Abstract
Salt plays a critical role in the physiological activities of cells. We show that ionic strength significantly affects the kinetics of noncovalent interactions in protein channels, as observed in stochastic studies of the transfer of various analytes through pores of wild-type and mutant α-hemolysin proteins. As the ionic strength increased, the association rate constant of electrostatic interactions was accelerated, whereas those of both hydrophobic and aromatic interactions were retarded. Dramatic decreases in the dissociation rate constants, and thus increases in the overall reaction formation constants, were observed for all noncovalent interactions studied. The results suggest that with the increase of salt concentration, the streaming potentials for all the protein pores decrease, whereas the preferential selectivities of the pores for either cations or anions drop. Furthermore, results also show that the salt effect on the rate of association of analytes to a pore differs significantly depending on the nature of the noncovalent interactions occurring in the protein channel. In addition to providing new insights into the nature of analyte-protein pore interactions, the salt-dependence of noncovalent interactions in protein pores observed provides a useful means to greatly enhance the sensitivity of the nanopore, which may find useful application in stochastic sensing.
INTRODUCTION
Stochastic sensing can detect analytes at the single-molecule level, in which a single transmembrane protein channel (or pore), most often the heptameric α-hemolysin (αHL), is embedded in a planar lipid bilayer (1). Stochastic sensing is achieved by monitoring the ionic current flowing through the single pore at a fixed applied potential. Individual binding events are detected as transient blockades in the recorded current. This approach shows both the concentration and the identity of an analyte: the former from the frequency of occurrence (1/τon) of the binding events and the latter by its characteristic current signature, typically the dwell time (τoff) of the analyte coupled with the extent of current block (amplitude) it creates.
The molecular transport and binding kinetics inside a channel are strongly dependent on the nature of the pore, the ion species passing through the channel, as well as the experimental conditions (e.g., pH, voltage, temperature, etc.) (2–5). Thus, the performance (e.g., resolution and sensitivity) of stochastic detection relies on the structural characteristics of the protein pore and the working conditions for the experiment. In an effort to develop improved nanopore sensor sensitivity, several critical issues can be addressed. These considerations include structural modification of the αHL and the regulation of ions on the performance of the protein pore. The former, in principle, provides a natural ground for designing single-molecule stochastic sensor, whereas the latter involves a combination of parameters that affect mass transport and possibly other dynamic factors associated with channels, for instance, pH, temperature, ionic strength, and applied potential.
It has been reported that mutating the amino acid residue methionine at position 113 of the wild-type αHL ((WT)7) protein to asparagine, tryptophan, tyrosine, or phenylalanine results in significantly longer dwell times for the noncovalent molecular adaptor β-cyclodextrin (βCD) inside the lumen of the mutant αHL (6). Additionally, a wide variety of substances have been identified and quantified by using properly engineered versions of the transmembrane protein αHL pore based on noncovalent (i.e., electrostatic, aromatic, and hydrophobic) interactions. These compounds include metal ions (7,8), anions (5), organic molecules (9,10), and proteins (11). It has also been shown that engineered αHL pores have the potential for use as a novel DNA sequencing technique (12–14).
In parallel to the research on structural modification of αHL, other fundamental studies to identify important parameters associated with channel transport and dynamic behaviors have also been investigated. For example, the binding of a molecule to an αHL pore can be enhanced by voltage-dependent electroosmosis (15). It was found that, if βCD was added to the trans compartment, with the increase of the applied potential, the association rate constant (kon) increased but the dissociation rate constants (koff) were almost unchanged for cation-selective pores. Conversely, kon decreased but koff increased for anion-selective pores (15). In terms of the interaction between the negatively charged phosphate and the (M113R)7 pore having a positively charged binding site, both kon and koff increased with an increase of applied potential (5). The temperature effect on stochastic sensing has also been investigated. It was reported that an increase in the experimental temperature would result in a larger open channel current, an increase in both kon and koff, and an overall smaller formation constant Kf (= kon/koff) (2).
Although bath concentration has been incorporated as an important parameter in the contribution of ion flux in simulation of channel properties (16,17), until now, relatively little work has been done on salt effects that may influence the noncovalent bonding interactions inside protein pores. It should be noted that salt effects play an important role in condensation of biomolecules (18), drug delivery (19), and chemical reactions involving ionic species (20). We report that salt effects significantly influence the association and dissociation rate constants as well as formation constants of the noncovalent interactions in αHL protein channels.
MATERIALS AND METHODS
Reagents
Peptide Y-P-F-F, and HIV-1 TAT protein peptide (TATp) were purchased from American Peptide (Sunnyvale, CA). The peptide Y-F-F, and organophosphates (2-hydroxy-adamantan-2-yl)-phosphonic acid diethyl ester (HAE) and adamantan-1-yl-phosphonic acid (APA) were obtained from Sigma (St. Louis, MO). All these analytes were dissolved in HPLC-grade water, and the concentrations of the stock solutions were 1 mM for TATp, 0.2 mM for both Y-P-F-F and Y-F-F, 5 mM for both HAE and APA, respectively. All other reagents were purchased from Sigma.
