Abstract
Stenotrophomonas maltophilia is an aerobic, non-fermentative Gram-negative bacterium widespread in the environment. S. maltophilia Sm777 exhibits innate resistance to multiple antimicrobial agents. Furthermore, this bacterium tolerates high levels (0.1 to 50 mM) of various toxic metals, such as Cd, Pb, Co, Zn, Hg, Ag, selenite, tellurite and uranyl. S. maltophilia Sm777 was able to grow in the presence of 50 mM selenite and 25 mM tellurite and to reduce them to elemental selenium (Se0) and tellurium (Te0) respectively. Transmission electron microscopy and energy dispersive X-ray analysis showed cytoplasmic nanometer-sized electron-dense Se0 granules and Te0 crystals. Moreover, this bacterium can withstand up to 2 mM CdCl2 and accumulate this metal up to 4% of its biomass. The analysis of soluble thiols in response to ten different metals showed eightfold increase of the intracellular pool of cysteine only in response to cadmium. Measurements by Cd K-edge EXAFS spectroscopy indicated the formation of Cd-S clusters in strain Sm777. Cysteine is likely to be involved in Cd tolerance and in CdS-clusters formation. Our data suggest that besides high tolerance to antibiotics by efflux mechanisms, S. maltophilia Sm777 has developed at least two different mechanisms to overcome metal toxicity, reduction of oxyanions to non-toxic elemental ions and detoxification of Cd into CdS.
Introduction
Stenotrophomonas maltophilia is an aerobic, non-fermentative Gram-negative bacterium widespread in the environment. This species constitutes one of the dominant rhizosphere inhabitant, frequently isolated from the rhizosphere of wheat, oat, cucumber, maize, oilseed rape, and potato [1]–[4]. S. maltophilia shows plant growth-promoting activity as well as antagonistic properties against plant pathogens. It is currently being studied for its biological control of plant pathogens and was therefore utilized for the development of biopesticides [5]. S. maltophilia is also able to degrade xenobiotic compounds [6], [7], to detoxify high molecular weight polycyclic aromatic hydrocarbons [8], possessing therefore a potential for soil decontamination (bioremediation). This bacterium was also increasingly described as an important nosocomial pathogen in debilitated and immunodeficient patients [9], [10], as well as associated with a broad spectrum of clinical syndromes, e.g. bacteraemia, endocarditis, respiratory tract infections [11]. S. maltophilia displays intrinsic resistance to many antibiotics, making selection of optimal therapy difficult. The mechanisms underlying this multiresistance to drugs seem to result from a combination of reduced permeability [12], and expression of efflux pumps. Two RND efflux systems have been identified, SmeABC [13] and SmeDEF [14], [15].
Considering on one hand the rhizospheric origin of various opportunistic pathogens [16] including S. maltophilia and, on the other hand, the description of horizontal gene transfers in the rhizosphere [17], the tolerance of this bacterium to a wide range of toxic oxianions and metals must be addressed.
In the present study, we evidenced the tolerance of the strain Sm777 that belongs to S. maltophilia species, to very high concentrations of various toxic metals, especially cadmium, selenium and tellurium, involving two different tolerance mechanisms.
Results and Discussion
The strain Sm777 was isolated as a culture contaminant associated to Pseudomonas strains and was revealed in a contest of heavy metal tolerance studies. This rod-shaped bacterium was persistent in cultures containing a high concentration of cadmium, and was identified as a Stenotrophomonas maltophilia by 16S rDNA sequencing. The sequence analysis (using the BLAST database of the National Center for Biotechnology Information; [http://www.ncbi.nlm.nih.gov]) showed that strain Sm777 matched 99.5% with 16S rDNA of the S. maltophilia LMG 958T (accession n° DQ469587).
