Abstract
The presence of one or more calcium-dependent ecto-ATPases (enzymes that hydrolyze extracellular 5′-triphosphates) in mammalian taste buds was first shown histochemically. Recent studies have established that dominant ecto-ATPases consist of enzymes now called nucleoside triphosphate diphosphohydrolases (NTPDases). Massively parallel signature sequencing (MPSS) from murine taste epithelium provided molecular evidence suggesting that NTPDase2 is the most likely member present in mouse taste papillae. Immunocytochemical and enzyme histochemical staining verified the presence of NTPDase2 associated with plasma membranes in a large number of cells within all mouse taste buds. To determine which of the three taste cell types expresses this enzyme, double label assays were performed using antisera directed against the glial glutamate/aspartate transporter, (GLAST), the transduction pathway proteins phospholipaseC β2 (PLCβ2) or the G protein subunit α-gustducin, and serotonin (5HT) as markers of type I, II or III taste cells, respectively. Analysis of the double labeled sections indicates that NTPDase2 immunoreactivity is found on cell processes that often envelop other taste cells, reminiscent of type I cells. In agreement with this observation, NTPDase2 was located to the same membrane as GLAST, indicating that this enzyme is present in type I cells. The presence of ecto-ATPase in taste buds likely reflects the importance of ATP as an intercellular signaling molecule in this system.
Keywords: Gustatory, ecto-ATPase, NTPDase2, taste cells, ATP signaling, mouse, taste bud
Taste buds, the sensory endorgans mediating taste, reside in the epithelium of the tongue, palate and larynx. In rodents, lingual taste buds occur in three types of papillae: fungiform papillae scattered over the anterior portion of the tongue, circumvallate papillae on the posterior dorsal surface, and foliate papillae along the posterior lateral surface. Taste buds also occur at the oral entrance to the nasoincisor ducts, soft palate, epiglottis and larynx.
Taste buds comprise several proliferative basal cells and 50–80 elongate epithelial cells which are heterogeneous in their morphologic and cytochemical characteristics. The differentiated taste cells in rodents are classified into three morphologic types: type I, type II, and type III as determined by cell structure, shape of the nucleus and apical processes (Murray and Murray, 1967; reviewed in Finger and Simon, 2000). These cell types usually correlate with particular histochemical features. For example, immunoreactivity for the G-protein subunit, a-gustducin, is localized to a subset of type II taste cells (Boughter et al., 1997; Yang et al., 2000b). This type of taste cell also expresses the transduction pathway proteins such as phospholipase C β2 and inositol 1,4,5-triphosphate receptor 3 (PLCβ2, IP3R3; Clapp et al., 2001) as well as the molecularly defined T1R (Hoon et al., 1999) and T2R taste receptors (Adler et al., 2000; Matsunami et al., 2000). In contrast, type III taste cells express none of the transduction-associated proteins but do exhibit several neuronal features including neural cell adhesion molecule (NCAM: Nelson and Finger, 1993), voltage-dependent calcium channels (Medler et al., 2003), synaptosomal-associated protein of 25kDa (SNAP-25: Yang et al., 2000a) and the ability to accumulate serotonin (Takeda et al., 1981; Yee et al., 2001). Though type I cells account for approximately half of the cells in each taste bud, this cell population is the least well characterized in terms of immunohistochemical markers but do express the glial glutamate/aspartate transporter GLAST (Lawton et al., 2000). The presence of this glial marker coupled with the observation that type I cells embrace the other cell types is strongly suggestive that these cells function like glia cells within taste buds.
The presence of calcium-dependent ecto-ATPases (enzymes that hydrolyze extracellular 5′-triphosphates such as ATP) in mammalian taste buds has long been known as demonstrated histochemically (Iwayama and Nada, 1967; Zalewski, 1968; Iwayama, 1969; Akisaka and Oda, 1977; Barry, 1992). Since these early reports, different families of these ecto-enzymes have been identified molecularly and functionally including the ecto-nucleoside triphosphate diphosphohydrolase (E-NTPDase) family (Zimmerman, 2001). Four members of this family, NTPDases 1, 2, 3, and 8 are anchored in the plasma membrane with catalytic sites facing the extracellular space and are implicated in the control of nucleotide levels at the cell surface (Zimmerman 2001; Bigonnesse et al., 2004; Kukulski et al., 2005). Conversely, NTPDases 4–7 mainly lie in the membranes of intracellular organelles and are not clearly involved in extracellular nucleotide signaling.
Previous studies showed gustatory nucleoside hydrolase activity with several nucleoside 5′-triphosphates (ATP, ITP, and GTP), but ATP was assumed to be the most relevant substrate based on its predominant abundance in physiological extracellular fluid (Barry, 1992). This activity most likely corresponds to NTPDases 1, 2, 3 or 8 based on their location and abilities to hydrolyze ATP: NTPDase1 hydrolyzes ATP and ADP equally well; NTPDase2 strongly prefers ATP; while NTPDase3 and NTPDase8 are functional intermediates between NTPDases 1 and 2 (Kukulski et al., 2005). The histochemical profile of taste bud ecto-ATPase reveals a strong preference for ATP over ADP (Iwayama, 1969), indicating that NTPDase2 is the most likely candidate.
The present study is aimed at molecularly identifying the predominant ecto-ATPase in taste buds, and characterizing which type of taste cell expresses this enzyme. We find that only NTPDase2 is highly abundant in taste buds and is associated with the plasma membrane of type I taste cells.
MATERIALS AND METHODS
Animals
Tissues of interest were obtained from either sex of C57B6 mice or transgenic GFP-gustducin mice on a C57B6 background in which the a-gustducin promoter drives expression of green fluorescent protein (GFP; Wong et al., 1999). The University of Colorado Health Sciences Center Institutional Animal Care and Use Committee approved the use of these animals for the following studies.
Massively Parallel Signature Sequencing (MPSS)
Total RNA was isolated from dissected taste buds or from lingual epithelium devoid of taste buds from C57B6 mice as previously described (LopezJimenez et al., 2005). Following DNAseI treatment, polyadenylated mRNA (poly (A)+) was isolated and used to generate cDNA libraries on microbeads using the Megaclone protocol (Brenner et al., 2000a). Sequence signatures were then generated by serial cutting and ligation of decoding adapters (Brenner et al., 2000b). Each signature was 17 bp in length and, due to the cloning protocol, generally located adjacent to the poly(A)+ proximal DpnII site. 2x106 and 4x106 signatures were sequenced respectively from the taste bud and lingual epithelium mRNA samples. The generation of the cDNA libraries and massively parallel signature sequencing (MPSS) analyses were performed by Solexa, Inc. (Hayward, CA). The inner ear libraries examined for comparisons were also generated by Solexa, Inc. using the same protocols. The GenBank accession numbers for the genes encoding NTPDase1, NTPDase2, NTPDase3, and NTPDase8 (genes are Entpd1, Entpd2, Entpd3 and, Entpd8) are NM_009848, NM_009849, NM_178676 and BC031143, respectively.
Enzyme histochemistry
Wild type mice (C57B6) were deeply anesthetized with 20% chloral hydrate (2.5 ml, intraperitoneally) and perfused transcardially with fixative consisting of 2% paraformaldehyde (PFA), 0.2% glutaraldehyde (Electron Microscope Sciences, Fort Washington, PA) and 2mM CaCl2 in 0.1M Tris-maleate buffer (pH 7.4). The tongue was postfixed in three changes of fixative for a total of one hour. Cryoprotection of the tongue was carried out overnight in 20% sucrose in 0.1M tris-maleate buffer at 4°C. The tongue was cut on a cryostat at 16μm and collected on Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA).
Ecto-ATPase or -ADPase activity was demonstrated via lead precipitation as detailed in previous publications (Barry, 1992; Kittel et al., 2002). For this method, the slides were first rinsed 2 x 10 minutes in 0.07M tris-maleate buffer (pH 7.4). The following incubation medium was applied to the slides for 30 minutes at room temperature: 2mM Pb(NO3)2 (captures the free phosphate), 1mM levamisole (inhibits alkaline phosphatases), 1mM ouabain (inhibits Na+, K+-ATPase), 50μM α,β-methylene ADP (inhibits 5′-nucleotidase), 5mM KCl, 2mM CaCl2 and 1mM of substrate, either ATP or ADP (all chemicals from Sigma, St. Louis, MO). The incubation was followed by three ten-minute washes in 0.07M tris-maleate buffer. The lead precipitate was visualized by treating the slides for one minute with 1% ammonium sulfide. After several rinses in distilled water, slides were counterstained with 0.25% thionin in acetate buffer, pH 4.0 (J.T. Baker Chemical CO., Phillipsburg, NJ) before dehydration through ethanol to xylene. The slides were coverslipped with Permount (Fisher). Applying the incubation medium without a substrate did not result in detectable lead precipitate.