Preparation and formation of protein pore
Mutant αHL M113F and M113E genes were constructed by side-directed mutagenesis (Mutagenex, Piscataway, NJ) with a WT αHL gene (21) in a T7 vector (pT7-αHL). Wild-type αHL and mutant αHL monomers were first synthesized by coupled in vitro transcription and translation (IVTT) using the Escherichia coli T7 S30 Extract System for Circular DNA from Promega (Madison, WI). Subsequently, they were assembled into homoheptamers by adding rabbit red cell membranes and incubating for 1 h (22). The heptamers were purified by SDS-polyacrylamide gel electrophoresis and stored in aliquots at −80°C.
Planar bilayer recording
A bilayer of 1,2-diphytanoylphosphatidylcholine (Avanti Polar Lipids; Alabaster, AL) was formed on an aperture (150 μm) in a Teflon septum (25 μm thick; Goodfellow, Malvern, PA) that divided a planar bilayer chamber into two compartments, cis and trans. The formation of the bilayer was monitored by using a function generator (BK precision 4012A; Yorba Linda, CA). The experiments were carried out under a series of symmetrical buffer conditions with a 2.0 mL solution comprising NaCl (ranging from 1 to 5 M), and 10 mM Tris · HCl (pH 7.5) at 22 ± 1°C. Unless otherwise noted, the αHL protein was added to the cis compartment, which was connected to “ground”, whereas the analyte was added to the trans compartment. In such a way, after the single-channel formation, the mushroom cap of the αHL channel would be located in the cis compartment, whereas the β-barrel of the αHL would insert into the lipid bilayer and connect with the trans of the chamber device. The final concentration of the αHL protein was 0.2–2.0 ng/mL, whereas those of βCD, Y-P-F-F, and TATp were 40 μM, 200 nM, and 25 μM, respectively. The applied potential was +40 mV for both (WT)7 and (M113E)7 proteins, and +50 mV for (M113F)7 pore, unless otherwise noted. Currents were recorded with a patch clamp amplifier (Axopatch 200B, Molecular Devices, Sunnyvale, CA). They were low-pass filtered with a built-in four-pole Bessel filter at 2 kHz and sampled at 10 kHz by a computer equipped with a Digidata 1322 A/D converter (Molecular Devices).
Data analysis
Data were analyzed with the following software: pClamp 9.2 (Molecular Devices) and Origin 7.0 (MicroCal, Northampton, MA). Conductance values were obtained from the amplitude histograms after the peaks were fit to Gaussian functions. Mean residence times (τ-values) for the analytes were obtained from dwell time histograms by fitting the distributions to single exponential functions by the Levenberg-Marquardt procedure. Kinetic constants were calculated by using koff = 1/τoff, kon =1/(Cτon), and Kf = kon/koff, where C is the concentration of the analyte.
The charge selectivity of a protein pore or the permeability ratios (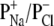 ) can be calculated by using the Goldman-Hodgkin-Katz equation (23): where αM is the activity of ion M, subscripts c and t represent cis and trans sides of the bilayer, Vr is the reversal potential, F the Faraday constant, and R the gas constant. For the measurement of reversal potential Vr, single-channel currents were recorded under asymmetric conditions with a 2.0 mL solution comprising NaCl (ranging from 1 to 5 M), 10 mM Tris · HCl (pH 7.5) (cis), and a 2.0 mL solution comprising NaCl (0.2 M), 10 mM Tris · HCl (pH 7.5) (trans). Vr was obtained by a polynomial fit of the current-voltage (I/V) data near zero current.
) can be calculated by using the Goldman-Hodgkin-Katz equation (23): where αM is the activity of ion M, subscripts c and t represent cis and trans sides of the bilayer, Vr is the reversal potential, F the Faraday constant, and R the gas constant. For the measurement of reversal potential Vr, single-channel currents were recorded under asymmetric conditions with a 2.0 mL solution comprising NaCl (ranging from 1 to 5 M), 10 mM Tris · HCl (pH 7.5) (cis), and a 2.0 mL solution comprising NaCl (0.2 M), 10 mM Tris · HCl (pH 7.5) (trans). Vr was obtained by a polynomial fit of the current-voltage (I/V) data near zero current.
To obtain the streaming potentials of protein pores (24,25), single-channel current recording experiments were carried out under asymmetric conditions: the cis chamber compartment contained a 2.0 mL solution comprising NaCl (ranging from 1 to 5 M), 10 mM Tris · HCl (pH 7.5), whereas the trans compartment contained 2.0 mL of the same buffer solution plus 1 M urea. Streaming potential Δφ for the protein pore was obtained by linearly fitting the I/V curves, which were recorded from ±20 mV to ±120 mV.