MICs of drugs and heavy metals
S. maltophilia Sm777 was able to grow during 16 h in the presence of 500 µM CdCl2, 20 mM tellurite or 50 mM selenite without any significant increase of the lag phase. It is worthnoting that strain Sm777 also grew to a high density (109 cfu.ml−1) in the presence of high concentrations of other heavy metals (0.1 mM CoCl2, 5 mM CuSO4, 4 mM ZnSO4, 10 mM NiSO4, 0.05 mM HgCl2, 0.02 mM AgNO3, >1 mM uranyl, and 5 mM Pb(NO3)2). Moreover, this bacterium was resistant to a wide range of antibiotics, such as kanamycin (50 µg.ml−1), gentamycin (100 µg.ml−1), tetracycline (50 µg.ml−1), and 50 µg.ml−1 of nalidixic acid. These may suggest that strain Sm777 overproduces some multidrug resistance (MDR) efflux pumps that are known to be involved in bacterial resistance to a wide range of compounds by extruding antibiotics and other toxic compounds.
Oxianions reduction
To verify the hypothesis of overexpression of efflux systems to get ride of drugs and heavy metals, we analysed and localized the elemental composition of bacteria grown in the presence of tellurite and selenite, by using Energy Dispersive X-ray Spectroscopy (EDX) in conjunction with Transmission Electron Microscopy (TEM) or Environmental Scanning Electron Microscopy (ESEM).
The chemical microanalysis (TEM-EDX) of reddish colonies of strain Sm777 grown in the presence of selenite revealed cytoplasmic electron-dense Se0 granules (Fig. 1A). No detectable extracellular particles were observed. The intracellular Se0 granules strongly suggest that selenite tolerance of strain Sm777 is not related to an efficient efflux system. On the contrary, a S. maltophilia strain isolated from a seleniferous agricultural drainage pond sediment was shown to transform selenate and selenite and to form spherical extracellular deposits consisting of Se [18]. TEM-EDX observations of black colonies of strain Sm777 grown in the presence of tellurite revealed the presence of Te0 crystals in the cytoplasm and proved that tellurite was taken up by the cells and was reduced into tellurium in the intracellular compartment (Fig. 1B).
Figure 1. ESEM-EDX and TEM-EDX observations.
Microscopic observations and representative energy-dispersive X-ray spectra of electron-dense particles of S. maltophilia Sm777 cells grown in ten fold-diluted TSB medium solidified with 15 g.l−1 agar, and supplemented with metals. (A & B) Colony shape, TEM-EDX micrographs and spectra of S. maltophilia Sm777 grown in the presence of selenite (10 mM) and tellurite (1 mM). (C) Colony shape, ESEM-EDX observation and analysis of cells grown in the presence of CdCl2 (500 µM). Arrows on micrographs indicate the presence of intracellularly localized electron-dense particles of Se and Te, and arrows on spectra indicate metal-specific peak detected.
Active efflux of the metal is a frequently utilized strategy to produce tolerance by lowering the intracellular concentration to subtoxic levels. However, our data showing intracellular nanometer-sized particles of elemental selenium or tellurium, suggest that MDR efflux pumps probably do not mediate the heavy metal tolerance mechanism in strain Sm777 since tellurite and selenite-tolerance was associated to an intracellular reduction of these oxyanions and then by their accumulation.
Tolerance of S. maltophilia to cadmium
ESEM observations coupled to EDX analysis of strain Sm777 grown in the presence of 500 µM CdCl2 revealed the presence of Cd associated to bacterial cells, but did not allow localizing it exactly (Fig. 1C). The bacterial Cd content was determined by ICP-AES as previously described [19]. This analysis revealed an accumulation of Cd strongly associated with the bacterial cell wall or incorporated into cells. Hence, this strain was able to accumulate Cd representing up to 4% of its dry mass. The presence of a cluster of genes from Gram-positive bacteria involved in both antibiotic and heavy metal resistance has been described in S. maltophilia D457R [20]. This cluster contains genes encoding a macrolide phosphotransferase (mphBM) and a cadmium efflux determinant (cadA). This study indicated a lateral gene transfer between Gram-positive and Gram-negative bacteria. The role of these genes in heavy metal tolerance of S. maltophilia has not been clearly evidenced yet.