Anti-NTPDase2 antibody production
The NTPDase2 polyclonal antibody was raised in rabbit by direct intramuscular and intradermal injection of the complementary DNA (cDNA), encoding the entire mouse Entpd2 gene (GenBank accession number AY376711) ligated into pcDNA3.1/V5-His (Invitrogen, Ontario, Canada). This plasmid expressing mouse Entpd2 has been described previously (Jhandier, et al., 2005; Kukulski et al., 2005). The antibody specificity was determined by both Western blot and immunocytochemistry.
Western blot
Transiet transfection of COS-7 cells with the murine NTPDase2 plasmid and protein extracts were prepared as previously described (Kaczmarek et al., 1996; Kukulski et al., 2005). Protein supernatants from homogenized cells were suspended in NuPAGE Lithium Dodecyl Sulfate (LDS) Sample Buffer (Invitrogen) under non-reducing conditions. Proteins were separated on a NuPAGE 4–12% Bis-Tris gel and transferred to Immobilon-P membrane (Millipore, Ontario, Canada) by electroblotting according to the manufacturer’s recommendation (Invitrogen). The membrane was then incubated overnight at 4°C with 2.5% non-fat milk in phosphate buffered saline-tween (PBST; (−10.1 mM Na2HPO4, 1.8 mM KH2PO4, 136.9 mM NaCl, 2.7 mM KCl, 0.15% (vol/vol) Tween 20, pH 7.4). After incubation with the rabbit anti-NTPDase2 antibody (dilution 1:3000) for 90 minutes, the bands were visualized using horseradish peroxidase-conjugated goat anti-rabbit IgG (dilution 1:10,000; GE healthcare, Quebec, Canada) for one hour and Lightning Western Blot Chemiluminescence Reagent Plus (Perkin Elmer, Boston, MA).
Immunocytochemistry on cell cultures
COS-7 cells were fixed in 10% phosphate-buffered formalin mixed with cold acetone. Briefly, cells were incubated for 30 minutes in a blocking solution of 7% normal goat serum in PBS and then incubated overnight at 4°C with the rabbit anti-NTPDase2 antibody (1:1000) or the pre-immune serum. The cells were incubated with 0.15% hydrogen peroxide in PBS for 10 minutes, incubated with Avidin/Biotin blocking kit (Vector Laboratories, Burlingame, CA) and then with a biotin-labeled goat anti-rabbit secondary antibody, followed by avidin/biotinylated horseradish peroxidase (HRP) complex (Vector Laboratories). Peroxidase activity was revealed using 3,3′-Diaminobenzidine (DAB; Sigma) as a substrate. Cells were counterstained with aqueous hematoxylin (Biomeda, Foster City, CA) as recommended. Cells were imaged on a Olympus BX51 microscope system and photographed with a QImaging MicroPublisher 3.3 RTV camera. Brightness and contrast was adjusted using Photoshop® software (Adobe Systems, San Jose, CA).
Immunohistochemistry on brain tissue
To test whether this antibody recognized the appropriate protein in intact tissue, the brain from a perfused C57B6 mouse (see below) was removed and postfixed in 4% paraformaldehyde for four hours. After cryoprotection in 20% sucrose, 16μm parasagittal sections were cut on a cryostat and collected on Superfrost Plus slides. Fluorescent immunocytochemistry was performed as previously described (Braun et al., 2003) using the rabbit anti-NTPDase2 (1:1000) followed by detection with Alexa568 anti-rabbit (1:400; Molecular Probes).
NTPDase2 immunocytochemistry in taste buds
Perfusion and fixation
In order to label serotonin-accumulating type III taste cells, mice were injected with 5-hydroxy-L-tryptophan (5HTP; 0.08 mg/g; Sigma, St. Louis, MO) intraperitoneally (i.p.) one hour prior to sacrifice (Takeda et al., 1981; Yee et al., 2001). Mice were deeply anesthetized with 20% chloral hydrate (2.5ml; i.p.) and perfused transcardially with 4% paraformaldehyde (PFA) in 0.1M phosphate buffer (PB; pH 7.2–7.4). The tissue was dissected, postfixed in PFA for 2 hours and cryoprotected in 20% sucrose in PB overnight at 4°C. After sectioning transversely or sagitally on a cryostat; 16μm sections were collected onto Superfrost Plus Microscope slides (Fisher Scientific) or 30μm free-floating sections were collected in PB.
Single-label staining for NTPDase2
After 3 x 10 minute washes in 0.1M phosphate buffered saline (PBS; pH 7.2–7.4), slides were incubated in blocking solution (3% normal goat serum, 1% bovine serum albumin, 0.3% triton in PBS) for one hour. Incubation with rabbit (rb) anti-NTPDase2 antibody (1:1000) diluted in blocking solution was carried out overnight. Three PBS washes were followed by two-hour incubation with Alexa568 anti-rb (1:400; Molecular Probes, Eugene, OR). The slides then were washed three times ten minutes in PB before coverslipping slides with Fluormount G (Southern Biotechnology Associates, Inc., Birmingham, AL). Omission of the primary antibody or incubation with the preimmune serum resulted in no apparent fluorescent signal.
Double-label staining with NTPDase2 and other cell type markers
Assays with NTPDase2 and the type III cell marker, serotonin (5HT), were carried out simultaneously using a mouse derived antibody against 5HT (1:1 dilution; immunogen: 5HT conjugated to Bovine serum albumin; catalog #066D; Biomeda) and the rabbit derived anti-NTPDase2 described above. In order to prevent the anti-mouse secondary from binding to endogenous tissue sites, the slides were first exposed for three hours in anti-mouse unconjugated F(ab)2 (1:50 in blocking solution; Jackson, West Grove, PA). The slides were washed three times in PBS, incubated with blocking solution for one hour, and both primary antibodies were diluted in the same blocking solution and allowed to incubate overnight. The slides were rinsed three times with PBS the following day. Secondary antibodies were diluted in the same blocking solution and incubated for two hours: Alexa568 anti-rabbit and Alexa488 anti-mouse (1:400; Molecular Probes, Eugene, OR). Three PB washes before mounting slides with Fluormount G. Elimination of one of the primary antibodies with application of both secondary antibodies confirmed specific binding of the secondary antisera.
Assays with NTPDase2 and the type II cell marker, PLCβ2 (immunogen: synthetic peptide from amino acids 1170–1181 of human origin; catalog #sc-206; Santa Cruz Biotechnology, Santa Cruz, CA), were carried out sequentially because both antibodies were raised in rabbit (adapted from Shindler and Roth, 1996). Briefly, the first antibody applied to the tissue is significantly diluted to below the detection limit with fluorescently labeled secondary antibodies. This highly diluted antibody is labeled with a biotinylated secondary which is subsequently amplified with an avidin-biotin complex conjugated to peroxidase. The peroxidase enzyme, in the presence of hydrogen peroxide and the Alexa568 conjugated tyramide signal amplification reagent (TSA), catalyzes the reaction that further amplifies the signal and results in detectable fluorescent precipitate. The second primary antibody is then applied at standard working concentrations and detected with a conventional FITC-fluorescently labeled secondary. Because the first antibody was diluted beyond the detection limit for direct visualization by the fluorescent labeled secondary (as determined by omission of the second primary antiserum), the FITC signal is entirely attributable to the second primary antisera.
The slides were washed three times in PBS, followed by a 10 min. incubation in 3% hydrogen peroxide (in PB) to block endogenous peroxidase activity. After three PBS washes, the slides were placed in blocking solution for one hour. The first rabbit antibody, PLCβ2, was significantly diluted in blocking solution (1:4000) and placed on slides overnight. The next day, slides were washed three times in PBS and incubated for two hours with biotin F(ab)2 anti-rb (1:1000; Jackson) followed by rinsing in PBS and two hour exposure to avidin-biotin-complex (ABC; Vector Laboratories). After three PBS washes, the tissue was reacted with tyramide signal amplification (TSA conjugated to Alexa568; Molecular Probes) for seven minutes before washing slides four times in PBS. To ensure that all rabbit IgG binding sites were blocked, the slides were incubated with unconjugated F(ab) anti-rb overnight (1:50 in blocking solution; Jackson). After three rinses in PBS, the slides were put in blocking solution for one hour. The second rabbit primary antibody, NTPDase2, was then applied at the standard immunohistochemical concentration (1:1000) and incubated on slides overnight. The slides were washed three times in PBS followed by incubation in Alexa488 anti-rabbit (1:400; Molecular Probes) for two hours. Three rinses in PB and the slides were then coverslipped with Fluormount G. To ensure the specificity of the second secondary antibody, controls were run in which PLCβ2 was detected as described but the NTPDase2 was omitted while it’s secondary (Alexa488 anti-rabbit) was still applied. These sections showed no green fluorescence confirming that the Alexa488 signal arose only from binding to the NTPDase2 antibody.