RESULTS AND DISCUSSION
The noncovalent interactions examined with αHL pores
It is generally accepted that ionic strength affects the rates of chemical reactions, particularly those involving charged particles. Ion permeation, gating or blocking through protein channels or solid-state nanopores have been studied extensively under low to moderate buffer concentrations, for instance, with the concentration of NaCl or KCl <1 M (23,26–28). However, until now, as far as we aware, there have been no systematic studies of the effects of ionic strength on noncovalent interactions in nanopores, especially at high salt concentrations. In this work, we investigated the effect of ionic strength (with NaCl at concentrations from 1 to 5 M) on three types of noncovalent interactions, specifically, electrostatic, aromatic, and hydrophobic interactions, the interactions used most frequently in the design of protein pore-based stochastic sensors. For example, the (WT)7 αHL and the neutral molecule βCD with a hydrophobic cavity were used to examine the hydrophobic interaction; the mutant (M113E)7 pore and the positively charged HIV-1 TAT protein peptide (TATp) with a sequence of Y-G-R-K-K-R-R-Q-R-R-R (29) were chosen to study the electrostatic interaction; the engineered (M113F)7 pore and the aromatic peptide Y-P-F-F were selected to examine the aromatic interaction. It should be noted that the mutant (M113E)7 pore has an electrostatic interaction site (containing seven negatively charged Glu amino acid residues) for positively charged compounds (X. Guan, X. F. Kang, S. Cheley, and H. Bayley, unpublished data), whereas the (M113F)7 pore contains an aromatic binding site (consisting of seven aromatic Phe side chains) for aromatic molecules (9).
Kinetics of association and dissociation
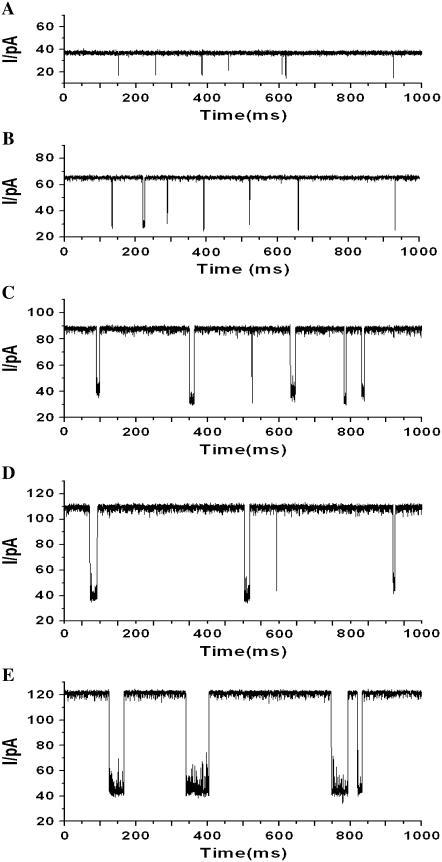
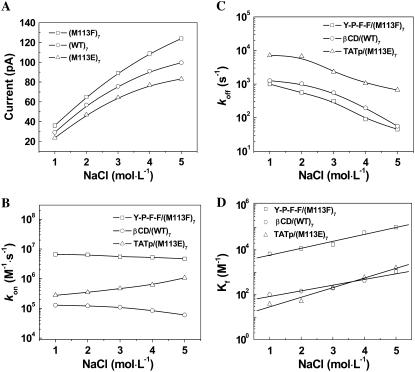
The typical electrical recordings with (M113F)7 pore and peptide Y-P-F-F, shown in Fig. 1, showed that both the open channel current and the mean dwell time of events increased with increasing concentration of NaCl added to the chamber compartments. Further data analysis (Fig. 2 A) showed that the open channel current did not increase linearly with the NaCl concentration. When the concentration of NaCl increased from 1 M to 5 M, the open channel conductance increased 3.4-fold (from 720 pS to 2480 pS), and the event residual conductance increased 2.0-fold (from 400 pS to 800 pS). Similar phenomena were also found in (WT)7 and (M113E)7 pore systems (Fig. 2 A, and Supplementary Material Figs. S1 and S2), where the open channel conductances increased 3.4-fold (from 728 pS to 2488 pS), and 3.5-fold (from 588 pS to 2078 pS), respectively, with the increase of NaCl concentration from 1 M to 5 M. It should be mentioned that a 3.2-fold (from 778 pS to 2478 pS, data not shown) conductance increase was observed for the (M113K)7 pore with the same NaCl solutions. The conductivity of the bulk solution increased 2.8-fold (from 83.4 pS × cm−1 to 236.4 pS × cm−1) with the increase of the concentration of NaCl from 1 M to 5 M (see Supplementary Material, Fig. S3). Therefore, the increased ratios of our experimental open channel conductances in the three different protein channels were in agreement with that of the measured conductivities in bulk solutions (note that conductance is proportional to the conductivity, i.e., G ∝ κ).
FIGURE 1.
Single-channel recordings showing the effect of ionic strength on Y-P-F-F binding to a (M113F)7 pore. The experiments were carried out at +50 mV (cis at ground) under a series of symmetrical buffer conditions with concentrations of NaCl ranging from 1 to 5 M, and 10 mM Tris · HCl (pH 7.5). (A) 1 M NaCl; (B) 2 M NaCl; (C) 3 M NaCl; (D) 4 M NaCl; and (E) 5 M NaCl.
FIGURE 2.
Dependence of the open channel currents of protein pores and the kinetic constants of the noncovalent interactions on the salt concentration. (A) Open channel currents; (B) association rate constants kon; (C) dissociation rate constants koff; and (D) reaction formation constants Kf.