Cysteine accumulation in response to cadmium
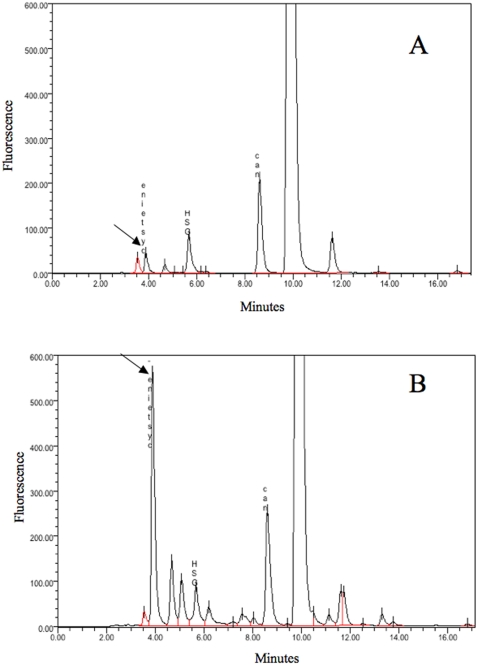
The role of thiol compounds in the protection against heavy metals is well known [21]. Moreover, the chemical sequestration of Cd is thought to occur by coordination of cysteine thiolate groups. For that reason, we determined the concentration of soluble thiol compounds of strain Sm777 cells in response to Cd. We noticed an increase of intracellular cysteine pool when bacteria were grown in the presence of 500 µM CdCl2 (Fig. 2). Unlike other bacteria or yeast, no modification of gluthatione content was observed [21]. Moreover, no modification of the intracellular pool of cysteine was observed in response to the following metals: NiSO4, CuSO4, Pb(NO3)2, ZnSO4, CoCl2, HgCl2, AgNO3, tellurite and selenite. The increase of intracellular pool of cysteine might reduce the bioavailability of Cd.
Figure 2. Soluble thiols analysis.
HPLC analysis of nonprotein thiols in S. maltophilia Sm777 grown in TSB/10 without (A) or supplemented (B) with 500 µM of CdCl2. The arrow indicates cysteine peack. N-acetyl-L-cysteine (NAC) was used as an internal standard.
Park and Imlay [22] have shown that high levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. They actually found that when cysteine homeostasis is disrupted, intracellular cysteine acts as an adventitious reductant of free iron and thereby promotes oxidative DNA damage.
The toxic effect of Cd is mainly mediated by its high degree of reactivity with S, O and N atoms in biomolecules. Cysteine promotes an oxidative stress in cells, however it also protects against Cd toxicity probably by chelating Cd. The resulting metal thiolate complex formation may neutralize the toxicity of heavy metal. To deal with this dilemma, increasing the intracellular cysteine pool, bacterial cells are potentially exposed to an oxidative stress, but these cysteine residues may be stabilized by formation of Cd-cysteine complex decreasing that way the amount of free Cd and free cysteine.
