Abstract
Differential scanning calorimetry provides a new window into the plasma proteome. Plasma from normal individuals yields a characteristic, reproducible thermogram that appears to represent the weighted sum of denaturation profiles of the most abundant constituent plasma proteins. Plasma from diseased individuals yields dramatically different signature thermograms. Thermograms from individuals suffering from rheumatoid arthritis, systemic lupus, and Lyme disease were measured. Each disease appears to have a distinctive and characteristic thermogram. The difference in thermograms between normal and diseased individuals is not caused by radical changes in the concentrations of the most abundant plasma proteins but rather appears to result from interaction of as yet unknown biomarkers with the major plasma proteins. These results signal a novel use for calorimetry as a diagnostic tool.
INTRODUCTION
The human plasma proteome is a complex fluid that contains more than 3000 individual proteins and peptides that are present in quantities that range from picograms to tens of milligrams per milliliter (1–3). The human plasma proteome holds great promise as a convenient specimen for disease diagnosis and therapeutic monitoring (4–10). Blood samples may be easily obtained from individuals who have given informed consent by minimally invasive, safe procedures. A number of FDA-approved plasma/serum diagnostic assays are routinely used. These include serum plasma electrophoresis (11) and a variety of immunochemical assays that can monitor the concentrations of specific proteins in plasma/serum. These existing low- to moderate-resolution assays have had a profound practical impact on medical diagnosis. For the medical practitioner, the measurement of plasma proteins can be a powerful clinical assessment tool for detecting, diagnosing, and monitoring diseases (12).
The recent explosion of proteomics has brought increased interest in the human plasma/serum proteome as a source for biomarkers of human disease. Higher-resolution methods such as two-dimensional electrophoresis (13,14) and mass spectrometry (5,15–18), coupled with often elaborate protocols for sample preparation and fractionation, have made it possible to identify apparent changes in the composition of the less abundant proteins and peptides in plasma that seem to correlate with particular diseases. Typically, no single protein emerges from such analyses as a wholly reliable biomarker, but instead, changes in the patterns of panels of proteins often serve as the best diagnostic for a particular malady. These patterns often involve protein or peptide components of plasma that are present in low concentrations.
Interest in the array of existing proteins in a patient's serum has thus evolved to a more detailed consideration of the low-molecular-weight peptides within serum, which represent a mixture of small intact proteins plus degradation fragments of larger proteins. This low-molecular-weight region of the serum proteome has been dubbed the “peptidome” (19) and has been touted as a “treasure trove of diagnostic information that has largely been ignored….” (20). Mass spectrometry, in particular SELDI methods, made the peptidome accessible for analysis. (Others, however, have considered the peptidome to be “unidentified flying peptides” and have questioned the reliability of peptidome SELDI patterns as a meaningful diagnostic until the functions of all of the peptide peaks in the peptidome have been properly identified (5).) Many components of the “peptidome” were found to be complexed with more abundant serum proteins, particularly human serum albumin (HSA) and immunoglobulins. This led to the concept of the “interactome” (21), which introduces the added complexity that serum/plasma may be “comprised of a ‘network’ of protein-protein and peptide-protein interactions,” in which potential biomarkers are bound to the more abundant proteins within the fluid. Interestingly, the article that introduced the “interactome” concept concluded by saying that “the discovery of novel biomarkers in serum/plasma requires new biochemical and analytical approaches, and, most importantly, it is clear that no single sample preparation or detection method will suffice if biomarker investigations are to be broadly successful using current technologies” (21). We describe here an entirely new technology for the analysis of the plasma/serum proteomes.
Ten proteins make up 90% of the mass of plasma proteins (by weight). These are, in order of abundance: albumin, IgG, fibrinogen, transferrin, IgA, α2-macroglobulin, α1-antitrypsin, complement C3, IgM, and haptoglobin. Another 12 proteins account for another 9% of the plasma mass, the three most abundant of which are the apolipoproteins A1 and B, and α1-acid glycoprotein. Twenty-two proteins thus comprise 99% of the mass of plasma, making it challenging to fractionate and quantify the remaining 1%. Existing useful assays monitor different aspects of the plasma proteome. The FDA-approved serum protein electrophoresis monitors changes in the most abundant protein population (11). More recent two-dimensional gel electrophoresis and mass spectrometry assays focus more on the least abundant components of plasma, following laborious prefractionation protocols to rid the plasma/serum of the proteins present in high concentrations. What is clear is that no single assay or technology can fully exploit plasma as a source for biomarkers. Multiple technologies are needed, each with a distinctive physical basis that can sense unique properties of the individual proteins within the plasma milieu. Electrophoresis and mass spectrometry, for example, both separate plasma based on protein size and charge. Other assays based on other physical properties of proteins would be useful adjuncts to current approaches for characterizing the plasma proteome.