Double-label assays with NTPDase2 and the type I cell marker GLAST (affinity-purified antibody directed against the rat carboxyl-terminal peptide sequence QLIAQDNEPEKPVADSETKM; catalog #AB1782; Chemicon, Temecula, CA), though the antibodies are derived from rabbit and guinea pig (gp) respectively, were carried out sequentially using the TSA method in order to amplify the GLAST signal. In addition, before the hydrogen peroxide treatment, 30μm free-floating sections were subjected to the following antigen retrieval method: tissue was placed in 10mM sodium citrate (dissolved in water, pH 9.0) at 80°C for 25 minutes then rinsed three times in PBS. The same TSA method detailed above was used with the following: the first primary antibody was GLAST (1:1000) followed by biotin anti-gp secondary (1:1000; Jackson). Blocking with unlabelled F(ab)2 was unnecessary since the two primary antisera were raised in different species. The second primary antibody, NTPDase2, was used as previously described and detected with indirect immunofluoresence. Elimination of one of the primary antibodies with application of both secondary antibodies confirmed specificity of secondary binding and detection systems.
Triple labeling
Gustducin GFP mice were injected with 5HTP, perfused, and the tissue collected onto slides as described earlier. In order to prevent the anti-mouse secondary from binding to endogenous tissue, the slides were blocked for three hours in unconjugated anti-mouse F(ab)2. The primary antibodies, mouse anti-5HT (1:1) and rabbit anti-NTPDase2 (1:1000) were diluted in the same blocking solution and incubated overnight. After three washes in PBS, the secondary antibodies were also applied simultaneously for two hours: CY5 anti-rabbit (1:400; Molecular Probes) and Alexa568 anti-mouse (1:400; Molecular Probes). Slides were rinsed in PB and mounted with fluormount G. Specific secondary binding was again confirmed by eliminating one of the primary antibodies and applying both of the secondary antibodies. In these preparations, GFP indicated gustducin (type II cells), Alexa568 showed 5HT (type III cells) and CY5 showed NTPDase2.
All images were collected using an Olympus Fluoview confocal laser-scanning microscope (LSCM) using 60x objective, 1.4 n.a. For each image, the channels were collected sequentially with single wavelength excitation and then merged to produce the composite image. This avoids the problem of bleed-through of images due to side-band excitation of the fluorochromes. Brightness and contrast were adjusted in Photoshop® (Adobe Systems, San Jose CA)
RESULTS
Massively parallel signature sequencing (MPSS)
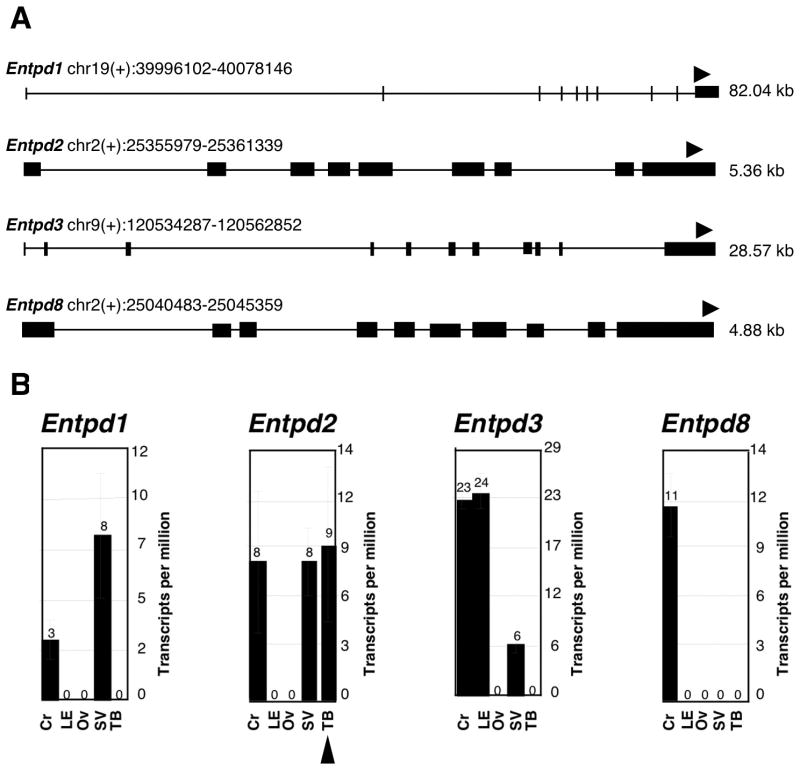
Massively parallel signature sequencing (MPSS; Brenner et al., 2000 a,b) was performed using RNA isolated from taste bud enriched tissue (TB), lingual epithelium devoid of taste buds (LE), inner ear derived tissue including the Organ of Corti (Cr), stria vascularis (SV) and otic vesicle (OV). Using MPSS, signature sequences can be obtained from millions of clones allowing for a full analysis of the mRNA population of a particular tissue. Examination of library signatures for those derived from E-NTPDase family members (Fig 1A) indicated that Entpd2 (gene encoding the NTPDase2 protein) was represented at 9 times per million transcripts (tpm) in the taste bud enriched library (Fig 1B; arrowhead at bottom). In contrast, it was not represented in the lingual epithelium library devoid of taste buds, suggesting that Entpd2 is selectively expressed in taste buds. Signatures derived from other enzyme gene members that readily hydrolyze extracellular ATP, namely Entpd1, Entpd3 and Entpd8 were not detected in the taste bud derived library, although they were present in one or more of the other libraries examined. The presence of NTPDase 1 & 2 in otic tissues is known from other studies (e.g. Vlajkovic et al., 2002) with NTPDase1 being expressed at high levels in the vasculature of the stria vascularis (SV) and NTPDase2 being more abundant in the Organ of Corti (Cr). Our results are consistent with these immunocytochemical findings but further indicate the presence of other ectoATPases in the ear tissues.
Figure 1.
Massively parallel signature sequencing (MPSS) A: The genomic locations and structures of Entpd1, Entpd2, Entpd3 and Entpd8 (genes encoding NTPDases 1, 2, 3 and 8) are shown. Gene chromosomal positions were obtained from the Build 33 assembly by NCBI. Exons are indicated by filled boxes, and the position and direction of the signature tags are indicated by arrowheads. As expected from the cloning procedures, each signature is associated with the 3′ most DpnII site of the coding sequence. B: Histograms display the abundance of each signature, expressed as transcripts per million (tpm), in libraries constructed from mRNA isolated from either taste bud enriched tissue (TB), lingual epithelium devoid of taste buds (LE), or inner ear derived tissues including the Organ of Corti (Cr), stria vascularis (SV) and otic vesicle (OV). Entpd2 mRNA is represented 9 times per million in the TB library (arrowhead below bar graph) but is not represented in the LE library, suggesting selective Entpd2 expression in taste buds. Taken together with the absence of Entpd1, Entpd3, and Entpd8 in the TB library, these results support that NTPDase2 is the primary ecto-ATPase expressed in taste buds. For sake of comparison, the taste receptor T1R2 is represented 10 tpm in the TB library and is absent in the LE library (data not shown).
Enzyme histochemistry
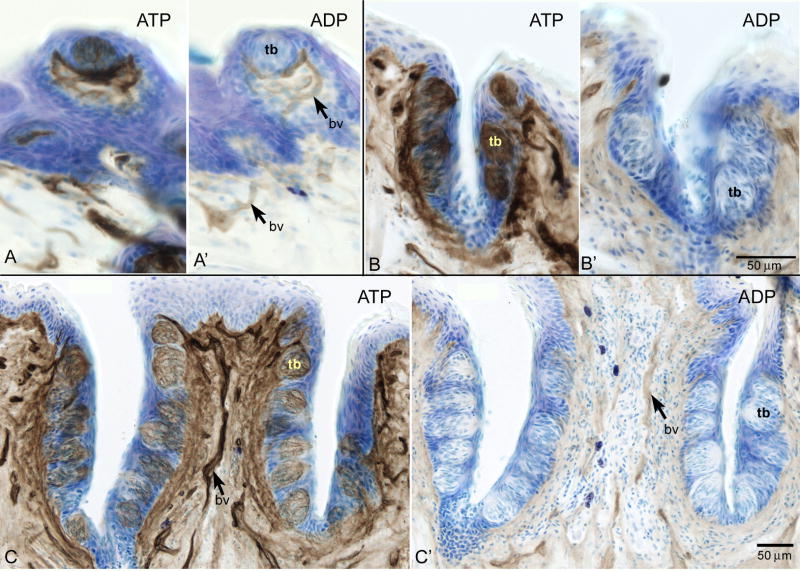
The above molecular data indicate the presence of NTPDase2 in taste buds. This enzyme strongly prefers ATP over ADP (Kegel et al., 1997; Mateo et al., 1999; Kukulski et al., 2005) and may be identified from this characteristic. We therefore evaluated substrate specificity in mouse taste buds by enzyme histochemistry utilizing pharmacological blockade of non-specific phosphatases and other nucleotidases. Utilizing ATP as a substrate in the assay reveals that ecto-ATPase activity is highly abundant in the fungiform, foliate, and vallate taste buds (tb) as well as the nearby nerve fibers (Fig 2A, B, C, respectively). Faint ecto-ADPase activity is present in surrounding blood vessels (bv) with no reactivity in the taste buds (Fig 2A, B, C’).