Formation of CdS particles
When strain Sm777 was grown under aerobic conditions on solid media containing 500 µM CdCl2, it formed yellow colonies (Fig. 1C). This observation suggested that bacterial cells may have transformed the Cd(II) into CdS as previously reported for Klebsiella pneumoniae [23], and for Klebsiella planticola [24]. To test this hypothesis, we used Cd K-edge EXAFS spectroscopy to probe the detailed coordination environment of the metal. The EXAFS spectrum was adjusted using different atomic neighbors around Cd. The nature, number and distances of atoms surrounding Cd in the sample are detailed in Table 1 and the calculated and experimental EXAFS curves are compared in Fig. 3A. EXAFS modeling indicated that the first coordination sphere of Cd was composed of four sulfur atoms at 2.50 and 2.64 Å and confirmed the formation of CdS compounds. EXAFS calculations also indicated the presence of Cd in the second coordination sphere at 3.42 and 3.68 Å. These Cd-Cd contributions indicated that CdS4 tetrahedra present in the cell bond to form Cd-S-Cd clusters. The low number of Cd atoms around each Cd (1.3) suggested that the size of the Cd-S-Cd clusters is small and can be a mixture of Cd dimers and trimers as illustrated in Fig. 3B. Thus, Cd-S clusters are formed in the cells and the low coordination number for the Cd-Cd contributions suggests that the product is less crystalline than the CdS reference compound. However, it is not possible to conclude whether these Cd-S clusters are surrounded by poly-thiols molecules or not.
Table 1. Cd atomic environment.
| Atomic pair | Interatomic distance R (Å) | Debye-Waller parameter (Å) | Number of atoms | Residue |
| Cd↔S | 2.50 | 0.090 | 3.1 | 0.02 |
| Cd↔S | 2.64 | 0.100 | 0.9 | |
| Cd↔Cd | 3.42 | 0.092 | 0.4 | |
| Cd↔Cd | 3.68 | 0.110 | 0.9 |
Structural parameters of the Cd atomic environment derived from EXAFS modeling of the Cd K edge EXAFS spectrum of S. maltophilia Sm777 bacterial cells.
Figure 3. Extended X-ray Absorption Fine Structure (EXAFS) Spectroscopy.
(A) Comparison between experimental and calculated EXAFS of S. maltophilia Sm777 strain cells. (B) Modeling of Cd dimers and trimers.
The mechanism underlying the formation of CdS by strain Sm777 remains unclear; it is obvious that strain Sm777 formed CdS under aerobic conditions, whereas the formation of CdS in Clostridium thermoaceticum is mediated by the production of H2S under stringent reductive conditions [25]. The aerobic sulfide production and Cd precipitation by Escherichia coli was possible by over-expression of the Treponema denticola cysteine desulfhydrase gene which product converts cysteine to sulfide under aerobic conditions. However, Cd precipitation as CdS was effective only when cysteine was added to the growth medium [26], whereas the production of CdS by strain Sm777 did not require any exogenous supply of cysteine. The high increase of intracellular pool of cysteine suggests that the bacterium reorients its metabolism to the production of cysteine that might be converted to sulfide used for CdS formation. Cysteine is able to form high-affinity metal ligand clusters and to promote the formation of CdS particles.
Alonso and colleagues [20] showed that a Stenotrophomonas strain has acquired a cluster of antibiotic and heavy metal resistance genes from Gram positive bacteria. Most of these genes are homologues of genes previously found on Staphylococcus aureus plasmids. In the present study, we evidenced the high tolerance to various heavy metals by S. maltophilia Sm777. To our knowledge, this is the first report indicating the high ability of a member of this species to tolerate and to detoxify several heavy metals. This bacterial species is also described as an opportunistic pathogen responsible for nosocomial infections. The severity of these infections is due to the virulence factors of the bacteria and to their occurrence in debilitated patients in whom invasive devices are used. To get more insight in the different mechanisms of heavy metals tolerance, and to identify pathogenesis related genes, it would be of great interest to perform a genome analysis and functional genomic studies of this species.
Materials and Methods
Growth conditions
S. maltophilia Sm777 was grown aerobically in an incubating shaker at 30°C in tenfold diluted tryptic soy broth (TSB/10) (DIFCO Laboratories, Detroit, USA). For growth on plates, media were solidified with 15 g.l−1 Bacto-agar (DIFCO Laboratories, Detroit, USA).