A novel calorimetric assay is described here that provides a new window through which to view the properties of the human plasma (or serum) proteome. Calorimetry provides a direct means for detecting what is perhaps the most fundamental property of chemical and biochemical reactions, heat changes. Biological calorimetry dates from the time of Lavoisier (1743–1794), who invented a calorimetric method for measuring the heats of metabolism of living animals (22). Our assay exploits the high sensitivity of modern microcalorimeters, which can reliably measure heat changes of 0.1 μcal. In particular, we use the technique of differential scanning calorimetry (DSC) (23–27).
A primary DSC thermogram is an extensive property of a protein solution, and as such it is directly proportional to the mass of the protein in solution. If the weight concentration of the protein is doubled, for example, the calorimetric heat response will double. Similarly, in a solution of mixtures of proteins, the heat response will be proportional to the mass of each protein component in the mixture. That is the fundamental basis of our assay and provides a distinct advantage. Mixtures of proteins may be resolved with respect to the fundamental characteristic denaturation profiles of their component proteins. Each protein in a noninteracting mixture will denature at its characteristic melting temperature (Tm) and with its characteristic melting enthalpy. The observed overall thermogram will be, in the absence of interactions, the weighted sum of all of the individual protein thermograms, weighted according to the mass of each component. DSC has an added advantage. Binding interactions can alter the thermal denaturation of proteins in ways that are now well understood (24,28). If the “interactome” hypothesis (21) is correct, calorimetry would be uniquely sensitive to the proposed biomarker-protein interactions in plasma in ways that electrophoresis and mass spectrometry are not.
We show that DSC provides an entirely new window into the plasma proteome. Plasma from normal individuals yields a reproducible and characteristic thermogram. Unique signature thermograms are obtained from the plasma from diseased individuals, indicating the potential of calorimetry as a new type of clinical diagnostic.
MATERIALS AND METHODS
Pure protein samples
HSA (lot No. 113K7601), immunoglobulin G (IGG) (lot No. 415781/1), immunoglobulin A (IGA) (lot No. 105K3777), α1-acid glycoprotein (AAG) (lot No. 073K7607), α1-antitrypsin (AAT) (lot No. 033K7603), fibrinogen (FIB) (lot No. 083K7604), transferrin (TRF) (lot No. 123K14511), haptoglobin (HPT) (lot No. 055K1664), and immunoglobulin M (IGM) (lot No. 016K4876) were purchased from Sigma-Aldrich (St. Louis, MO). α1-Antichymotrypsin (ACT) (lot No. B58700), complement C3 (C3) (lot No. D33204), complement C4 (C4) (lot No. D34721), ceruloplasmin (CER) (lot No. B70322), α2-macroglobulin (A2M) (lot No. B73605), and prealbumin (PRE) (lot No. B68296) were purchased from Calbiochem (LaJolla, CA). C-reactive protein (CRP) (lot No. 32F0305FP) was purchased from Life Diagnostics (West Chester, PA).
Standard reference serum
A serum reference material (sample No. 16910) was purchased from Sigma-Aldrich.
Plasma samples
Normal plasma samples (lot Nos. JA053759, JA053761, JA053763, JA053764, JA053765, JA053766, JC014372, JM034968, JM034969, JM034970, JM034971) were purchased from Innovative Research (Southfield, MI) and were also obtained from the Gynecological Cancer Repository of the James Graham Brown Cancer Center. Plasma from individuals suffering from Lyme disease (lot Nos. BM146897, BM140032, BM140031, BM140028), systemic lupus erythematosis (lot Nos. BM142168, BM142160), and rheumatoid arthritis (lot Nos. BM204810, BM205222, BM203373, BM202803, BM200182) were purchased from BBI Diagnostics (West Bridgewater, MA).
Institutional approval
All studies utilized deidentified plasma samples using protocols and procedures reviewed and approved by the University of Louisville Human Subjects Protection Program Office (studies 177.07 and 608.03).
