a component of TFIIH, forming the pre-initiation complex (for details see Proudfoot et al. 2002; Soutoglou and Talianidis 2002; Boehm et al. 2003). Phosphorylation of NF-Y by PKAc enhances NF-Y binding to the CAT-box and NF-Y recruitment of the co-activator RNF4. Through interactions with NF-Y (Wu et al. 2004) and TBP (Moilanen et al. 1998), RNF4 enhances TBP binding to the weak GCH1 TATA box, thereby serving along with CBP to tether the transcriptional machinery to the promoter. Transcription elongation then proceeds after phosphorylation of the CTD of Pol II at Serine 2 by an unidentified kinase. In the late cAMP response, C/EBPβ is phosphorylated by PKAc in the cytoplasm and is then shuttled to the nucleus. The length of the temporal delay between when C/EBPβ is phosphorylated and when it enters the nucleus is critical for determining the duration of the cAMP response. Upon entering the nucleus, phosphorylated C/EBPβ displaces CREB from the CRE and the co-activator CBP from the promoter. In the process, NF-Y and RNF4 are also displaced, the holoenzyme complex is released and transcription subsides. The early cAMP response thus proposes a mechanism for activation of GCH1 transcription from the CRE and CAT-box that requires CREB and NF-Y while the late cAMP response proposes a mechanism for terminating cAMP-dependent transcription through the displacement of CREB by C/EBPβ.
Fig. 6.
A model of cAMP-dependent GCH1 transcription. Early cAMP response. The early cAMP response describes a mechanism for cAMP-dependent transcription that is mediated through the CRE and CCAAT-box by CREB and NF-Y. In response to increased cytoplasmic levels of cAMP, PKA is activated and PKAc is shuttled to the nucleus. CREB already bound to the CRE is phosphorylated at the critical regulatory site Ser133 by PKAc, resulting in the recruitment of the co-activator CBP to the promoter. CBP interacts with the holoenzyme complex, bringing Pol II and the general transcriptional machinery into contact with the promoter. In response, the CTD of Pol II is phosphorylated at serine 5 by Cdk7, a component of TFIIH, generating the pre-initiation complex. Phosphorylation of NF-Y by PKAc promotes NF-Y binding to the CCAAT-box and NF-Y recruitment of the co-activator RNF4. Through its interactions with NF-Y and TBP, RNF4 enhances TBP binding to the weak GCH1 TATA box, thus serving with CBP to tether the transcriptional machinery. Transcription elongation then proceeds following phosphorylation of the Pol II CTD at Serine 2 by an unknown kinase. Late cAMP response: The late cAMP response describes a mechanism for termination of cAMP-dependent transcription by recruitment of C/EBPβ to the CRE. C/EBPβ is phosphorylated by PKAc in the cytoplasm and then shuttled to the nucleus. Upon entering the nucleus, phosphorylated C/EBPβ displaces CREB from the CRE and the co-activator CBP from the promoter. NF-Y and RNF4 are also displaced from the CCAAT-box. Following promoter depletion of CREB, NF-Y and associated co-activators, the holoenzyme complex is released and transcription subsides.
A number of conclusions can also be reached regarding the role of histone acetylation in GCH1 transcription. First, proximal promoter poised nucleosomes are not acetylated at K9,14 of histone H3 or K5,8,12,16 of histone H4 as part of the transcriptional program initiated in response to cAMP. Second, the acetylation of histone H3 and H4 does not correlate with basal levels of transcription. Third, pharmacological inhibition of HDAC enhances acetylation of histone H4 at proximal promoter nucleosomes, demonstrating that HAT complexes are part of the transcriptional machinery. Fourth, the effect of pharmacological inhibition of HDAC on transcription is dependent upon cell type and the basal rate of transcription. Fifth, persistent maintenance of the pre-initiation complex might play a critical role in cells where GCH1 is rapidly induced by inflammatory cytokines in concert with nitric oxide synthase (Werner et al. 1990; D’Sa et al. 1996; Vann et al. 2000, 2002). Sixth, persistent maintenance of histone H3 and H4 acetylation and the preinitiation complex at the proximal promoter might prevent permanent silencing of the GCH1 locus during periods of gene repression.