The plots of kon, koff and Kf versus NaCl concentration are shown in Fig. 2 B–D, respectively. When the concentration of NaCl increased from 1 M to 5 M, the values of association rate constants kon for Y-P-F-F in (M113F)7 pore and for βCD in (WT)7 αHL decreased by 29% (from 6.59 × 106 M−1 × s−1 to 4.66 × 106 M−1 × s−1), and 53% (from 1.28 × 105 M−1 × s−1 to 6.02 × 104 M−1 × s −1), respectively, whereas that for positively charged TATp in (M113E)7 pore increased by 2.8-fold (from 2.77 × 105 M−1 × s−1 to 1.06 × 106 M−1 × s−1). However, unlike kon, the values of all the dissociation rate constants koff (=1/τoff) decreased for all the noncovalent interactions studied. For example, a 96% decrease (from 1.0 × 103 s−1 to 45.4 s−1) for Y-P-F-F in the (M113F)7 pore, a 96% reduction (from 1.27 × 103 s−1 to 56.2 s−1) for βCD in (WT)7 αHL, and a 91% decrease (from 7.14 × 103 s−1 to 6.67 × 102 s−1) for TATp in (M113E)7 pore were observed with increased salt concentration. The overall reaction formation constants Kf for all the noncovalent interactions examined increased with the increase of salt concentration added. For example, when the concentration of NaCl increased from 1 M to 5 M, Kf of βCD in the (WT)7 αHL pore increased 10.6-fold (from 1.01 × 102 M−1 to 1.07 × 103 M−1), that of Y-P-F-F in (M113F)7 pore increased 15.5-fold (from 6.59 × 103 M−1 to 1.02 × 105 M−1), whereas that of TATp in (M113E)7 increased 41.2-fold (from 38.7 M−1 to 1.60 × 103 M−1). From these results, we can conclude that the ionic strength affects koff more significantly than kon. Thus, the effect of ionic strength on the reaction formation constant (Kf) was due predominantly to the salt effect on koff, a result that is clearly different from the effect of voltage dependence (15).
To investigate whether the salt effect on the binding kinetics is dependent on the charge selectivity of the pore or determined by the noncovalent interactions inside the pore, a series of cross-check experiments were carried out, including the aromatic peptide Y-P-F-F in the cation-selective (M113E)7 pore; the hydrophobic compound βCD in the anion-selective (M113K)7 pore; and the positively charged TATp in weakly anion-selective (M113F)7 pore. It should be noted that, the general hydrophobic interactions occur in all these experiments, although some of the protein pores used have binding sites designed specifically for positively or negatively charged compounds. The results showed that both kon and koff for all these three experiments decreased with the increase of NaCl concentration in the buffer. As the salt concentration increased from 1 M to 5 M, kon decreased by 11% (from 8.82 × 106 M−1 × s−1 to 7.84 × 106 M−1 × s−1), by 66% (from 6.42 × 104 M−1 × s−1 to 2.20 × 104 M−1 × s−1), and by 77% (from 4.87 × 105 M−1 × s−1 to 1.10 × 105 M−1 × s−1), respectively, for the sequence noted above, whereas koff decreased by 94% (from 6.67 × 103 s−1 to 3.85 × 102 s−1), by 75% (from 9.10 × 102 s−1 to 2.32 × 102 s−1), and by 99% (from 1.92 × 103 s−1 to 13.0 s−1), respectively. These phenomena were similar to those of βCD in (WT)7, and Y-P-F-F in (M113F)7 pore described above, where a hydrophobic or aromatic interaction occurred, but different from that of TATp in (M113E)7 pore, in which an electrostatic interaction happened. Combined these results show that the salt effect on the association rate constant is dependent on the nature of the noncovalent interaction inside the pore, rather than on the charge selectivity of the pore.