Sample preparation
IGM, C3, C4, and CRP were purchased as solutions in buffer, lyophilized to dryness, and then reconstituted in a smaller volume of ultrapure water (18.2 MΩ-cm) to yield a concentration suitable for DSC. PRE, A2M, CER, and ACT were purchased as powders lyophilized from buffer and were reconstituted with ultrapure water. HSA, IGG, IGA, AAG, AAT, FIB, TRF, and HPT were reconstituted with 10 mM potassium phosphate, 150 mM NaCl, pH 7.5. Reference serum was reconstituted according to the guidelines. Pure proteins and reference serum were dialyzed for 24 h at 4°C against 10 mM potassium phosphate, 150 mM NaCl, pH 7.5, to ensure complete solvent exchange. Pure protein samples were diluted with dialysate to a concentration suitable for DSC. Reference serum was diluted 25-fold with the dialysate. Plasma samples (100 μl) were dialyzed for 24 h at 4°C against 10 mM potassium phosphate, 150 mM NaCl, 0.38% (w/v) sodium citrate, pH 7.5, to ensure complete solvent exchange and then diluted 25-fold with the same buffer. All samples (0.45 μm, cellulose acetate or polyethersulfone) and buffers (0.22 μm, polyethersulfone) were filtered before use. Pure protein concentrations were quantified spectrophotometrically using the following extinction coefficients (ɛ280; liter−1·g·cm−1): HSA, 0.53; IGG, 1.38; IGA, 1.32; AAG, 0.89; AAT, 0.53; FIB, 1.55; TRF, 1.12; HPT, 1.2; IGM, 1.18; ACT, 0.62; C3, 0.97; C4, 0.92; CER, 1.49; A2M, 0.893; PRE, 1.41; CRP, 1.95.
DSC protocol
Scans were performed using an automated capillary DSC (MicroCal, Northampton, MA). Samples and dialysate were stored in 96-well plates at 5°C until being loaded into the calorimeter by the robotic attachment. Scans were recorded from 20°C to 110°C at 1°C/min using the mid feedback mode, a filtering period of 2 s, and with a prescan thermostat of 15 min. Data were analyzed using Origin 7.0. Sample scans were first corrected for the instrument baseline by subtracting an appropriate buffer scan. Nonzero baselines were then corrected by applying a linear baseline fit. Scans were finally normalized for the gram concentration of protein. For the pure protein samples, protein concentrations were determined spectrophotometrically as outlined above. Total protein concentrations of the reference serum and plasma samples were measured by the bicinchoninic acid method (Pierce, Rockford, IL). Thermograms were plotted as excess specific heat capacity (cal/°C·g) versus temperature.
Clinical assay methods
Protein electrophoresis was performed on agarose gels using the SPIFE 3000 and scanned with the QUICKSCAN 2000 (Helena Laboratories, Beaumont, TX). Total protein was measured by the biuret method on the Ortho Vitros 950 (Vitros) (Ortho-Clinical Diagnostic, Rochester, NY) chemistry analyzer. Albumin was measured on the Vitros by the bromocresol green dye binding assay or by an immunoturbidometric assay on the Cobas Integra 800 (Roche, Indianapolis, IN). Albumin concentrations were also determined from the fraction percentage on the protein electrophoresis assay along with the total protein concentration. Specific serum proteins (IGG, IGA, TRF, HPT, IGM, C3, C4, PRE, CRP) were measured by immunoturbidimetry on the Integra.
Column depletion experiments
Reference serum was depleted of HSA using the SwellGel Blue albumin removal kit with some minor modifications to the manufacturer's protocol (Pierce, Rockford, IL). The serum sample was diluted 10-fold into 10 mM potassium phosphate, pH 7.5, to achieve salt conditions and albumin concentrations required for good column binding. Diluted serum (200 μl) was applied to a column containing two SwellGel disks. An HSA-depleted fraction was obtained following the standard protocol. A single 200-μl volume of the supplied binding/wash buffer was used to obtain a wash fraction. Finally, an eluted HSA fraction was obtained from a single 200-μl addition of the supplied elution buffer. To obtain a greater volume of each fraction for subsequent experiments, multiple columns were run using an identical protocol, and all of the fractions were pooled. Fractions for DSC analysis were dialyzed for 24 h at 4°C against 10 mM potassium phosphate, 150 mM NaCl, pH 7.5, and diluted as necessary with dialysate. DSC scans were performed on an N-DSC II instrument (Calorimetry Sciences, Provo, UT) from 20°C to 110°C at 1°C/min with a prescan equilibration time of 10 min. Data were analyzed using Origin 7.0 as described above.