Figure 2.
Enzyme histochemical analysis revealing ecto-ATPase and ecto-ADPase activity via lead sulfide precipitate (brown) with thionin couterstain (blue) in taste buds in fungiform (A,A’), foliate (B, B’) and circumvallate (C, C’) papillae. With ATP as a substrate (A, B, C) strong ecto-ATPase activity is present in all taste buds (tb). Substituting ADP as a substrate results in lack of activity in the taste buds (A’, B’, C’) though some ecto-ADPase activity is present in nearby blood vessels (bv) (A’, C’). Scale bar in B’ also applies to A, A’, B. Scale bar in C applies to C’.
Anti-NTPDase2 antibody specificity
Protein samples from COS-7 cells transfected with NTPDase2 and untransfected COS-7 cells were incubated with the rabbit anti-NTPDase2 antibody. The antibody detected a protein band of the expected size, ~75 kDa (Sévigny et al., 2002; Braun et al., 2003), in the transfected cell lysate (Fig 3A). The specificity of the anti-NTPDase2 antibody was also tested by immunocytochemistry. Incubation with NTPDase2 antibody in untransfected cells (Fig 3B3) does not reveal any immunoreactive cells while antibody incubation with transfected cells (Fig 3B4) shows cellular staining. Conversely, incubation with the pre-immune serum on both untransfected (B1) and transfected (B3) cells does not show immunoreactivity. Anti-NTPDase2 antibody was also tested on Cos7 cells expressing the other membrane bound mouse NTPDases (NTPDase 1, 3, and 8) and no cross reactivity was found (data not shown). To verify protein specificity in tissue, this anti-NTPDase2 antibody was incubated on parasagittal brain sections. This staining (Fig 3C) revealed NTPDase2 immunoreactivity in the rostral migratory stream (RMS) as previously described for a different NTPDase2 antibody, one directed against rat NTPDase2 (Braun et al., 2003). Taken together, these experiments show that the antiserum is specific for mouse NTPDase2.
Figure 3.
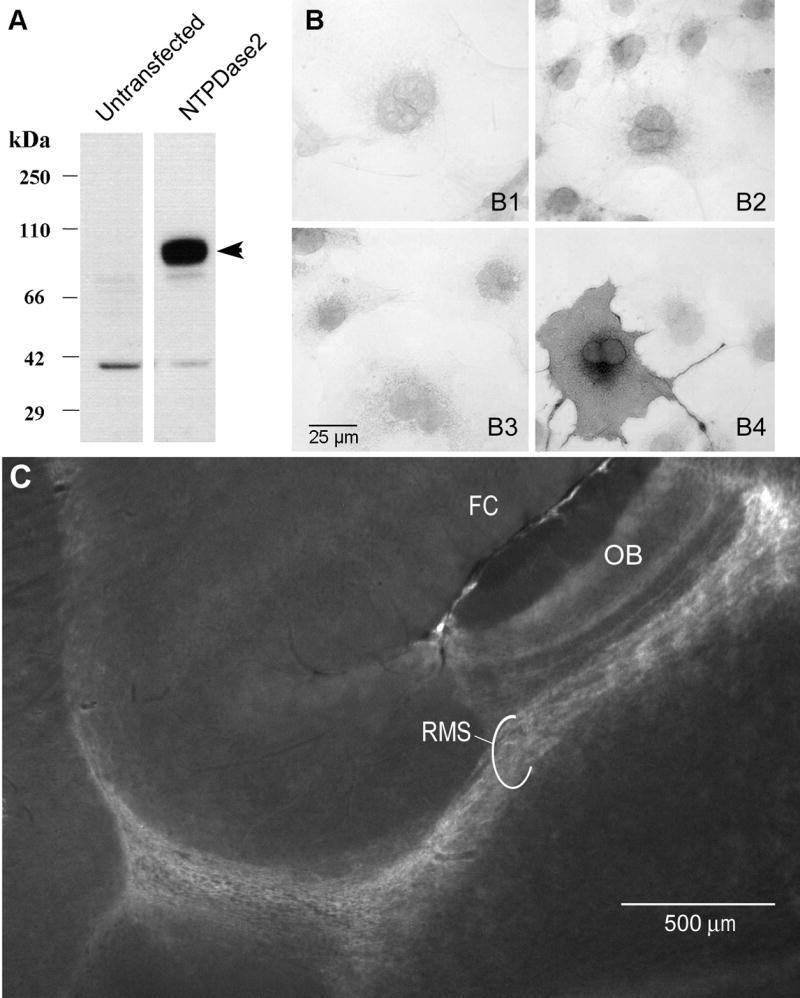
Anti-NTPDase2 polyclonal antibody specificity A: Western blot analysis of lysate from untransfected COS-7 cells compared to lysate from COS-7 cells transfected with the expression vector plasmid containing the entire NTPDase2 cDNA. As expected, the transfected cells show a prominent band of ~75 kDa (arrowhead) confirming the presence of NTPDase2. B: Immunocytochemistry on intact COS-7 cells transfected (B3,B4) or untransfected (B1,B2) with NTPDase2 cDNA. Cells were incubated with the pre-immune serum (B1,B3) or the anti-NTPDase2 antibody (B2,B4). Immunoreactivity is present only with transfected cells incubated with the NTPDase2 antibody (B4). C: Immunofluorescence for NTPDase2 in a parasagittal brain section (anterior to the right) showing strong expression of NTPDase2 in the rostral migratory stream (RMS) connecting to the olfactory bulb (OB). Staining with this antibody correlates with the previously described distribution of NTPDase2 in the RMS (Braun et al., 2003). FC = frontal cortex.
NTPDase2 immunocytochemistry in taste buds
As revealed by immunohistochemistry, NTPDase2 expression is robust in lingual (Fig 4A–C), palatal (Fig 4E, F) and laryngeal (Fig 4F insert) taste buds. This reactivity appears to outline immunoreactive cells suggesting a plasma membrane localization of this enzyme. NTPDase2 immunoreactivity also appears as an annulus around the nerve fibers near the taste buds (Fig 5A’). Incubation with the pre-immune serum results in no apparent fluorescent signal (Fig 4D; taken at eight times the exposure of the other panels in this figure in order to see the underlying tissue structure).
Figure 4.
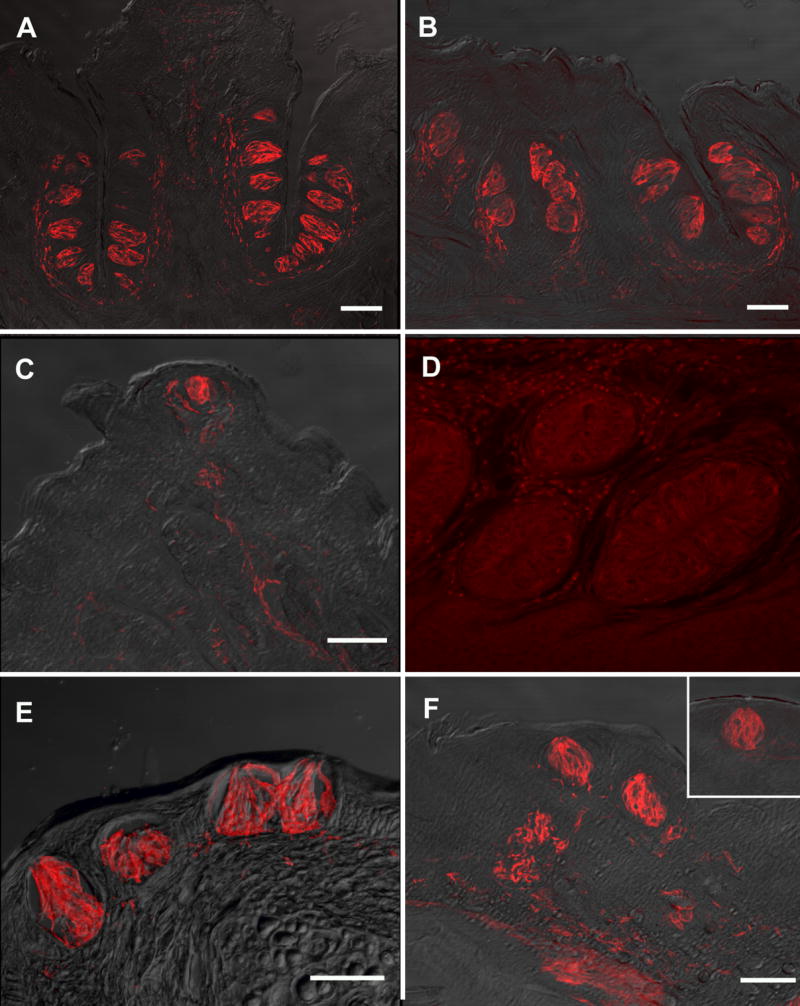
Nomarski images overlaid with images showing for NTPDase2 in mouse taste buds and surrounding fibers. A: circumvallate papillae, B: foliate papillae, C: fungiform papillae, E: naso-incisor ducts, F: palate, insert: larynx. Incubation with the preimmune serum did not reveal any significant fluorescent signal in the foliate papillae shown in D (taken at eight times the exposure of the other panel figures). Scale bars = 50μm. Scale bar in F also applies to F insert and D.