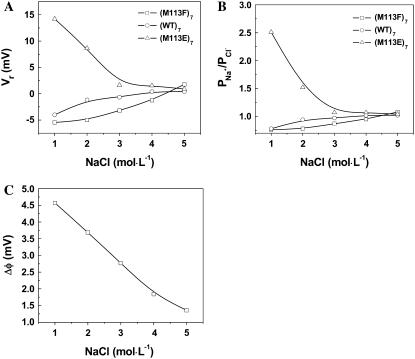
An increased transport of analyte and water coupled with ions inside a protein pore is assumed to be driven on application of the transmembrane field (30). This suggests that the binding of analytes toward the pore is affected by both the solvent flow and the diffusion of target molecules. Under a certain applied voltage, the diffusion coefficients of ions can be reflected from charge selectivity, which is dependent largely on the dimension of a pore and the spatial distribution of charges at the entrance to and within the lumen of the channel. In general, the narrower the pores are, the higher the charge selectivity of the channels (31). The charge selectivity of an ion channel can also be estimated from the ratio of the Debye length to the pore dimension. It was found that for small ratios, i.e., the pore size much greater than the Debye length, the channel would have low or no charge selectivity. In contrast, the pore showed high charge selectivity if the Debye length was comparable with or greater than the pore dimension (32). In the case of αHL pores, the narrowest internal diameter is ∼14 Å near Met-113. In 1 M NaCl solution, the Debye length of the αHL pore was 3.7 Å (see supplementary materials), which is equivalent to 26.4% of the αHL pore dimension. When the salt concentration increased to 5 M, the Debye length decreased from 3.7 Å to 1.3 Å, thus accounting for only 9.3% of the pore radius. This indicates that the charge selectivities or permeability ratios (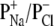 ) of the protein pores may be altered by the change of the ionic strength in the buffer solution. As shown in Fig. 3 A, with the increase of NaCl concentration, the reversal potentials of weakly anion selective pores ((M113F)7 and (WT)7) increased, whereas those of the cation selective (M113E)7 pore decreased, so that the preferential charge selectivities of the pores for either cations or anions dropped. As shown in Fig. 3 B, with the increase of NaCl concentration from 1 M to 5 M, the charge selectivity for aromatic (M113F)7 pore increased from 0.76 to 1.08, that of the hydrophobic (WT)7 pore increased from 0.78 to 1.02, whereas the ion selectivity for the charged (M113E)7 pore decreased from 2.51 to 1.04. These results suggest that ionic strength significantly influences the permeability ratio (
) of the protein pores may be altered by the change of the ionic strength in the buffer solution. As shown in Fig. 3 A, with the increase of NaCl concentration, the reversal potentials of weakly anion selective pores ((M113F)7 and (WT)7) increased, whereas those of the cation selective (M113E)7 pore decreased, so that the preferential charge selectivities of the pores for either cations or anions dropped. As shown in Fig. 3 B, with the increase of NaCl concentration from 1 M to 5 M, the charge selectivity for aromatic (M113F)7 pore increased from 0.76 to 1.08, that of the hydrophobic (WT)7 pore increased from 0.78 to 1.02, whereas the ion selectivity for the charged (M113E)7 pore decreased from 2.51 to 1.04. These results suggest that ionic strength significantly influences the permeability ratio (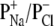 ) in protein pores so that all the tested protein channels (weakly anion selective or cation selective in 1 M NaCl) have no preferential charge selectivity to either Na+ or Cl− in 5 M NaCl. Furthermore, because an increase of NaCl concentration from 1 M to 5 M results in a more even transport of solvent by cations and anions, their effects on solvent flow essentially cancel one another, and thus streaming potentials will become smaller. This concept is clearly supported by our experimental results (Fig. 3 C).
) in protein pores so that all the tested protein channels (weakly anion selective or cation selective in 1 M NaCl) have no preferential charge selectivity to either Na+ or Cl− in 5 M NaCl. Furthermore, because an increase of NaCl concentration from 1 M to 5 M results in a more even transport of solvent by cations and anions, their effects on solvent flow essentially cancel one another, and thus streaming potentials will become smaller. This concept is clearly supported by our experimental results (Fig. 3 C).
FIGURE 3.
Effect of NaCl concentration on the (A) reversal potentials, (B) charge selectivities, and (C) streaming potentials of αHL pores, showing that with the increase of salt concentration, the streaming potentials for all the protein pores decrease, whereas the preferential selectivities of the pores for either cations or anions drop. The obtained values of streaming potentials for all the three protein pores were very similar at each salt concentration.
Application of salt dependence of noncovalent interactions
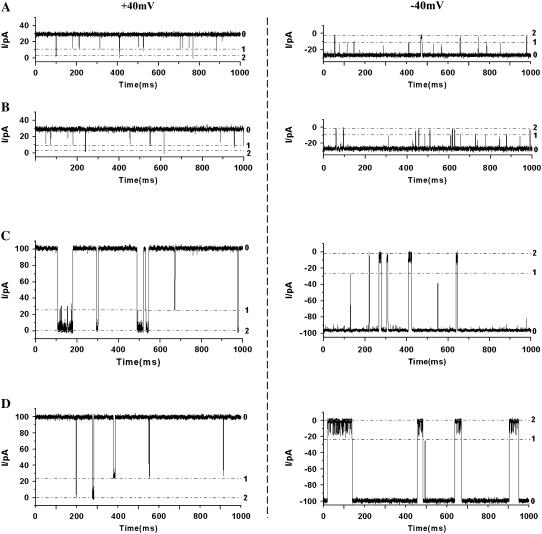