RESULTS AND DISCUSSION
Thermogram for normal plasma
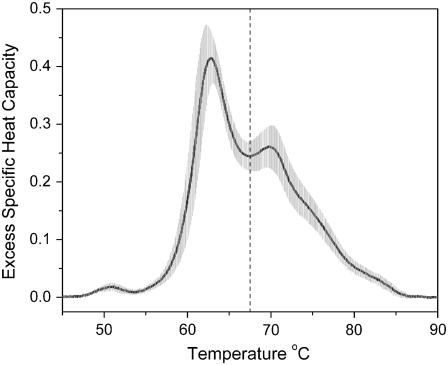
Fig. 1 shows an average thermogram obtained from plasma samples from 15 normal individuals. The thermogram displays multiple peaks and shoulders yet is surprisingly simple, given the complexity of the plasma proteome. The standard deviation of the data (shaded area in Fig. 1) is low and is comparable to the range in values observed in normal individuals for the concentrations of individual plasma proteins (12). Human serum albumin, for example, has a normal reference range of ∼35–55 g/liter, dependent on age and gender (12). The average normal thermogram in Fig. 1 shows clear peaks at 50.8°, 62.8°, and 69.8°C. The area under the thermogram is 5.02 ± 0.23 cal g−1 and defines the specific enthalpy for the denaturation of normal plasma over the range 45–90°C. The first moment of the thermogram with respect to the temperature axis is 67.4 ± 0.8°C. The sample size used in these studies is appropriate for exploratory preclinical studies and, indeed, is on a par with the numbers expected for a Phase I clinical trial (29), although that is not our current aim.
FIGURE 1.
DSC thermogram of normal plasma. The solid line is the average of 15 thermograms of plasma samples from 15 individuals (9 male, 6 female; ages 22–50). The shaded area is the standard deviation at each temperature. The vertical dashed line is the first moment of the thermogram at 67.4°C.
The normal plasma thermogram is the weighted sum of the denaturation of individual plasma proteins
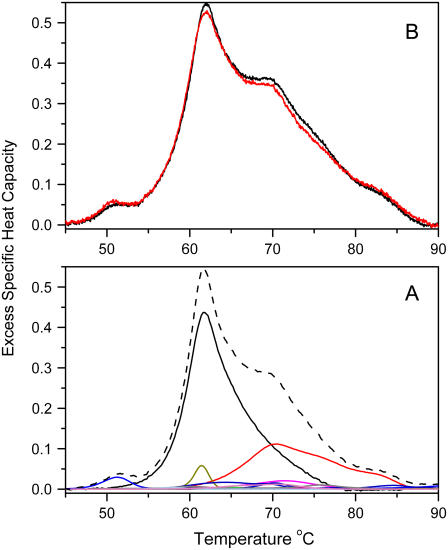
We hypothesize that the thermogram seen in Fig. 1 arises from the denaturation of the individual proteins within plasma and represents the sum of individual protein denaturation reactions weighted according to their concentrations within plasma. We tested this hypothesis in two ways, as shown in Fig. 2. First, individual thermograms for the denaturation of the 16 most abundant plasma proteins were determined (Supplementary Fig. 1). These thermograms display a range of denaturation temperatures and differences in the complexities of their denaturation reactions. Many of these thermograms show multiple peaks, indicative of complex denaturation reactions, whereas other thermograms are consistent with simple two-state melting behavior. Fig. 2 A shows the calculated plasma thermogram obtained by simple summation of the individual thermograms for the 16 most abundant plasma proteins after their contributions were weighted according to their known average concentrations in normal plasma (12). A tacit assumption in this exercise is that there are no interactions among these proteins that might alter their thermal denaturation. The resultant shape of the calculated thermogram mimics that of the experimental one seen in Fig. 1, in support of our hypothesis. As a second test, mixtures of pure individual plasma proteins were prepared, and their thermograms determined by DSC (Fig. 2 B). A mixture containing the 16 most abundant plasma proteins at their average concentrations found in normal plasma yields a thermogram whose shape mimics that of actual plasma (Fig. 2 B). A mixture with only the four major components (HSA, IgG, fibrinogen, and transferrin) yields a thermogram that closely matches the observed normal but lacks subtle features (Fig. 2 B). The data in Fig. 2 show that the normal thermogram is dominated by contributions from those four proteins. The small peak at 50.8°C can be unambiguously assigned to a transition in fibrinogen. The major peak at 62.8°C primarily reflects the denaturation of unligated HSA, with a contribution from haptoglobin. The peak at 69.8°C and the shoulders at higher temperature arise primarily from IgG.