Figure 5.
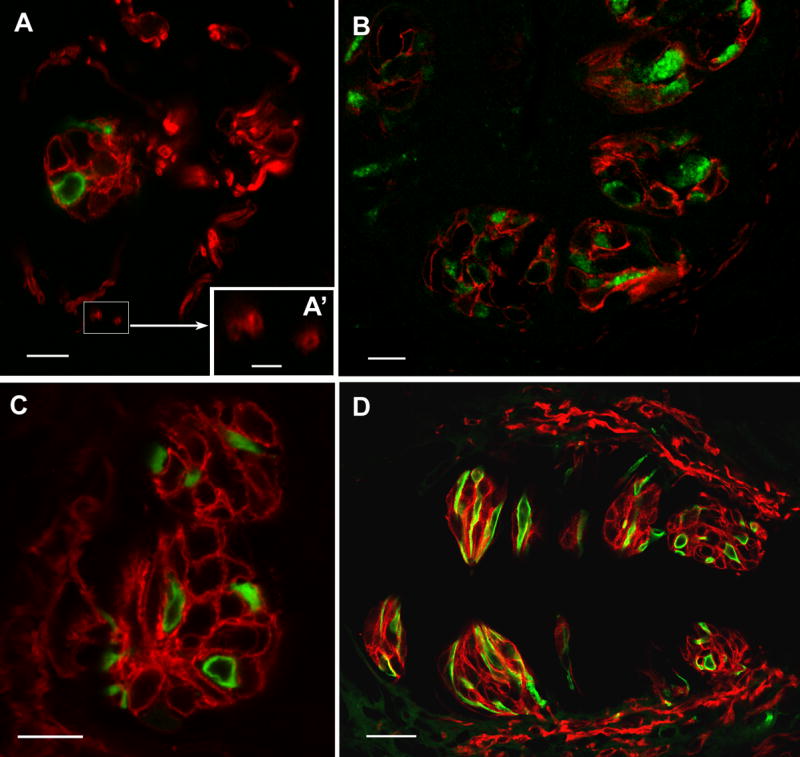
LSCM images of double label immunocytochemistry with NTPDase2 and the type II cell marker, PLCβ2 or the type III cell marker, 5HT. A, B: Staining for NTPDase2 (red) and PLCβ2 (green) in oblique sections cut through the fungiform (A) and foliate papillae (B). The NTPDase-immunoreactive cells are different from those exhibiting reactivity to either PLCβ2 or 5HT. NTPDase2 reactivity is also associated with nerve processes. Close observation of cross sections of this staining reveals that NTPDase2 immunoreactivity is limited to the periphery of the nerve profile while the core of the fiber remains void of staining, indicative either of membrane-associated neural reactivity or of glial cells circling the fiber (A’, scale bar 2.5μm) C, D: NTPDase2 (red) and 5HT (green) in foliate papillae (C) and cirumvallate papillae (D). Scale bars in A, B and C are 10μm; D scale bar is 25μm.
NTPDase2 and taste-cell types
Type II cells (PLCβ2 or gustducin) and Type III cells (5HT)
The large majority of type II taste cells are reactive for PLCβ2 (Clapp et al., 2001). The typical morphology of this cell population is spindle-shaped or pyriform with a large round nucleus. Taste cells that display PLCβ2 immunoreactivity (green) do not express NTPDase2 (red, Fig 5A, B). Rather the NTPDase2 cells envelop the type II cells.
Following injection of the serotonin precursor, 5HTP, serotonin staining is specific to a subset of type III cells as previously described (Takeda et al., 1981; Yee et al., 2001); this staining is not necessarily indicative of the endogenous 5HT but rather an indication of the cells’ ability to concentrate biogenic amines.
Serotonin immunoreactive cells are thin and spindle-shaped with an elongate nucleus, characteristic of the type III population. Type III cells that are immunoreactive for 5HT in the circumvallate and foliate papillae (Fig 5C, D green) are not reactive for NTPDase2; instead, the NTPDase2 cells (red) enwrap the type III cells in the same manner that the NTPDase2 reactive cells envelope the type II cells. Figure 5D shows double-label for NTPDase2 and 5HT in two taste buds of the circumvallate papillae cut in cross and longitudinal sections in the same section. In viewing taste buds in cross sections, it is obvious that there is a distinct gap between the cytoplasmic immunoreactivity of type III cells and the NTPDase2 immunoreactivity of type I cells while longitudinal sections are ambiguous for this spatial relationship.
Triple label staining in foliate papillae was achieved using gustducin-GFP tissue. The GFP expression is driven by the α-gustducin promoter and is present in a subset of type II taste cells (Fig 6 green). 5HT staining was used to label type III cells (Fig 6 red). This assay offers more comprehensive evidence for the enveloping nature of the NTPDase2 reactive cells (Fig 6 blue). NTPDase2 reactivity is also more abundant than the other cell type markers, commensurate with the larger number of type I taste cells than either type II or type III cells.
Figure 6.
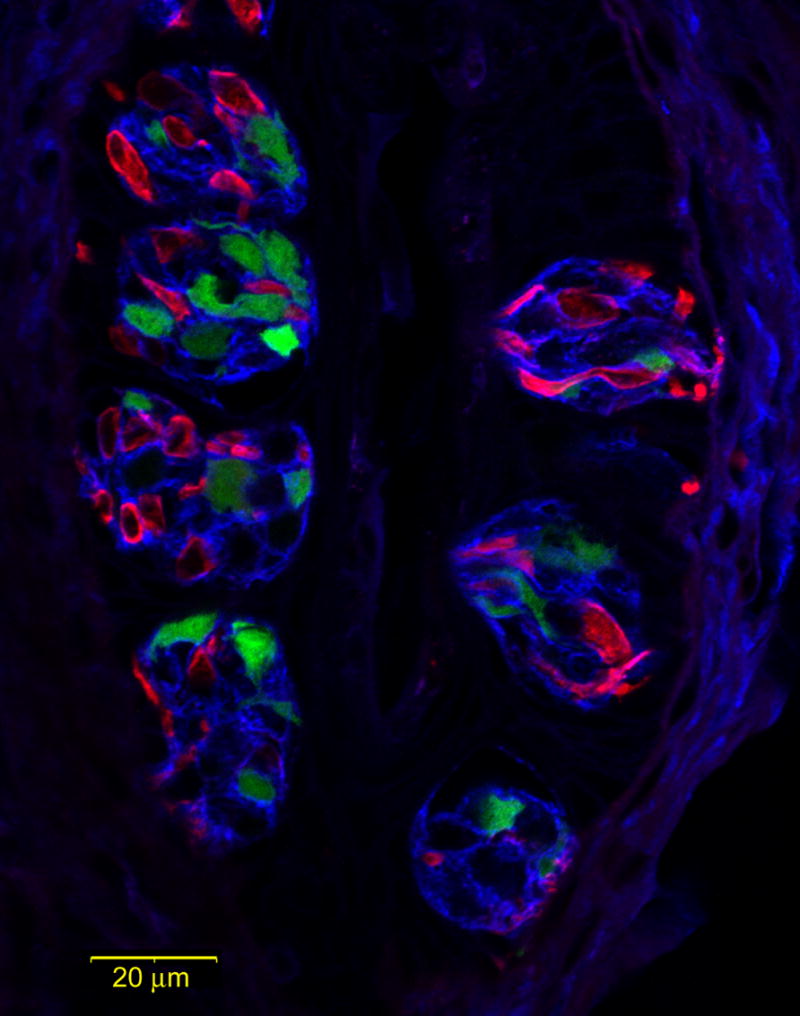
LSCM images of triple label staining in the foliate papillae with NTPDase2 in blue, the type II cell marker, gustducin in green (GFP), and the type III cell marker, 5HT, in red. NTPDase2-immunoreactive (-ir) cells wrap around the other two cell types, strongly reminiscent of type I cells. NTPDase2-ir type I cells also account for a greater number of cells compared to the other cell types.