Our experimental results show that, with an increase in salt concentration, the dissociation rate constants decrease, whereas the formation constants Kf increase significantly, for all three noncovalent interactions in the protein pores. This suggests that, in general, the sensitivity or the resolution of the nanopore increases with increasing salt concentration. Therefore, the effect of ionic strength on noncovalent interactions may find useful application in nanopore stochastic sensing. To date, the engineered αHL protein pores have been used successfully as stochastic sensors for a variety of compounds, whereas the artificial pores have achieved only limited success (28). Artificial pore technology available currently provides a very poor resolution, which allows only large molecules such as DNA to be detected by using the electrophoretic effect to drive these molecules through the pore due to the lack of surface functionality (33), although a recent study showed that it was possible to impart solid-state nanopores with selectivity (34). The much improved salt dependence sensitivity discovered in this work may not only provide the potential for WT αHL and artificial pores to be used for the identification of small molecules, but may also permit use of engineered αHL for the differentiation, and possibly simultaneous detection, of a mixture of compounds. The latter analysis is difficult, or even sometimes impossible to achieve, in low salt concentration solutions. For example, it is well accepted that βCD can be used as an adaptor to sense organic molecules. When βCD is bound to the αHL pore, the channel is partially blocked and the open channel current drops. When a guest molecule is captured by βCD, and the resultant complex is bound to the pore, the current is further decreased (35). Our experiments (Fig. 4) showed that in 1 M NaCl solution, βCD bound to (WT)7 protein very weakly, with a dwell time of 0.79 ms at an applied +40 mV potential. With βCD binding to the pore, the channel current dropped from 26.5 pA to 8.8 pA. If a mixture of βCD and guest molecules organophosphate (2-hydroxy-adamantan-2-yl)-phosphonic acid diethyl ester (HAE) or a mixture of βCD and organophosphate APA was added to the (WT)7 αHL pore, the blocking event by the βCD · HAE or βCD · APA complex further reduced the current to 2.0 pA. The dwell times of βCD · HAE and βCD · APA were 1.4 ms and 0.85 ms, respectively at +40 mV, and 2.2 ms and 3.1 ms, respectively at −40 mV (Fig. 4). Although the identification of either HAE or APA could be achieved in 1 M NaCl due to the small difference in their dwell times, the sensitivity or the resolution would not allow accurate differentiation and quantification between them if a mixture of HAE and APA were present. However, in sharp contrast, in 5 M NaCl solution, βCD, βCD · HAE, and βCD · APA interacted with (WT)7 protein pore much more strongly (Fig. 4). For example, the formation constant Kf for αHL · βCD · HAE increased from 39 M−1 (in 1 M NaCl) to 598 M−1 (in 5 M NaCl), suggesting that βCD · HAE bound to (WT)7 αHL pore 15 times more strongly in 5 M NaCl than in 1 M NaCl. Furthermore, the effects of ionic strength on βCD · HAE and βCD · APA differed significantly, resulting in their quite different signatures, with dwell times of βCD · HAE and βCD · APA at 35.3 ms and 4.4 ms, respectively at +40 mV, as well as 7.7 ms and 60.7 ms, respectively at −40 mV, thus allowing the accurate differentiation and even quantification of these two different organophosphates.
FIGURE 4.
Single-channel recordings showing the application of salt effect for the differentiation of organophosphates HAE and APA in a (WT)7 pore using βCD as an adaptor. (Left) Traces recorded at +40 mV. (Right) Traces recorded at −40 mV. (A) HAE in 1 M NaCl; (B) APA in 1 M NaCl; (C) HAE in 5 M NaCl; and (D) APA in 5 M NaCl. The experiments were carried out at ±40 mV (cis at ground) under symmetrical buffer conditions with the concentration of NaCl at 1 M (or 5 M), and 10 mM Tris · HCl (pH 7.5). αHL protein was added in the cis compartment of the chamber, whereas 40 μM βCD and 80 μM HAE (or APA) were added in the trans compartment of the chamber. The dashed lines 0, 1, and 2 indicate the current levels for open channel, βCD · αHL, and HAE · βCD · αHL (or APA · βCD · αHL), respectively.
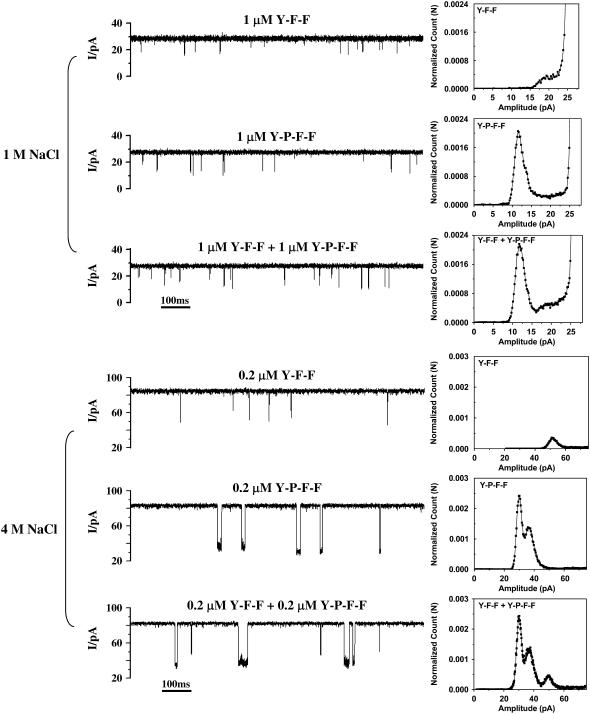
Another significant example is that the effect of ionic strength can be employed for the identification, and even simultaneous quantification, of structurally similar peptides. As shown in Fig. 5, the events of peptide Y-P-F-F in the (M113F)7 pore exhibited a wide range of amplitudes in 1 M NaCl. The amplitudes of the long events were in the range from 10 to 16 pA, whereas that of the short events were between 16–24 pA. The short events of Y-P-F-F had similar amplitudes to those of peptide Y-F-F (with a small shoulder at ∼20 pA). Therefore, if a mixture of Y-P-F-F and Y-F-F was present in the solution, Y-F-F might not be identified because the signature of Y-F-F was hidden by that of Y-P-F-F. This would not allow the simultaneous detection and quantification of these two peptides, though it was possible to identify and even quantify Y-P-F-F by considering only the long events. However, with the increase of the ionic strength, e.g., in 4 M NaCl solution, the aromatic interactions between peptides (i.e., Y-F-F and Y-P-F-F) and the (M113F)7 protein became much stronger, resulting in the significant difference between the signatures of Y-P-F-F and Y-F-F. As shown in Fig. 5, the events of Y-F-F showed a major amplitude at 52.3 pA, whereas those of Y-P-F-F had two amplitudes with a major peak at 28.8 pA and a shoulder at 35.3 pA. Thus, the separation of the events of Y-P-F-F and those of Y-F-F allowed their convenient discrimination and even concurrent quantification. It should be noted that the two different amplitude events observed for Y-P-F-F in 4 M NaCl solution may be attributed to the different directions in which Y-P-F-F entered the (M113F)7 pore. Our results show clearly that for a certain αHL pore (including both WT and mutants), regulation of ionic strength influences the sensitivity of stochastic sensing significantly, and thus can be used as a useful means for the detection and differentiation of various analytes that can not otherwise be achieved.