FIGURE 2.
Origin of the plasma thermogram. (A) Calculated thermogram (dashed line) obtained from the sum of the weighted contributions of the 16 most abundant plasma proteins. Thermograms for the individual pure plasma proteins are shown in Fig. 3 of the Supplementary Material). (B) Thermograms obtained from mixtures of pure plasma proteins, mixed at concentrations that mimic their known average concentrations in normal plasma. The red curve is a mixture of HSA, IgG, fibrinogen, and transferrin. The black curve is a mixture of the 16 most abundant plasma proteins.
Thermograms of HSA-depleted serum
Supplementary Fig. 2 shows the results from experiments in which albumin was removed from serum by affinity chromatography. (Serum differs from plasma primarily by the absence of fibrinogen, which is removed when plasma is allowed to clot.) Supplementary Fig. 2 A shows the expected thermogram obtained by calculating the weighted sum of the most abundant proteins minus HSA and fibrinogen. Supplementary Fig. 2 B shows the observed experimental thermogram for albumin-depleted serum. The agreement between the shape of the calculated and observed thermograms is excellent. Apart from confirming the major contribution by HSA to the peak at 62.8°C in plasma thermograms, these data show that the contributions of other plasma proteins to thermograms can be amplified for more detailed study.
Plasma thermograms from diseased individuals differ dramatically from those of normal individuals
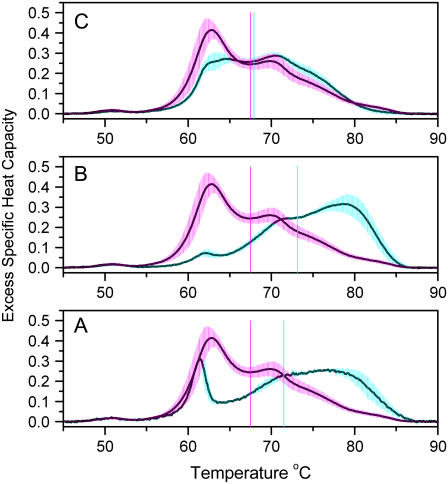
Fig. 3 compares average thermograms for individuals with three different diseases (rheumatoid arthritis, Lyme disease, systemic lupus) with the average normal thermogram. In all cases, dramatic differences from normal are evident. In addition, each disease seems to display a signature thermogram that differs from those of other diseases. In all cases, the 62.8°C peak associated with HSA is greatly diminished, and the thermograms are shifted to higher temperatures. Fig. 3 A shows the thermogram for lupus. The first moment shifts from the normal value of 67.5°C to a value of 71.5°C. A sharp peak near 61°C is evident that would be consistent with an elevation in haptoglobin concentration. The thermogram for Lyme disease (Fig. 3 B) is distinct from that seen for systemic lupus. The first moment at 73.15°C is higher still, and the shape of the thermogram clearly differs from both normal and lupus thermograms. Fig. 3 C shows yet another distinctive thermogram for individuals suffering from rheumatoid arthritis. That thermogram is characterized by a first moment of 67.9°C, only slightly higher than normal, but with distinct changes in the shape relative to normal that are well beyond the standard deviations in the two thermograms. These collective results indicate the potential of DSC as a clinical diagnostic tool, providing a unique window into the plasma proteome. Thermograms can at a glance distinguish diseased states from normal and can potentially provide signatures for specific diseases. The latter possibility, of course, can not be confirmed until a more systematic and comprehensive study of multiple diseases is undertaken. The sample sizes used in these studies conform to the accepted standards for exploratory preclinical studies (29).
FIGURE 3.
Thermograms of plasma from diseased individuals. Each panel compares normal plasma (magenta shading) with diseased plasma (cyan shading). The average thermogram is shown as a solid line; the shading indicates the standard deviation. (A) Plasma from individuals with systemic lupus. The average was obtained from four thermograms, duplicate DSC runs on samples from two diseased individuals. (B) Plasma from individuals with Lyme disease. The average was obtained from eight thermograms, duplicate DSC runs on samples from four diseased individuals. (C) Plasma from individuals with rheumatoid arthritis. The average was obtained from 10 thermograms, duplicate DSC runs on samples from five diseased individuals. Each panel shows the first moment of the thermogram as vertical solid lines—magenta for normal, and cyan for the disease.