Type I cells (GLAST)
As previously reported, immunoreactivity for the glial glutamate transporter GLAST is largely membrane-associated with bright puncta visible along the type I taste cell margins (Fig 7B, E; Lawton et al., 2001). Merged images of the GLAST and NTPDase2 staining confirm that these two proteins co-localize (Fig 7C, F). GLAST reactivity is also associated with surrounding nerve fibers, but not all nerve processes are double labeled with NTPDase2 as indicated with the arrow in figure 7F.
Figure 7.
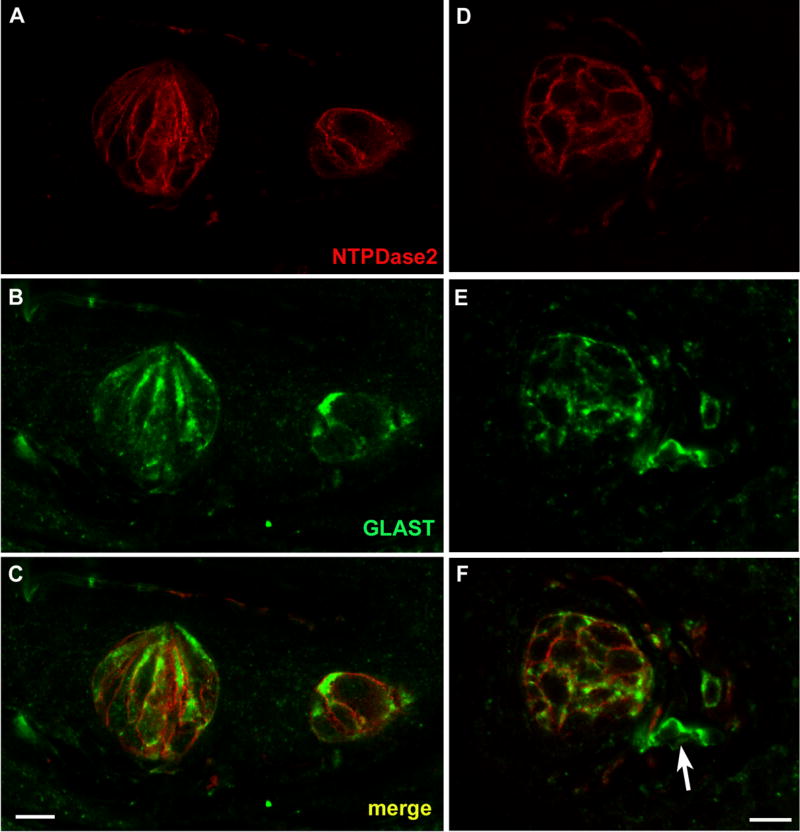
LSCM images of double label assays with NTPDase2 and the type I cell marker, GLAST. A, D: NTPDase2 reactive cells in foliate and fungiform papillae, respectively. B, E: GLAST reactive cells in the same sections of foliate and fungiform papillae. C, F: Merged images of NTPDase2 and GLAST revealing colocalization of these markers to the same cellular membranes. The GLAST reactivity extends deeper into the cytoplasm than does the NTPDase2, and so appears as a green halo inside of the double-label (yellow) plasma membrane. GLAST reactivity is also associated with nerve fibers, though not all GLAST positive nerve fibers are double labeled with NTPDase2 as indicated with the arrow in F.
DISCUSSION
The presence of ecto-ATPase activity in mammalian taste buds was noted nearly forty years ago on the basis of enzyme histochemistry on albino rats, rabbits, and Syrian golden hamsters (Iwayama and Oda, 1967; Zalewski, 1968; Iwayama, 1969; Barry, 1992). This characteristic was insufficient to identify the enzyme responsible for the ATPase activity in taste buds as we now know that various ecto-nucleotidases are capable of catalyzing the hydrolysis of ATP and other nucleotides. The family of ecto-nucleoside triphosphate diphosphohydrolase (E-NTPDase) includes enzymes previously called calcium-dependent ecto-ATPase, ecto-ATP diphosphohydrolase, ecto-apyrase, and CD39. Eight members of this family have been cloned and characterized functionally (Zimmerman, 2001; Kukulski et al., 2005). Since NTPDases 1, 2, 3, and 8 are located at the plasma membrane with extracellular active sites, these four members are relevant to controlling the levels of extracellular nucleotides. Though these four enzymes are capable of dephosphorylating a variety of nucleoside triphosphates and diphosphates, we concentrated on their affinities for adenine nucleotides: NTPDase1 hydrolyzes ATP and ADP equally well; NTPDase2 strongly prefers ATP; while NTPDase3 and NTPDase8 are functional intermediates with some preference for ATP over ADP (Zimmerman, 2001; Kukulski et al., 2005).
To further confound the process of identifying the ecto-ATPase of taste buds, the ectonucleotide pyrophosphatase/phosphodiesterase family (E-NPP) consists of various members capable of hydrolyzing ATP (Goding, 2000; Zimmerman, 2001). Likewise, alkaline phosphatase can also degrade extracellular ATP. It was therefore necessary to employ various assays to assess the identity of the ecto-enzyme present in taste buds.
Massively parallel signature sequencing (MPSS) was performed using RNA isolated from taste bud enriched tissue and lingual epithelium devoid of taste buds. Signature sequences were obtained from millions of clones allowing for a full analysis of the mRNA population within these tissues where the number of times per million a clone is represented in the library is a direct reflection of its abundance. Our search for various NTPDase enzymes revealed expression of Entpd2 (gene encoding NTPDase2) in the taste bud library with 9 transcripts per million (tpm), and no expression detected in the surrounding lingual epithelium library (see Fig 1). Conversely, Entpd1, Entpd3 and Entpd8 were not represented in the taste bud library. Entpd2 shares a similar prevalence in taste buds as other taste-specific genes, such as the taste receptor T1R2 (10 tpm), which is also absent in the surrounding lingual tissue. In comparison, genes that are not taste specific occur in both libraries; for example, ENaC-alpha is found 86 tpm in the taste bud library and 138 tpm in the non-taste lingual library.
As another measure to verify the presence of NTPDase2, we employed enzyme histochemical assays. The high specificity for ATP over ADP in taste buds in consistent with the known specificity of the NTPDase2 enzyme (Kegel et al., 1997; Mateo et al., 1999; Kukulski et al., 2005).
While NTPDase2 reactivity within the taste cells ceases with the replacement of ATP with ADP, not all enzymatic activity is eliminated in the tissue. ADP reaction product can be seen, albeit faintly, immediately surrounding the taste buds in what appear to be blood vessels. Given the localization of NTPDase1 at the surface of blood vessels (Kaczmarek et al., 1996; Sévigny et al., 1997) it is likely that this enzyme, which only slightly prefers ATP to ADP, is responsible for the product observed. This distribution is also similar to the ADPase activity reported in rat taste tissue (Zalewski, 1968; Iwayama, 1969). These investigators described ADP reaction product at the blood capillaries, as well as on the taste “hairs” (apical microvilli extending to the taste pore), though we did not find the latter activity in our assays. Our histochemical assay differed from the previous studies in that we inhibited 5′-nucleotidases and utilized a different fixation protocol, which may explain the slight difference of results. Alternatively, the difference may be due to a variation in enzymatic distribution between species.
Immunocytochemical staining further verified the abundance of NTPDase2 within taste buds of the tongue, palate and larynx. In keeping with NTPDase2 being a transmembrane protein (Zimmerman, 2001), immunoreactivity is strongest in association with plasma membranes of taste cells. Additionally, NTPDase2 staining is associated with nearby nerve fibers. Close examination of the immunopositive nerve fibers reveals that NTPDase2 reactivity is limited to the periphery of the fiber with the core being devoid of staining (Fig 5A’). It may be that the enzyme is localized to the outer membranes of the nerve fiber itself, yet recent studies report the presence of NTPDase2 in glial cell populations within both the central (Braun et al., 2003) and peripheral nervous systems (Braun et al., 2004). It is likely that Schwann cells wrapping the nerve fibers express NTPDase2, though ultrastructural studies would be necessary to resolve this issue.
The earlier histochemical ecto-ATPase studies also examined taste cells ultrastructurally whereby it was determined that the “dark” cell population expressed this enzyme. The dark cell population is now classified as type I cells and are seen to form lamellar processes which embrace the other two taste cell types (Pumplin et al., 1997; Finger and Simon, 2000). In order to determine the cellular localization of NTPDase2, we relied upon immunohistochemical assays. These did not show any co-localization of NTPDase2 with type II (PLCβ2 and α-gustducin) and type III (5HT) cell markers. Rather, NTPDase2 reactive cells wrap around the type II and type III cells. It is also evident that NTPDase2 immunoreactive cells account for a large number of taste cells within each taste bud, correlating with previous quantitative studies showing that type I cells constitute about half of taste cells within the taste bud (Farbman, 1965; Murray, 1973; Delay et al., 1986). Further, we find NTPDase2 co-localizes to the same membranes as GLAST, confirming that this enzyme is specific to the type I cells.