Determination of metals and antibiotics maximum tolerance concentrations
To determine the MTCs (maximal tolerated concentration) for different heavy metals, bacteria were grown on 10 ml of TSB/10 in the presence of different concentrations of different metals, CdCl2, NiSO4, CuSO4, Pb(NO3)2, ZnSO4, CoCl2, HgCl2, uranyl acetate and AgNO3, at 30°C under shaking. The MTCs corresponded to the highest concentration of each metal at which growth was still observed [27]. The MTCs for the four antibiotics, kanamycin, gentamycin, nalidixic acid, and tetracycline were also determined, and are expressed in µg.ml−1. Experiments were performed in triplicate for each condition.
Analysis of cadmium accumulation
To determine the Cd content of bacterial cells grown in TSB/10 supplemented with 500 µM CdCl2 for 48 h cells were harvested, rinsed three times using TSB/10 and dried at 55°C for 24 h . Following addition of 5 ml HNO3 (70%), mineralization was carried out in a microwave oven (Mars X; CEM Corp., Matthews, N.C.). Metal content was determined using an inductively coupled plasma atomic emission spectrometry (ICP-OES device; Varian); standard solutions were supplied by Merck.
Soluble thiols analysis
Cells were harvested and rinsed with TSB/10 and stored at −80°C until analysis. Nonprotein thiols were extracted by disruption of cells by sonication of 5 to 7 mg of frozen bacteria in 0.5 to 0.7 ml of extraction buffer (6.3 mM diethylenetriamine pentaacetic acid [DTPA]−0.1% [vol/vol] trifluoroacetic acid). Thirty microliters of 100 µM N-acetyl-L-cysteine was added as an internal standard. The homogenate was centrifuged at 10,000× g for 15 min at 4°C (Centromix 1236 V, Rotor 20RT) and the supernatant was filtered (0.22 µm). The derivatization procedure was modified from Rijstenbil and Wijnholds [28]. Filtered extracts (125 µl) were mixed with 225 µl of reaction buffer [0.2 M 4-(2-hydroxy-ethyl)-piperazine-1-propanesulfonic acid pH 8.2 containing 6.3 mM DTPA] and 5 µl of 25 mM monobromobimane dissolved in acetonitrile. Following 15 min of incubation in the dark at room temperature, the reaction was stopped by adding 150 µl of 1 M methane sulfonic acid. The samples were stored at 4°C in the dark until high-performance liquid chromatography (HPLC) analysis. The bimane derivatives were separated on a reversed-phase Nova-Pak C18 analytical column (pore size, 60 Å; particle size, 4 µm; dimensions, 3.9 by 300 mm; Waters catalog no. 11695) using two eluents (0.1% [vol/vol] trifluoroacetic acid in water and acetonitrile) at a flow rate of 1 ml.min−1. Fluorescence was monitored by a Waters 464 detector (λexcitation = 380 nm; λemission = 470 nm). Calibration curves of glutathione were used in all measurements. Cysteine, GSH and -glutamylcysteine (-EC) (from Sigma) were used as standard.
ESEM-EDX and TEM-EDX observations
For transmission electron microscopy (TEM), bacterial cells were harvested from TSA/10 plates containing tellurite (1 mM) or selenite (10 mM). Cells were then fixed in 2.5% glutaraldehyde and postfixed with osmium tetroxide in sodium cacodylate buffer. Dehydration was performed in ethanol and inclusion in epoxy resin. Ultrathin sections were made using a Reichert ultramicrotome. Electron micrographs and chemical microanalyses were obtained with a Jeol (Tokyo, Japan) 100CX transmission electron microscope coupled with an energy dispersive X-ray spectrometer (EDX). Environmental scanning electron microscopy (ESEM) microscope coupled with an energy dispersive X-ray spectrometer (EDX) observations were realized on colonies grown on TSA/10 containing 500 µM CdCl2.