FIGURE 5.
Simultaneous detection of a mixture of two structure-related peptides in a (M113F)7 pore. (Left) Current traces. (Right) Event histograms. The experiments were carried out at +40 mV (cis at ground) under symmetrical buffer conditions with 1 M (or 4 M) NaCl, and 10 mM Tris · HCl (pH 7.5). αHL protein was added in the cis compartment, whereas 1 μM Y-P-F-F or 1 μM Y-F-F was added in the trans compartment of the chamber.
CONCLUSIONS
The work presented shows that the salt effect can be used as an effective approach to significantly increase the resolution and sensitivity of stochastic sensing. The salt dependence of noncovalent interactions in protein pores greatly enhances the potential sensor application of nanopores. For example, it may provide the potential for permitting more frequent use of WT αHL and artificial pores as stochastic sensing elements, and hence be used as an alternative approach to protein engineering. The salt dependence may also be combined with the modified pore, thus simplifying protein engineering because the development of a highly sensitive nanopore sensor is not required. These findings may have implications in studies of the fundamental functional properties of ion channels, e.g., to provide evidence that the noncovalent interactions occurring in a channel play a significant role in facilitating or retarding the translocation of molecules through the pore. In addition, the results of this investigation should stimulate additional theoretical studies and molecular dynamics simulations.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Dr. Richard B. Timmons (University of Texas at Arlington) for invaluable comments on the manuscript. The WT αHL plasmid was made in Dr. Hagan Bayley's laboratory at Texas A&M University.
Financial support for this work was provided by the Defense Advanced Research Projects Agency.
Editor: David S. Weiss.
References
- 1.Bayley, H., and P. S. Cremer. 2001. Stochastic sensors inspired by biology. Nature. 413:226–230. [DOI] [PubMed] [Google Scholar]
- 2.Kang, X. F., L. Q. Gu, S. Cheley, and H. Bayley. 2005. Single protein pores containing molecular adapter at high temperature. Angew. Chem. Int. Ed. 44:1495–1499. [DOI] [PubMed] [Google Scholar]
- 3.Jung, Y., H. Bayley, and L. Movileanu. 2006. Temperature-responsive protein pores. J. Am. Chem. Soc. 128:15332–15340. [DOI] [PubMed] [Google Scholar]
- 4.Kuyucak, S., and T. Bastug. 2003. Physics of ion channels. J. Biol. Phys. 29:429–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheley, S., L. Q. Gu, and H. Bayley. 2002. Stochastic sensing of nanomolar inositol 1,4,5-trisphosphate with an engineered pore. Chem. Biol. 9:829–838. [DOI] [PubMed] [Google Scholar]
- 6.Gu, L. Q., S. Cheley, and H. Bayley. 2001. Prolonged residence time of a noncovalent molecular adapter, β-cyclodextrin, within the lumen of mutant α-hemolysin pores. J. Gen. Physiol. 118:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braha, O., L. Q. Gu, L. Zhou, X. Lu, S. Cheley, and H. Bayley. 2000. Simultaneous stochastic sensing of divalent metal ions. Nat. Biotechnol. 18:1005–1007. [DOI] [PubMed] [Google Scholar]
- 8.Braha, O., B. Walker, S. Cheley, J. J. Kasianowicz, L. Song, J. E. Gouaux, and H. Bayley. 1997. Designed protein pores as components for biosensors. Chem. Biol. 4:497–505. [DOI] [PubMed] [Google Scholar]
- 9.Guan, X., L. Q. Gu, S. Cheley, O. Braha, and H. Bayley. 2005. Stochastic sensing of TNT with a genetically engineered pore. ChemBioChem. 6:1875–1881. [DOI] [PubMed] [Google Scholar]
- 10.Kang, X. F., S. Cheley, X. Guan, and H. Bayley. 2006. Stochastic detection of enantiomers. J. Am. Chem. Soc. 128:10684–10685. [DOI] [PubMed] [Google Scholar]
- 11.Movileanu, L., S. Howorka, O. Braha, and H. Bayley. 2000. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 18:1091–1095. [DOI] [PubMed] [Google Scholar]
- 12.Howorka, S., S. Cheley, and H. Bayley. 2001. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat. Biotechnol. 19:636–639. [DOI] [PubMed] [Google Scholar]
- 13.Ashkenasy, N., J. Sánchez-Quesada, H. Bayley, and M. R. Ghadiri. 2005. Recognizing a single base in an individual DNA strand: a step toward DNA sequencing in nanopores. Angew. Chem. Int. Ed. 44:1401–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee, M., and M. A. Burns. 2006. Nanopore sequencing technology: research trends and applications. Trends Biotechnol. 24:580–586. [DOI] [PubMed] [Google Scholar]