NTPDase2 seems to be crucial in regulating extracellular ATP signaling in various systems including the kidney (Kishore et al., 2005), cochlea (Vlajkovic et al., 2002; 2004) and cardiac vasculature (Enjyoji et al., 1999; Sévingy et al., 2002). The presence of the ionotropic ATP receptors (P2X2 and P2X3) on taste fibers (Bo et al., 1999), and various metabotropic ATP receptors (P2Y) on taste cells (Baryshnikov et al., 2003; Kataoka et al., 2004) suggests the possibility that ATP serves as a transmitter or co-transmitter in this system. In that case, NTPDase2 may serve to clear the neurotransmitter from the extracellular synaptic space and prevent receptor desensitization, as in other neural systems (Abbracchio and Burnstock, 1998).
Because ADP is only a marginal substrate of NTPDase2, the action of this enzyme on ATP will increase extracellular levels of ADP (Kukulski et al., 2005). While ADP can exert effects via various P2 receptors (Muller, 2002), the complete breakdown of ATP may occur through the action of other enzymes. While our investigations didn’t find the presence of other E-NTPDase enzymes that readily hydrolyze both ATP and ADP (NTPDases 1, 3, or 8), it may be that members of another enzyme family (e.g. E-NPP) occur in taste buds. Similarly, alkaline phosphatase may act to regulate increased ADP levels. The resulting AMP could ultimately be dephosphorylated by ecto-5′-nucleotidases (Zimmerman, 2001), giving rise to adenosine which is also a biologically relevant signaling molecule acting via its own receptors (Fredholm et al., 2001; Klinger et al., 2002). As the extracellular signaling of nucleotides is a complex cascade, further studies are needed to investigate the broader implications of ecto-ATP in the taste system.
In summary, our data show that NTPDase2 is the predominant ecto-ATPase in mouse taste buds. Further, NTPDase2 appears to be selectively expressed within the type I cells of taste buds which surround the type II and type III taste cells. Thus, NTPDase2 may play an important role in the control of nucleotide-mediated transmission of taste information (Finger et al., 2005) by destruction of ATP released by taste cells to activate P2X receptors on the afferent nerves (Bo et al., 1999). Type II and Type III taste cells frequently contact intragemmal nerve processes without there being intervening ATPase reactive processes of Type I cells (Iwayama, 1969; Kinnamon et al., 1985). Thus ATP released by either Type II or Type III taste cells can stimulate neuronal P2X receptors before being inactivated by the ectoATPase of Type I cells.
Acknowledgments
The authors thank Robert Margolskee (Mount Sinai School of Medicine and HHMI, New York, NY) for use of the gustducin-GFP transgenic mice. This work was supported by NIH grants DC006070 and P30 DC04657 to T.E.F., and grants from the Canadian Institutes of Health Research (CIHR) to J.S.. E.G.L. was a scholarship recipient from funds on arthritis and rheumatic diseases from Université Laval and J.S. of a New Investigator award from CIHR. This research was also supported in part by the Intramural Research Program of the NIH, NIDCD (S.L.S).
References
- Abbracchio MP, Burnstock G. Purinergic signaling-pathophysiological roles. Jpn J Pharmacol. 1998;78:113–145. doi: 10.1254/jjp.78.113. [DOI] [PubMed] [Google Scholar]
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Akisaka T, Oda M. The fine structural localization of adenosine triphosphatase activity on the taste bud in the fungiform papillae of the rat. Arch Histol Jpn. 1977;40:63. doi: 10.1679/aohc1950.40.63. [DOI] [PubMed] [Google Scholar]
- Barry M. Ecto-calcium-dependent ATPase activity of mammalian taste buds cells. J Histochem Cytochem. 1992;40:1919–1928. doi: 10.1177/40.12.1453008. [DOI] [PubMed] [Google Scholar]
- Baryshnikov SG, Rogachevskaja OA, Kolesnikov SS. Calcium signaling mediated by P2Y receptors in mouse taste cells. J Neurophysiol. 2003;90:3283–3294. doi: 10.1152/jn.00312.2003. [DOI] [PubMed] [Google Scholar]
- Bigonnesse F, Lévesque SA, Kukulski F, Lecka J, Robson SC, Fernandes MJG, Sévigny J. Cloning and characterization of mouse nucleoside triphosphate diphosphohydrolase-8. Biochem. 2004;43:5511–5519. doi: 10.1021/bi0362222. [DOI] [PubMed] [Google Scholar]
- Bo X, Alavi A, Xiang Z, Oglesby I, Ford A, Burnstock G. Localization of ATP-gated P2X2 and P2X3 receptor immunoreactive nerves in rat taste buds. Neuroreport. 1999;10:1107–1111. doi: 10.1097/00001756-199904060-00037. [DOI] [PubMed] [Google Scholar]
- Boughter JD, Jr, Pumplin DW, Yu C, Christy RC, Smith DV. Differential expression of alpha-gustducin in taste bud populations of the rat and hamster. J Neurosci. 1997;17:2852–2858. doi: 10.1523/JNEUROSCI.17-08-02852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, Sévigny J, Mishra SK, Robson SC, Barth SW, Gerstberger R, Hammer K, Zimmerman H. Expression of the ecto-ATPase NTPDase2 in the germinal zones of the developing and adult rat brain. Eur J Neurosci. 2003;17:1355–1364. doi: 10.1046/j.1460-9568.2003.02567.x. [DOI] [PubMed] [Google Scholar]
- Braun N, Sévigny J, Robson SC, Hammer K, Hanani M, Zimmerman H. Association of the Ecto-ATPase NTPDase2 with glial cells of the peripheral nervous system. Glia. 2004;45:124–132. doi: 10.1002/glia.10309. [DOI] [PubMed] [Google Scholar]
- Brenner S, Williams SR, Vermaas EH, Storck T, Moon K, McCollum C, Mao JI, Luo S, Kirchner JJ, Eletr S, DuBridge RB, Burcham T, Albrecht G. In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs. Proc Natl Acad Sci USA. 2000a;97:1665–1670. doi: 10.1073/pnas.97.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nature Biotech. 2000b;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Clapp TR, Stone LM, Margolskee RF, Kinnamon SC. Immunocytochemical evidence for coexpression of type III IP3 receptor with signaling components of bitter taste transduction. BMC Neurosci. 2001;2:6. doi: 10.1186/1471-2202-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay RJ, Kinnamon JC, Roper SD. Ultrastructure of mouse vallate taste buds: II. Cell types and cell lineage. J Comp Neurol. 1986;253:242–252. doi: 10.1002/cne.902530210. [DOI] [PubMed] [Google Scholar]
- Enjyoji K, Sévigny J, Lin Y, Frenette PS, Christie PD, Esch JS, II, Imai M, Edelberg JM, Rayburn H, Lech M, Beeler DL, Csizmadia E, Wgner DD, Robson SC, Rosenberg RD. Targeted disruption of cd39/ATP diphosphohydrolase results in disordered hemostasis and thromboregulation. Nature Medicine. 1999;5:1010–1017. doi: 10.1038/12447. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Fine structure of the taste bud. J Ultrastruc Res. 1965;12:328–350. doi: 10.1016/s0022-5320(65)80103-4. [DOI] [PubMed] [Google Scholar]
- Finger TE, Danilova V, Barrows J, Bartel DL, Vigers AJ, Stone L, Hellekant G, Kinnamon SC. ATP Signaling Is Crucial For Communication From Taste Buds To Gustatory Nerves. Science. 2005;310:1495–9. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- Finger TE, Simon SA. Cell biology of taste epithelium. In: Finger TE, Silver WL, Restrepo D, editors. The neurobiology of taste and smell. New York: Wile-Liss; 2000. pp. 287–314. [Google Scholar]
- Fredholm BB, IJzerman AP, Jacobson KA, Klotz K, Linden J. International union of pharmacology. XXV. Nomenclature and classification of adenosine receptors. Pharmacol Rev. 2001;53:527–552. [PMC free article] [PubMed] [Google Scholar]
- Goding JW. Ecto-enzymes: physiology meets pathology. J Leukoc Bil. 2000;67:285–311. doi: 10.1002/jlb.67.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJ, Zuker CS. Putative mammalian taste receptors: A class of taste-specific GPCRs with distinct topographic selectivity. Cell. 1999;96:541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- Iwayama T. Nucleoside phosphatase activities and submicroscopic localization of adenosine triphosphatase activity in gustatory epithelium. J Histochem Cytochem. 1969;17:724–733. doi: 10.1177/17.11.724. [DOI] [PubMed] [Google Scholar]