Origin of the altered thermograms
What causes the dramatic alterations in thermograms seen in Fig. 3? One possibility is that the concentrations of the major proteins in plasma are changed. This possibility was tested by experiments shown as Supplementary Material that indicate that such is not the case. Supplementary Fig. 3 shows the concentrations of the major plasma proteins for the same samples shown in Fig. 3. The data show that the protein composition of plasma from diseased individuals is in most case indistinguishable from normal concentration values. Plasma from lupus patients represents a slight exception, with samples showing elevated concentrations of haptoglobin, IgA, and IgM. Notably, albumin concentrations are normal for all of the diseased states, even though the thermogram peak at 62.8°C that is characteristic of albumin is absent or greatly diminished in diseased samples (Fig. 3). Supplementary Fig. 4 shows protein electrophoresis patterns for normal plasma and the diseased states. Only subtle variations can be seen when these traces are compared, in contrast to the dramatic shifts in thermograms seen in Fig. 3. These data reveal a distinct advantage of our calorimetric approach. Although whatever is present in plasma in the diseased state that differentiates samples from normal does not seem to drastically alter the concentrations or the sizes and charges of the plasma proteins (as revealed by electrophoresis), it does exert dramatic effects on the thermal properties of the proteins.
The most likely explanation for the shifts in the thermograms in Fig. 3 is that they result from binding interactions that involve the most abundant plasma proteins, particularly albumin. This view is consistent with the “interactome” hypothesis, which suggests that peptide and protein biomarkers specific for a particular disease are not free in plasma but rather are bound to albumin or the immunoglobins. Such binding would result in thermal stabilization of the protein to which the biomarkers are bound and a drastic alteration of the plasma thermogram with respect to normal. That is exactly what is seen in Fig. 3. A definitive test of this model is prohibitively difficult at this point. In principle, one could isolate the ligated proteins from diseased plasma, wash off the bound biomarkers, and purify them. These purified biomarkers could then be added to normal plasma to demonstrate the characteristic alteration in the thermogram. Similar laborious fractionation procedures are routinely attempted in mass spectrometry studies that attempt to probe the plasma “interactome” in search of biomarkers, but on a preparative scale that yields amounts of material that are orders of magnitude lower than would be needed for calorimetric studies.
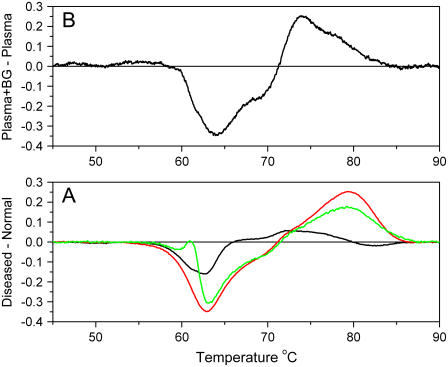
To test the hypothesis that shifted thermograms result from interactions, we designed an alternative demonstration. Bromocresol green is known to bind to Site I of HSA with a binding constant of 7 × 105 M−1 (30). Its binding to HSA within normal plasma ought to mimic the effects of putative biomarker binding. Fig. 4 shows the results of the experiment. Fig. 4 A shows difference thermograms for diseased states, obtained by subtracting the normal thermograms from the average thermograms seen in Fig. 3. These difference plots feature a negative peak near 62°C, attributable to a shift in HSA denaturation to higher temperatures. Positive difference peaks are evident at 70°C and higher, attributable to denaturation of ligated HSA (or other proteins). Such behavior can be mimicked by addition of bromocresol green (Fig. 4 B). Fig. 4 B shows a difference thermogram calculated from normal plasma samples with and without added bromocresol green. (More details of experiments showing the effects of bromocresol green on plasma and pure HSA are shown in Supplementary Fig. 5.) The shape of the difference thermogram is qualitatively similar to those seen for diseased plasma samples, suggesting that the “interactome” hypothesis has merit and provides a plausible explanation for shifts in thermograms observed in Fig. 3.
FIGURE 4.
Difference plots of plasma samples. (A) The differences between the average thermograms of diseased plasma and normal are shown: lupus (green), Lyme disease (red), and arthritis (black). (B) Difference in thermograms between normal plasma and normal plasma to which bromocresol green was added to a final concentration of 686 μM.