It is well established that promoter-associated chromatin can be remodeled by protein complexes that covalently modify histone tails in nucleosomes wrapping promoter DNA (Jenuwein and Allis, 2002; Turner 2002). In this broadly accepted model, the acetylation of N-terminal lysines in the core histones H3 and H4 is associated with the recruitment of HAT-containing protein complexes and enhanced gene expression. The acetylation of histone tails not only decreases nucleosome interactions with DNA but, also supplies docking sites for DNA remodeling complexes which are recruited to acetylated H3 and H4 via protein bromodomains, thereby spreading acetylation among adjacent nucleosomes (Grant et al. 1997; Syntichaki et al. 2000). Complicating this model, however, are recent studies showing that histone deacetylation is also involved in the enhancement of gene expression (Fass et al. 2003; Rascle et al. 2003; Xu et al. 2003). This has lead to the proposal that cAMP and CREB-responsive genes can be divided into two groups based upon their response to HDAC inhibition, those that are activated, such as c-fos, and those that are inhibited, such as ICER (Fass et al. 2003). For genes activated by HDAC inhibition the actions of HAT-containing promoter complexes are potentiated by inhibiting the turnover of histone N-terminal acetyl groups and the subsequent recruitment of bromodomain-containing enhancer complexes. Repression of transcription by HDAC inhibition is more difficult to reconcile with this model but, might involve acetylation-dependent blockade of some critical aspect of transcription activation. CAMP stimulation of GCH1 transcription in PC12 cells does not appear to require HAT activity and so does not fit into either of these models. On the contrary, both basal and cAMP-dependent transcription in C6 cells are potentiated by HDAC inhibition, leaving us to conclude that GCH1 transcription in these cells can be grouped with genes that are activated by HAT-containing promoter complexes.
Studies of GCH1 gene transcription are important because tetrahydrobiopterin is an essential cofactor for the synthesis of clinically important signaling molecules (Thony et al. 2000). Just one example of this importance is the childhood movement disorder known as DOPA-responsive dystonia (DRD). Heterozygous mutations in GCH1 often produce DRD (Ichinose et al. 1994) which is characterized by severe decrements of dopamine in otherwise normal nigrostriatal dopamine neurons (Rajput et al. 1994; Furukawa et al. 2002). Nigrostriatal dopamine neurons are peculiar because they respond to cAMP with an increase in GCH1 transcription (Zhu et al., 1993; Bauer et al. 2002) yet in rodents and humans maintain low basal levels of GCH1 transcription (Lentz and Kapatos 1996; Hirayama and Kapatos 1998; Shimoji et al. 1999; Hirayama and Kapatos 2001). DRD might therefore be thought of as a GCH1 haplo-insufficiency that is specific to nigrostriatal dopamine neurons (Kapatos and Hirayama 2002). Approximately half the DRD patients with GCH1 deficiencies have no detectable mutation in the GCH1 open reading frame and might instead have mutations in regulatory elements which then suppress transcription from one or both alleles (Inagaki et al. 1999). Our research has identified the CRE and CAT-box as highly conserved regulatory elements that are likely sites through which genetic variation would impact GCH1 transcription (Kapatos et al. 2000; Hirayama et al. 2001). Knowledge of the transacting factors recruited by these cis-elements, such as C/EBPβ and NF-Y, is equally important because these proteins might ultimately become pharmacological targets for manipulating GCH1 transcription in dopamine neurons (Tanaka et al. 1999; Cieslik et al. 2002).
Acknowledgments
This work was supported by a grant from the NINDS (RO1-NS26081).