- Iwayama T, Nada O. Histochemically demonstrable ATPase activity in the taste buds of the rat. Exp Cell Res. 1967;46:607–608. doi: 10.1016/0014-4827(67)90388-6. [DOI] [PubMed] [Google Scholar]
- Jhandier MN, Kruglov EA, Lavoie EG, Sévigny J, Dranoff JA. Portal fibroblasts regulate the proliferation of bile duct epithelia via expression of NTPDase2. J Biol Chem. 2005;280:22986–22992. doi: 10.1074/jbc.M412371200. [DOI] [PubMed] [Google Scholar]
- Kaczmarek E, Koziak K, Sévigny J, Siegel JB, Anrather J, Beaudoin AR, Back FH, Robson SC. Identification and characterization of CD39/vascular ATP diphosphohydrolase. J Biol Chem. 1996;271:33116–33122. doi: 10.1074/jbc.271.51.33116. [DOI] [PubMed] [Google Scholar]
- Kataoka S, Toyono T, Seta Y, Ogura T, Toyoshima K. Expression of P2Y1 receptors in rat taste buds. Histochem Cell Biol. 2004;121:419–426. doi: 10.1007/s00418-004-0647-3. [DOI] [PubMed] [Google Scholar]
- Kegel B, Braun N, Heine P, Maliszewski CR, Zimmerman H. An ecto-ATPase and an ecto-ATP diphosphohydrolase are expressed in rat brain. Neuropharm. 1997;36:1189–1200. doi: 10.1016/s0028-3908(97)00115-9. [DOI] [PubMed] [Google Scholar]
- Kinnamon JC, Taylor BJ, Delay RJ, Roper SD. Ultrastructure of mouse vallate taste buds. I. Taste cells and their associated synapses. J Comp Neurol. 1985;235:48–60. doi: 10.1002/cne.902350105. [DOI] [PubMed] [Google Scholar]
- Kishore BK, Isaac J, Fausther M, Tripp SR, Shi H, Gill PS, Braun N, Zimmermann H, Sévigny J, Robson SC. Expression of NTPDase1 and NTPDase2 in murine kidney: relevance to regulation of P2 receptor signaling. AJP-Renal. 2005;288:1032–1043. doi: 10.1152/ajprenal.00108.2004. [DOI] [PubMed] [Google Scholar]
- Kittel A, Garrido M, Varga G. Localization of NTPDase1/CD39 in normal and transformed human pancreas. J Histochem Cytochem. 2002;50:549–555. doi: 10.1177/002215540205000412. [DOI] [PubMed] [Google Scholar]
- Klinger M, Freissmuth M, Nanoff C. Adenosine receptors: G protein-mediated signaling and the role of accessory proteins. Cell Signal. 2002;14:99–108. doi: 10.1016/s0898-6568(01)00235-2. [DOI] [PubMed] [Google Scholar]
- Kukulski F, Levesque SA, Lavoie EG, Lecka J, Bigonnesse F, Knowles AF, Robson SC, Kirley TL, Sévigny J. Comparative hydrolysis of P2 receptor agonists by NTPDases 1, 2, 3 and 8. Purinergic Signalling. 2005;1:193–204. doi: 10.1007/s11302-005-6217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton DM, Furness DN, Lindemann B, Hackney CM. Localization of the glutamate-aspartate transporter, GLAST, in rat taste buds. Eur J Neurosci. 2000;12:3163–3171. doi: 10.1046/j.1460-9568.2000.00207.x. [DOI] [PubMed] [Google Scholar]
- LopezJimenez ND, Sainz E, Cavenagh MM, Cruz-Ithier MA, Blackwood CA, Battey JF, Sullivan SL. Two novel genes, Gpr113, which encodes a family 2 G-protein-coupled receptor, and Trcg1, are selectively expressed in taste receptor cells. Genomics. 2005;85:472–482. doi: 10.1016/j.ygeno.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Mateo J, Harden TK, Boyer JL. Functional expression of a cDNA encoding a human ecto-ATPase. Br J Pharmacol. 1999;128:396–402. doi: 10.1038/sj.bjp.0702805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- Medler KF, Margolskee RF, Kinnamon SC. Electrophysiological characterization of voltage-gated currents in defined taste cell types of mice. J Neurosci. 2003;23(7):2608–2617. doi: 10.1523/JNEUROSCI.23-07-02608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CE. P2-pyrimidinergic receptors and their ligands. Curr Pharm Des. 2002;8:2353–2369. doi: 10.2174/1381612023392937. [DOI] [PubMed] [Google Scholar]
- Murray RG. The ultrastructure of taste buds. In: Friedman, editor. The ultrastructure of sensory organs. Amsterdam: North-Holland; 1973. pp. 3–37. [Google Scholar]
- Murray RG, Murray A. Fine structure of taste buds of rabbit foliate papillae. J Ultrastruct Res. 1967;19:327–53. doi: 10.1016/s0022-5320(67)80224-7. [DOI] [PubMed] [Google Scholar]
- Nelson GM, Finger TE. Immunolocalization of different forms of neural cell adhesion molecule (NCAM) in rat taste buds. J Comp Neurol. 1993;336:507–516. doi: 10.1002/cne.903360404. [DOI] [PubMed] [Google Scholar]
- Pumplin DW, Yu C, Smith DV. Light and dark cells of rat vallate taste buds are morphologically distinct cell types. J Comp Neurol. 1997;378:389–410. doi: 10.1002/(sici)1096-9861(19970217)378:3<389::aid-cne7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Sévigny J, Levesque FP, Grondin G, Beaudoin AR. Purification of the blood vessel ATP diphosphohydrolase, identification and localization by immunological techniques. Biochim Biophys Acta. 1997;1334:73–88. doi: 10.1016/s0304-4165(96)00079-7. [DOI] [PubMed] [Google Scholar]
- Sévigny J, Sundberg C, Braun N, Guckelberger O, Csizmadia E, Qawi I, Imai M, Zimmerman H, Robson SC. Differential catalytic properties and vascular topography of murine nucleoside triphosphate diphosphohydrolase (NTPDase1) and NTPDase2 have implications for thromboregulation. Blood. 2002;99:2801–2809. doi: 10.1182/blood.v99.8.2801. [DOI] [PubMed] [Google Scholar]
- Shindler KS, Roth KA. Double Immunoflorescent staining using two unconjugated primary antisera raised in the same species. J Histochem Cytochem. 1996;44:1331–1335. doi: 10.1177/44.11.8918908. [DOI] [PubMed] [Google Scholar]
- Takeda M, Shishido Y, Kitao K, Suzuki Y. Biogenic monoamines in developing taste buds of mouse circimvallate papillae. Arch Histol Jpn. 1981;44:485–495. doi: 10.1679/aohc1950.44.485. [DOI] [PubMed] [Google Scholar]
- Vlajkovic SM, Thorne PR, Sévigny J, Robson SC, Housley GD. NTPDase1 and NTPDase2 immunolocalization in mouse cochlea: implications for regulation of P2 receptor signaling. J Histochem Cytochem. 2002;50:1435–1441. doi: 10.1177/002215540205001102. [DOI] [PubMed] [Google Scholar]
- Vlajkovic SM, Housley GD, Muwoz DJB, Robson SC, Sévigny J, Wang CJH, Thorne PR. Noise exposure induces up-regulation of ecto-nucleoside triphosphate diphosphohydrolases 1 and 2 in rat cochlea. Neurosci. 2004;126:763–773. doi: 10.1016/j.neuroscience.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Wong GT, Ruiz-Avila L, Margolskee RF. Directing gene expression to gustducin-positive taste receptor cells. J Neurosci. 1999;19:5802–5809. doi: 10.1523/JNEUROSCI.19-14-05802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Crowley HH, Rock ME, Kinnamon JC. Taste cells with synapses in rat circumvallate papillae display SNAP-25-like immunoreactivity. J Comp Neurol. 2000a;424:205–215. doi: 10.1002/1096-9861(20000821)424:2<205::aid-cne2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Yang R, Tabata S, Crowley HH, Margolskee RF, Kinnamon JC. Ultrastructural localization of gustducin immunoreactivity in microvilli of type II taste cells in the rat. J Comp Neurol. 2000b;425:139–151. doi: 10.1002/1096-9861(20000911)425:1<139::aid-cne12>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Yee CL, Yang R, Böttger B, Finger TE, Kinnamon JC. “Type III” cells of rat taste buds: immunohistochemical and ultrastructural studies of neuron-specific enolase, protein gene product 9.5, and serotonin. J Comp Neur. 2001;440:97–108. doi: 10.1002/cne.1372. [DOI] [PubMed] [Google Scholar]
- Zalewski AA. Changes in phosphatase enzymes following denervation of the vallate papilla of the rat. Exptl Neurol. 1968;22:40–51. doi: 10.1016/0014-4886(68)90018-6. [DOI] [PubMed] [Google Scholar]
- Zimmerman H. Ectonucleotidases: Some recent developments and a note on nomenclature. Drug Dev Res. 2001;52:44–56. [Google Scholar]