- 15.Gu, L. Q., S. Cheley, and H. Bayley. 2003. Electroosmotic enhancement of the binding of a neutral molecule to a transmembrane pore. Proc. Natl. Acad. Sci. USA. 100:15498–15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corry, B., S. Kuyucak, and S. H. Chung. 1999. Test of Poisson-Nernst-Planck theory in ion channels. J. Gen. Physiol. 114:597–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao, R., and N. R. Aluru. 2003. Ion concentrations and velocity profiles in nanochannel electroosmotic flows. J. Chem. Phys. 118:4692–4701. [Google Scholar]
- 18.Conwell, C. C., I. D. Vilfan, and N. V. Hud. 2003. Controlling the size of nanoscale toroidal DNA condensates with static curvature and ionic strength. Proc. Natl. Acad. Sci. USA. 100:9296–9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalia, Y. N., A. Naik, J. Garrison, and R. H. Guy. 2004. Iontophoretic drug delivery. Adv. Drug Deliv. Rev. 56:619–658. [DOI] [PubMed] [Google Scholar]
- 20.Tan, Z. J., and S. J. Chen. 2006. Ion-mediated nucleic acid helix-helix interactions. Biophys. J. 91:518–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song, L., M. R. Hobaugh, C. Shustak, S. Cheley, H. Bayley, and J. E. Gouaux. 1996. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 274:1859–1866. [DOI] [PubMed] [Google Scholar]
- 22.Cheley, S., O. Braha, X. Lu, S. Conlan, and H. Bayley. 1999. A functional protein pore with a “retro” transmembrane domain. Protein Sci. 8:1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hille, B. 2001. Ion Channels of Excitable Membranes, 3rd Ed. Sinauer, Sunderland, MA.
- 24.Levitt, D. G., S. R. Elias, and J. M. Hautman. 1978. Number of water molecules coupled to the transport of sodium, potassium and hydrogen ions via gramicidin nonactin or valinomycin. Biochim. Biophys. Acta. 512:436–451. [DOI] [PubMed] [Google Scholar]
- 25.Tripathi, S., and S. B. Hladky. 1998. Streaming potentials in gramicidin channels measured with ion-selective microelectrodes. Biophys. J. 74:2912–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nonner, W., D. P. Chen, and B. Eisenberg. 1998. Anomalous mole fraction effect, electrostatics, and binding in ionic channels. Biophys. J. 74:2327–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nadler, B., Z. Schuss, U. Hollerbach, and R. S. Eisenberg. 2004. Saturation of conductance in single ion channels: the blocking effect of the near reaction field. Phys. Rev. E. 70:051912.1–051912.11. [DOI] [PubMed] [Google Scholar]
- 28.Smeets, R. M. M., U. F. Keyser, D. Krapf, M. Y. Wu, N. H. Dekker, and C. Dekker. 2006. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 6:89–95. [DOI] [PubMed] [Google Scholar]
- 29.Torchilin, V. P., T. S. Levchenko, R. Rammohan, N. Volodina, B. Papahadjopoulos-Sternberg, and G. G. M. D'Souza. 2003. Cell transfection in vitro and in vivo with nontoxic TAT peptide-liposome-DNA complexes. Proc. Natl. Acad. Sci. USA. 100:1972–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer, W. R., and W. Nadler. 2006. Molecular transport through channels and pores: effects of in-channel interactions and blocking. Proc. Natl. Acad. Sci. USA. 103:11446–11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu, L. Q., M. Dalla Serra, J. B. Vincent, G. Vigh, S. Cheley, O. Braha, and H. Bayley. 2000. Reversal of charge selectivity in transmembrane protein pores by using noncovalent molecular adapters. Proc. Natl. Acad. Sci. USA. 97:3959–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kienker, P. K., and J. D. Lear. 1995. Charge selectivity of the designed uncharged peptide ion channel Ac-(LSSLLSL)3-CONH2. Biophys. J. 68:1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt, J. 2005. Stochastic sensors. J. Mater. Chem. 15:831–840. [Google Scholar]
- 34.Iqbal, S. M., D. Akin, and R. Bashir. 2007. Solid-state nanopore channels with DNA selectivity. Nature Nanotechnol. 2:243–248. [DOI] [PubMed] [Google Scholar]
- 35.Gu, L. Q., O. Braha, S. Conlan, S. Cheley, and H. Bayley. 1999. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature. 398:686–690. [DOI] [PubMed] [Google Scholar]