Abbreviations used
- C/EBPβ
CCAAT enhancer-binding protein beta
- CAT-box
CCAAT-box
- CBP
CREB binding protein
- CRE
cAMP response element
- CREB
cAMP regulatory element binding protein
- GCHI
GTP cyclohydrolase I
- MAPK
mitogen activated protein kinase
- NF-Y
nuclear factor Y
- PI3 kinase
phosphatidyl inositol 3-kinase
- PKA
protein kinase A
References
- Abou-Donia MM, Wilson SP, Zimmerman TP, Nichol CA, Viveros OH. Regulation of guanosine triphosphate cyclohydrolase and tetrahydrobiopterin levels and the role of the cofactor in tyrosine hydroxylastion in primary cultures of adrenomedullary chromaffin cells. J Neurochem. 1986;46:1190–1199. doi: 10.1111/j.1471-4159.1986.tb00637.x. [DOI] [PubMed] [Google Scholar]
- Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Rogers L, Kahn L, Niellez S, Fafet P, Cerulis S, Blanchard JM, Hipskind RA, Vignais ML. Cell type-dependent control of NF-Y activity by TGF-β. Oncogene. 2006;10:3387–3396. doi: 10.1038/sj.onc.1209385. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Bezin L, Gordon LJ, Imerman B, Blitz J, Levine RA. Vasoactive intestinal peptide induces both tyrosine hydroxylase activity and tetrahydrobiopterin biosynthesis in PC12 cells. Neuroscience. 1998;86:179–189. doi: 10.1016/s0306-4522(97)00611-8. [DOI] [PubMed] [Google Scholar]
- Baler R, Covington S, Klein DC. The rat arylalkylamine N-acetyltransferase gene promoter-cAMP activation via a cAMP-responsive element-CCAAT complex. J Biol Chem. 1997;272:6979–6985. doi: 10.1074/jbc.272.11.6979. [DOI] [PubMed] [Google Scholar]
- Bauer M, Suppman S, Meyer M, Hesslinger C, Gasser T, Widner HR, Ueffing M. Glial cell line-derived neurotrophic factor up-regulates GTP-cyclohydrolase I activity and tetrahydrobiopterin levels in primary dopaminergic neurones. J Neurochem. 2002;82:1300–1310. doi: 10.1046/j.1471-4159.2002.01074.x. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on hdsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boularand S, Darmon MC, Ravassard P, Mallet J. Characterization of the human tryptophan hydroxylases gene promoter. J Biol Chem. 1995;270:3757–3764. doi: 10.1074/jbc.270.8.3757. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cao W, Medvedev AV, Daniel KW, Collins S. β-Adrenergic activation of p38 MAP kinase in adipocytes. J Biol Chem. 2001;276:27 022–27 082. doi: 10.1074/jbc.M101049200. [DOI] [PubMed] [Google Scholar]
- Carey M. The enhancesome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RPS, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Cieslik K, Zhu Y, Wu KK. Salicylate suppresses macrophage nitric-oxide synthase-2 and cyclo-oxygenase-2 expression by inhibiting CCAAT/Enhancer-binding protein-β via a common signaling pathway. J Biol Chem. 2002;277:49 314–49 310. doi: 10.1074/jbc.M205030200. [DOI] [PubMed] [Google Scholar]
- Cote F, Schussler N, Boularand S, Peirotes A, Thevenot E, Mallet J, Vodjdani G. Involvement of NF-Y and Sp1 in basal and cAMP-stimulated transcriptional activation of the tryptophan hydroxylase (TPH) gene in the pineal gland. J Neurochem. 2002;81:673–685. doi: 10.1046/j.1471-4159.2002.00890.x. [DOI] [PubMed] [Google Scholar]
- Currie RA. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- D’Sa C, Hirayama K, West A, Hahn M, Zhu M, Kapatos G. Tetrahydrobiopterin biosynthesis in C6 cells: induction of GTP cyclohydrolase I gene expression by lipopolysaccharide and cytokine treatment. Molec Brain Res. 1996;41:105–110. doi: 10.1016/0169-328x(96)00073-3. [DOI] [PubMed] [Google Scholar]
- De Rooij J, Zwartkruis FJT, Verheijen MHG, Cool RH, Nijman SMB, Wittinghofer A, Bos JL. Epac is a Rap1 guanine-nucl;eotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:472–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- Faniello MC, Bevilacqua MA, Condorelli G, de Crombrugghe B, Maity SN, Avvedimento VE, Cimino F, Costanzo F. The B subunit of the CAAT-binding factor NFY binds the central segment of the co-activator p300. J Biol Chem. 1999;274:7623–7626. doi: 10.1074/jbc.274.12.7623. [DOI] [PubMed] [Google Scholar]