The shifts in denaturation transition curves that accompany ligand binding to protein are well understood and have been explained by a number of specific statistical mechanical and thermodynamic models (24,28). The effects of binding on the magnitude and exact shape of a melting transition curve depend precisely on the ligand binding affinity, enthalpy, and stoichiometry. Complex multiphasic transition curves can result from partial saturation. It is easy to imagine that peptide biomarkers in plasma might produce a myriad of thermogram shapes, depending on the exact proteins (and protein binding sites) that they occupy and their affinities. The interactions of multiple unique biomarkers with different plasma proteins could produce unique, characteristic thermograms that reflect the underlying complexity of the interactions. Although calorimetry may not sense signals arising from the denaturation of the biomarkers themselves, it would be uniquely sensitive to interactions of these biomarkers with the more abundant plasma proteins.
CONCLUSIONS
DSC provides a new window into the plasma proteome. Thermograms obtained for plasma samples from normal individuals yield a reproducible, characteristic signature that represents the weighted sum of denaturation profiles of the constituent proteins. Thermograms of plasma from diseased individuals differ dramatically from normal, and from one another, for the three diseases examined (arthritis, Lyme disease, and lupus). The radically different thermograms seen for diseased states are not consequences of major changes in the concentrations of the major constituent plasma proteins but, rather, seem to arise from binding interactions involving as yet unknown biomarkers with the more abundant plasma proteins, particularly albumin. Such behavior is consistent with the “interactome” hypothesis. These results portend a novel application of calorimetry as a clinical diagnostic tool.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
This work was supported by a subcontract awarded to J.B.C. from National Cancer Institute grant R44 CA103437 and by a grant to J.B.C. from the Elsa U. Pardee Foundation.
Editor: Kathleen B. Hall.
References
- 1.Anderson, N. L., and N. G. Anderson. 2002. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 1:845–867. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, N. L., M. Polanski, R. Pieper, T. Gatlin, R. S. Tirumalai, T. P. Conrads, T. D. Veenstra, J. N. Adkins, J. G. Pounds, R. Fagan, and A. Lobley. 2004. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol. Cell. Proteomics. 3:311–326. [DOI] [PubMed] [Google Scholar]
- 3.Omenn, G. S., D. J. States, M. Adamski, T. W. Blackwell, R. Menon, H. Hermjakob, R. Apweiler, B. B. Haab, R. J. Simpson, J. S. Eddes, E. A. Kapp, R. L. Moritz, D. W. Chan, A. J. Rai, A. Admon, R. Aebersold, J. Eng, W. S. Hancock, S. A. Hefta, H. Meyer, Y. K. Paik, J. S. Yoo, P. Ping, J. Pounds, J. Adkins, X. Qian, R. Wang, V. Wasinger, C. Y. Wu, X. Zhao, R. Zeng, A. Archakov, A. Tsugita, I. Beer, A. Pandey, M. Pisano, P. Andrews, H. Tammen, D. W. Speicher, and S. M. Hanash. 2005. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 5:3226–3245. [DOI] [PubMed] [Google Scholar]
- 4.Aebersold, R., L. Anderson, R. Caprioli, B. Druker, L. Hartwell, and R. Smith. 2005. Perspective: a program to improve protein biomarker discovery for cancer. J. Proteome Res. 4:1104–1109. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, N. L. 2005. The roles of multiple proteomic platforms in a pipeline for new diagnostics. Mol. Cell. Proteomics. 4:1441–1444. [DOI] [PubMed] [Google Scholar]
- 6.Ebert, M. P., M. Korc, P. Malfertheiner, and C. Rocken. 2006. Advances, challenges, and limitations in serum-proteome-based cancer diagnosis. J. Proteome Res. 5:19–25. [DOI] [PubMed] [Google Scholar]
- 7.Mor, G., I. Visintin, Y. Lai, H. Zhao, P. Schwartz, T. Rutherford, L. Yue, P. Bray-Ward, and D. C. Ward. 2005. Serum protein markers for early detection of ovarian cancer. Proc. Natl. Acad. Sci. USA. 102:7677–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenblatt, K. P., P. Bryant-Greenwood, J. K. Killian, A. Mehta, D. Geho, V. Espina, E. F. Petricoin 3rd, and L. A. Liotta. 2004. Serum proteomics in cancer diagnosis and management. Annu. Rev. Med. 55:97–112. [DOI] [PubMed] [Google Scholar]
- 9.Wulfkuhle, J. D., L. A. Liotta, and E. F. Petricoin. 2003. Proteomic applications for the early detection of cancer. Nat. Rev. Cancer. 3:267–275. [DOI] [PubMed] [Google Scholar]
- 10.Wulfkuhle, J. D., C. P. Paweletz, P. S. Steeg, E. F. Petricoin 3rd, and L. Liotta. 2003. Proteomic approaches to the diagnosis, treatment, and monitoring of cancer. Adv. Exp. Med. Biol. 532:59–68. [DOI] [PubMed] [Google Scholar]
- 11.O'Connell, T. X., T. J. Horita, and B. Kasravi. 2005. Understanding and interpreting serum protein electrophoresis. Am. Fam. Physician. 71:105–112. [PubMed] [Google Scholar]
- 12.Craig, W., T. Ledue, and R. Ritchie. 2004. Plasma Proteins: Clinical Utility and Interpretation. Foundation for Blood Research, Scarborough, ME.