Extended X-ray Absorption Fine Structure (EXAFS) Spectroscopy
Cd K-edge XAS experiments were carried out at the European Synchrotron Radiation Facility (ESRF, Grenoble-France) on the FAME (BM30-b) beamline with Si (220) monochromator crystals using the fluorescence detection mode. The storage ring was operated at 6 GeV with a current of 200 mA. XAS spectra were scanned from 100 eV below to 800 eV above the Cd K-edge. The pre-edge part was extracted from the XANES (X-ray Absorption Near Edge Structure) region (extended from 26600 eV to 26650 eV). XANES spectra intensity was normalized by fitting the photoelectric background above the absorption edge with a 2nd order polynomial function. The EXAFS (Extended X-ray Absorption Fine Structures) data reduction was done using a series of programs developed by Michalowicz [29] based on standard procedures [30]. The extracted EXAFS was k2 weighted (with k = wave vector) to enhance the high-k region and Fourier transformed over the k range 2.4 to 14–15 Å−1, to R space using a kaiser apodization window with t = 2.5. The resulting pseudo-Radial Distribution Functions (RDF) are uncorrected for phase shift leading to a shift of the peaks by 0.3–0.4 Å. Separate peaks in the RDF corresponding to successive shells of neighboring atoms around Cd were isolated by Back-Fourier Transformation (BFT) for single or multiple shell analysis. The analysis of partial c(k) was based upon the curved wave EXAFS formalism [31] in the single scattering approximation. Curve fitting was performed with a non linear least-square procedure, and phase (fbackscatterer(k), dcentral atom(k) ) and amplitude (|fbackscatterer(q, k, R)|) functions used were calculated with FEFF8 32]. Phase and amplitude functions of Cd-S and Cd-Cd atomic pairs were tested on reference compounds (Cd(OH)2, CdS).
Acknowledgments
We are grateful to M. Lesourd for the contribution to TEM observations and EDX analysis, to I. Felines for the contribution to ESEM observations and to A de Groot for critical reading of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the French ‘Toxicologie Nucléaire Environnementale’ program (2001–06). The funder did not contribute to the design and conduct of the study, in the collection, analysis, and interpretation of the data, and in the preparation, review, or approval of the manuscript.
References
- 1.Berg G, Knaape C, Ballin G, Seidel D. Biological control of Verticillium dahliae KLEB by naturally occurring rhizosphere bacteria. Arch Phytopathol Dis Prot. 1994;29:249–262. [Google Scholar]
- 2.Debette J, Blondeau R. Presence of Pseudomonas maltophilia in the rhizosphere of several cultivated plants. Can J Microbiol. 1980;26:460–463. [PubMed] [Google Scholar]
- 3.Heuer H, Smalla K. Bacterial phyllosphere communities of Solanum tuberosum L and T4-lysozyme producing genetic variants. FEMS Microbiol Ecol. 1999;28:357–371. [Google Scholar]
- 4.Lambert B, Joos H. Fundamental aspects of rhizobacterial plant growth promotion research. Trends Biotechnol. 1989;7:215–219. [Google Scholar]
- 5.Whipps J. Microbial interactions and biocontrol in the rhizosphere. J Exp Bot. 2001;52:487–511. doi: 10.1093/jexbot/52.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- 6.Binks PR, Nicklin S, Bruce NC. Degradation of RDX by Stenotrophomonas maltophilia PB1. Appl Environ Microbiol. 1995;61:1813–1822. doi: 10.1128/aem.61.4.1318-1322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee EY, Jun YS, Cho KS, Ryu HW. Degradation characteristics of toluene, benzene, ethylbenzene, and xylene by Stenotrophomonas maltophilia T3-c. J Air Waste Manag Assoc. 2002;52:400–406. doi: 10.1080/10473289.2002.10470796. [DOI] [PubMed] [Google Scholar]