- Fass DM, Butler JEF, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J Biol Chem. 2003;44:43 014–43 019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Kapatos G, Haycock JW, Worsley J, Wong H, Kish SJ, Nygard TG. Brain biopterin and tyrosine hydroxylase in asymptomatic dopa-responsive dystonia. Ann Neurol. 2002;51:637–641. doi: 10.1002/ana.10175. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones:characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;13:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grewal SS, Horgan AM, York RD, Withers GS, Banker GA, Stork PJS. Neuronal calcium activates a Rap1 and B-Raf signaling pathway via the cyclic adenosine monophosphate-dependent protein kinase. J Biol Chem. 2000;275:3722–3728. doi: 10.1074/jbc.275.5.3722. [DOI] [PubMed] [Google Scholar]
- Hirayama K, Kapatos G. Nigrostriatal dopamine neurons express low levels of GTP cyclohydrolase I protein. J Neurochem. 1998;70:164–170. doi: 10.1046/j.1471-4159.1998.70010164.x. [DOI] [PubMed] [Google Scholar]
- Hirayama K, Kapatos G. Human Nigrostrial Dopamine Neurons Express Low Levels of GTP Cyclohydrolase I mRNA. In: Milstien S, Kapatos G, Levine RA, Shane B, editors. Chemistry and Biology of Pteridines and Folates, Proceedings of the 12th International Symposium on Pteridines and Folates. Kluwer Academic Publishers; Boston/Dordrecht/London: 2001. pp. 291–295. [Google Scholar]
- Hirayama K, Lentz SI, Kapatos G. Tetrahydrobiopterin cofactor biosynthesis: GTP cyclohydrolase I mRNA expression in rat brain and superior cervical ganglia. J Neurochem. 1993;61:1006–1014. doi: 10.1111/j.1471-4159.1993.tb03614.x. [DOI] [PubMed] [Google Scholar]
- Hirayama K, Shimoji M, Swick L, Meyer A, Kapatos G. Characterization of GTP cyclohydrolase I gene expression in the human neuroblastoma SKN-BE(2)M17: enhanced transcription in response to cAMP is conferred by the proximal promoter. J Neurochem. 2001;79:576–587. doi: 10.1046/j.1471-4159.2001.00583.x. [DOI] [PubMed] [Google Scholar]
- Ichinose H, Ohye T, Takahashi E, et al. Hereditary progressive dystonia with marked diurnal fluctuation caused by mutations in GTP cyclohydrolase I gene. Nat Genet. 1994;8:236–242. doi: 10.1038/ng1194-236. [DOI] [PubMed] [Google Scholar]
- Inagaki H, Ohye T, Suzuki T, Segawa N, Nomura Y, Nagatsu T, Ichinose H. Decrease in GTP cyclohydrolase I gene expression caused by inactivation of one allele in hereditary progressive dystonia with marked diurnal fluctuation. Biochem Biophys Res Comm. 1999;260:747–751. doi: 10.1006/bbrc.1999.0976. [DOI] [PubMed] [Google Scholar]
- Ito T, Suzuki T, Ichinose H. Nerve growth factor-induced expression of the GTP cyclohydrolase I gene via Ras/MEK pathways in PC12D cells. J Neurochem. 2005;95:563–569. doi: 10.1111/j.1471-4159.2005.03414.x. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kapatos G, Hirayama K. GTP Cyclohydrolase I Gene Expression and Catecholamine Synthesis. In: Nagatsu T, Nabeshima T, McCarty R, Goldstein DS, editors. Catecholamine Research: From Molecular Insights to Clinical Medicine. Kluwer Academic Publishers; Boston/Dordrecht/London: 2002. pp. 143–146. [Google Scholar]
- Kapatos G, Kaufman S, Weller JL, Klein DC. Biosynthesis of biopterin: adrenergic cyclic adenosine monophosphate-dependent inhibition in the pineal gland. Science. 1981;213:1129–1131. doi: 10.1126/science.6168019. [DOI] [PubMed] [Google Scholar]
- Kapatos G, Stegenga SL, Hirayama K. Identification and characterization of basal and cyclic AMP response elements in the promoter of the rat GTP cyclohydrolase I gene. J Biol Chem. 2000;275:5947–5957. doi: 10.1074/jbc.275.8.5947. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Springett GM, Mochizuki N, Toki S, Nakaya M, Matsuda M, Housman DE, Graybiel AM. A family of cAMP-binding proteins that directly activate Rap 1. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- Kim IS, Sinha S, de Crombrugghe B, Maity SN. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol Cell Biol. 1996;16:4003–4013. doi: 10.1128/mcb.16.8.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Berrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz SI, Kapatos G. Tetrahydrobiopterin biosynthesis in rat brain: heterogeneity of GTP cyclohydrolase I mRNA expression in monoamine-containing neurons. Neurochem Int. 1996;28:569–582. doi: 10.1016/0197-0186(95)00124-7. [DOI] [PubMed] [Google Scholar]