- 13.Anderson, L., and N. G. Anderson. 1977. High resolution two-dimensional electrophoresis of human plasma proteins. Proc. Natl. Acad. Sci. USA. 74:5421–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson, N. L., and N. G. Anderson. 1991. A two-dimensional gel database of human plasma proteins. Electrophoresis. 12:883–906. [DOI] [PubMed] [Google Scholar]
- 15.Gygi, S. P., and R. Aebersold. 2000. Mass spectrometry and proteomics. Curr. Opin. Chem. Biol. 4:489–494. [DOI] [PubMed] [Google Scholar]
- 16.Liotta, L. A., E. C. Kohn, and E. F. Petricoin. 2001. Clinical proteomics: personalized molecular medicine. JAMA. 286:2211–2214. [DOI] [PubMed] [Google Scholar]
- 17.Yates 3rd, J. R. 2000. Mass spectrometry. From genomics to proteomics. Trends Genet. 16:5–8. [DOI] [PubMed] [Google Scholar]
- 18.Adkins, J. N., S. M. Varnum, K. J. Auberry, R. J. Moore, N. H. Angell, R. D. Smith, D. L. Springer, and J. G. Pounds. 2002. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics. 1:947–955. [DOI] [PubMed] [Google Scholar]
- 19.Liotta, L. A., and E. F. Petricoin. 2006. Serum peptidome for cancer detection: spinning biologic trash into diagnostic gold. J. Clin. Invest. 116:26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liotta, L. A., M. Ferrari, and E. Petricoin. 2003. Clinical proteomics: written in blood. Nature. 425:905. [DOI] [PubMed] [Google Scholar]
- 21.Zhou, M., D. A. Lucas, K. C. Chan, H. J. Issaq, E. F. Petricoin 3rd, L. A. Liotta, T. D. Veenstra, and T. P. Conrads. 2004. An investigation into the human serum “interactome.” Electrophoresis. 25:1289–1298. [DOI] [PubMed] [Google Scholar]
- 22.Fenby, D. V. 1987. Heat: its measurment from Balileo to Lavosier. Pure Appl. Chem. 59:91–100. [Google Scholar]
- 23.Biltonen, R. L., and E. Freire. 1978. Thermodynamic characterization of conformational states of biological macromolecules using differential scanning calorimetry. CRC Crit. Rev. Biochem. 5:85–124. [DOI] [PubMed] [Google Scholar]
- 24.Brandts, J. F., and L. N. Lin. 1990. Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry. 29:6927–6940. [DOI] [PubMed] [Google Scholar]
- 25.Bruylants, G., J. Wouters, and C. Michaux. 2005. Differential scanning calorimetry in life science: thermodynamics, stability, molecular recognition and application in drug design. Curr. Med. Chem. 12:2011–2020. [DOI] [PubMed] [Google Scholar]
- 26.Freire, E. 1995. Differential scanning calorimetry. Methods Mol. Biol. 40:191–218. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Ruiz, J. M. 1995. Differential scanning calorimetry of proteins. Subcell. Biochem. 24:133–176. [DOI] [PubMed] [Google Scholar]
- 28.Schellman, J. A. 1958. The factors affecting the stability of hydrogen-bonded polypeptide structures in solution. J. Phys. Chem. 62:1485–1494. [Google Scholar]
- 29.Motulsky, H. 1995. Intuitive Biostatistics. Oxford University Press, New York.
- 30.Peters, T., Jr. 1996. All About Albumin: Biochemistry, Genetics and Medical Applications. Academic Press, San Diego.