- Liang G, Lin JCY, Wei V, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Prenger MS, Norton DD, Mei L, Kusiak JW, Bai G. Nerve growth factor uses ras/ERK and phosphatidylinositol 3-kinase cascades to up-regulate the N-methyl-D-aspartate receptor 1 promoter. J Biol Chem. 2001;276:45 372–45 379. doi: 10.1074/jbc.M105399200. [DOI] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Mantovani R, Li X-Y, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogues of NF-YA. J Biol Chem. 1994;269:20 340–20 346. [PubMed] [Google Scholar]
- Mei FC, Qiao J, Tsygankova OM, Meinkoth JL, Quilliam LA, Chemg X. Differential signaling of cyclic AMP. J Biol Chem. 2002;277:11 497–11 504. doi: 10.1074/jbc.M110856200. [DOI] [PubMed] [Google Scholar]
- Metz R, Ziff E. cAMP stimulates the C/EBP-related transcription factor rNFIL-6 to translocate to the nucleus and induce c-fos transcription. Genes & Develop. 1991;5:1754–1766. doi: 10.1101/gad.5.10.1754. [DOI] [PubMed] [Google Scholar]
- Mink S, Haenig B, Klempnauer KH. Interaction and functional collaboration of p300 and C/EBPβ. Mol and Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moilanen AM, Poukka H, Karvonen U, Hakli M, Janne OA, Palvimo JJ. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol Cell Biol. 1998;18:5128–5139. doi: 10.1128/mcb.18.9.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro AF, Bernath VA, Kornblihtt AR. Interaction of the −170-cyclic AMP response element with the adjacent CCAAT box in the human fibronectin gene promoter. J Biol Chem. 1992;267:12767–12774. [PubMed] [Google Scholar]
- Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M, Krylov D, Echlin DR, Gardner K, Taparowsky E, Vinson C. A dominant-negative to AP1 that abolished DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- Osawa H, Robey RB, Printz RL, Granner DK. Identification and characterization of basal and cyclic AMP response elements in the promoter of the rat hexokinase II genes. J Biol Chem. 1996;271:17 296–17 303. doi: 10.1074/jbc.271.29.17296. [DOI] [PubMed] [Google Scholar]
- Pinaud S, Mirkovitch J. Regulation of c-fos expression by RNA polymerase elongation competence. J Mol Biol. 1988;280:785–798. doi: 10.1006/jmbi.1998.1905. [DOI] [PubMed] [Google Scholar]
- Pluss C, Werner ER, Blau N, Wachter H, Pfeilschifter J. Interleukin 1β and cAMP trigger the expression of GTP cyclohydrolase I in rat renal mesangial cells. Biochem J. 1996;318:665–671. doi: 10.1042/bj3180665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H, Karvonen U, Yoshikawa N, Tanaka H, Palvimo JJ, Janne OA. The RING finger protein SNURF modulates nuclear trafficking of the androgen receptor. J Cell Sci. 2000;113:2991–3001. doi: 10.1242/jcs.113.17.2991. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Pugazhenthi S, Boras T, O’Connor D, Meintzer MK, Heidenreich KA, Reusch JEB. Insulin-like growth factor I-mediated activation of the transcription factor cAMP response element-binding protein in PC12 cells. J Biol Chem. 1999;274:2829–2837. doi: 10.1074/jbc.274.5.2829. [DOI] [PubMed] [Google Scholar]
- Rajput AH, Gibb WRG, Zhong XH, Shannak KS, Kish S, Chang LG, Hornykiewicz O. Dopa-responsive dystonia: pathological and biochemical observations in a case. Ann Neurol. 1994;35:396–402. doi: 10.1002/ana.410350405. [DOI] [PubMed] [Google Scholar]
- Rangan VS, Oskouian B, Smith S. Identification of an inverted CCAAT-box motif in the fatty-acid synthase gene as an essential element for mediation of transcriptional regulation by cAMP. J Biol Chem. 1996;271:2307–2312. doi: 10.1074/jbc.271.4.2307. [DOI] [PubMed] [Google Scholar]
- Rascle A, Johnston JA, Amati B. Deacetylase activity is required for recruitment of the basal transcriptional machinery and transactivation by STAT5. Mol Cell Biol. 2003;23:4162–4173. doi: 10.1128/MCB.23.12.4162-4173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sarraj JA, Vinson C, Han J, Thiel G. Regulation of GTP cyclohydrolase I gene transcription by basic region leucine zipper transcription factors. J Cell Biochem. 2005;96:1003–1020. doi: 10.1002/jcb.20580. [DOI] [PubMed] [Google Scholar]
- Shimoji M, Hirayama K, Hyland K, Kapatos G. GTP cyclohydrolase I gene expression in the brains of male and female hph-1 mice. J Neurochem. 1999;72:757–764. doi: 10.1046/j.1471-4159.1999.0720757.x. [DOI] [PubMed] [Google Scholar]
- Sinha S, Maity SN, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the 3rd subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodeling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Okshima N, Hidaka H. Isolation of cDNAs encoding cellular drug-binding proteins using a novel expression cloning procedure: Drug-Western. Mol Pharm. 1999;55:356–363. doi: 10.1124/mol.55.2.356. [DOI] [PubMed] [Google Scholar]
- Thony B, Auerbach G, Blau N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J. 2000;347:1–16. [PMC free article] [PubMed] [Google Scholar]
- Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Van Buskirk R, Corcoran T, Wagner JA. Clonal variation of PC12 pheochromocytoma cells with defects in cAMP-dependent protein kinases induces ornithine decarboxylase in response to nerve growth factor but not to adenosine agonists. Mol Cell Biol. 1985;5:1984–1992. doi: 10.1128/mcb.5.8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann LR, Twitty S, Spiegel S, Milstien S. Divergence in regulation of nitric-oxide synthase and its cofactor tetrahydrobiopterin by tumor necrosis factor-α. J Biol Chem. 2000;275:13275–13281. doi: 10.1074/jbc.275.18.13275. [DOI] [PubMed] [Google Scholar]
- Vann LR, Payne SG, Edsall LC, Twitty S, Spiegel S, Milstien S. Involvement of sphingosine kinase in TNF-α-stimulated Tetrahydrobiopterin biosynthesis in C6 glioma cells. J Biol Chem. 2002;277:12 649–12 656. doi: 10.1074/jbc.M109111200. [DOI] [PubMed] [Google Scholar]
- Werner ER, Werner-Felmayer G, Fuchs D, Hausen A, Reibnegger G, Yim JJ, Pfleiderer W, Wachter H. Tetrahydrobiopterin biosynthetic activities in human macrophages, fibroblasts, THP-1, and T 24 cells: GTP cyclohydrolase I is stimulated by interferon-gamma, and 6-pyruvoyl tetrahydrobiopterin synthase and sepiapterin reductase are constitutively present. J Biol Chem. 1990;265:3189–3192. [PubMed] [Google Scholar]
- West AB, Kapatos G, O’Farrell C, Gonzalez-de-Chavez F, Chiu K, Farrer MJ, Maidment NT. N-myc regulates parkin expression. J Biol Chem. 2004;279:28 896–28 902. doi: 10.1074/jbc.M400126200. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Roesler WJ. CCAAT/enhancer binding proteins: do they possess intrinsic cAMP-inducible activity? Mol Cell Endocrinol. 2002;188:15–20. doi: 10.1016/s0303-7207(01)00754-7. [DOI] [PubMed] [Google Scholar]
- Wu S-M, Kuo W-C, Hwu W-L, Hwa K-Y, Mantovani R, Lee YM. RNF4 is a coactivator for nuclear factor Y on GTP cyclohydrolase I proximal promoter. Mol Pharm. 2004;66:1317–1324. doi: 10.1124/mol.66.5.. [DOI] [PubMed] [Google Scholar]
- Xing J, Kornhauser JM, Xia Z, Thiele EA, Greenberg ME. Nerve growth factor activates extracellular signal-regulated kinase and p38 mitogen-activated kinase pathways to stimulate CREB serine 133 phosphorylation. Mol Cell Biol. 1998;18:1946–1955. doi: 10.1128/mcb.18.4.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Nie L, Kim SH, Sun XH. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPβ. EMBO J. 2003;22:893–904. doi: 10.1093/emboj/cdg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Hirayama K, Kapatos G. Regulation of tetrahydrobiopterin biosynthesis in cultured dopamine neurons by depolarization and cAMP. J Biol Chem. 1994;269:11825–11829. [PubMed] [Google Scholar]