- 8.Juhasz AL, Stanley GA, Britz ML. Microbial degradation and detoxification of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia strain VUN 10,003. Lett Appl Microbiol. 2000;30:396–401. doi: 10.1046/j.1472-765x.2000.00733.x. [DOI] [PubMed] [Google Scholar]
- 9.Quinn JP. Clinical problems posed by multiresistant nonfermenting gram-negative pathogens. Clin Infect Dis. 1998;27:117–124. doi: 10.1086/514912. [DOI] [PubMed] [Google Scholar]
- 10.Valdezate S, Vindel A, Loza E, Baquero F, Canton R. Antimicrobial susceptibilities of unique Stenotrophomonas maltophilia clinical strains. Antimicrob Agents Chemother. 2001;45:1581–1584. doi: 10.1128/AAC.45.5.1581-1584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignani MC, Grazziutti M, Anaissie E. Stenotrophomonas maltophilia infections. Semin Respir Crit Care Med. 2003;24:89–98. doi: 10.1055/s-2003-37920. [DOI] [PubMed] [Google Scholar]
- 12.Yamazaki E, Ishii J, Sato K, Nakae T. The barrier function of the outer membrane of Pseudomonas maltophilia in the diffusion of saccharides and beta-lactam antibiotics. FEMS Microbiol Lett. 1989;51:85–88. doi: 10.1016/0378-1097(89)90082-7. [DOI] [PubMed] [Google Scholar]
- 13.Li XZ, Zhang L, Poole K. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2002;46:333–43. doi: 10.1128/AAC.46.2.333-343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonso A, Martinez JL. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2000;44:3079–3086. doi: 10.1128/aac.44.11.3079-3086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Li XZ, Poole K. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2001;45:3497–3503. doi: 10.1128/AAC.45.12.3497-3503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol. 2005;7:1673–85. doi: 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- 17.Knudsen GR, Walter MV, Porteous LA, Prince VJ, Amstrong JL, et al. Predictive model of conjugated plasmid transfer in the rhizosphere and phyllosphere. Appl Environ Microbiol. 1988;54:343–347. doi: 10.1128/aem.54.2.343-347.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dungan RS, Yates SR, Frankenberger WT., Jr Transformations of selenate and selenite by Stenotrophomonas maltophilia isolated from a seleniferous agricultural drainage pond sediment. Environ Microbiol. 2003;5:287–295. doi: 10.1046/j.1462-2920.2003.00410.x. [DOI] [PubMed] [Google Scholar]
- 19.Sauge-Merle S, Cuine S, Carrier P, Lecomte-Pradines C, Luu DT, et al. Enhanced toxic metal accumulation in engineered bacterial cells expressing Arabidopsis thaliana phytochelatin synthase. Appl Environ Microbiol. 2003;69:490–494. doi: 10.1128/AEM.69.1.490-494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alonso A, Sanchez P, Martinez JL. Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob Agents Chemother. 2000;44:1778–1782. doi: 10.1128/aac.44.7.1778-1782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, et al. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell. 2002;9:713–723. doi: 10.1016/s1097-2765(02)00500-2. [DOI] [PubMed] [Google Scholar]
- 22.Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J Bacteriol. 2003;185:1942–50. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes JD, Richardson DJ, Saed S, Evans-Gowing R, Russell DA, et al. Cadmium-specific formation of metal sulfide ‘Q-particles’ by Klebsiella pneumoniae. Microbiology. 1997;143:2521–2530. doi: 10.1099/00221287-143-8-2521. [DOI] [PubMed] [Google Scholar]
- 24.Sharma PK, Balkwill DL, Frenkel A, Vairavamurthy MA. A new Klebsiella planticola strain (Cd-1) grows anaerobically at high cadmium concentrations and precipitates cadmium sulfide. Appl Environ Microbiol. 2000;66:3083–3087. doi: 10.1128/aem.66.7.3083-3087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kredich NM, Foote LJ, Keenan BS. The stoichiometry and kinetics of the inducible cysteine desulfhydrase from Salmonella typhimurium. J Biol Chem. 1973;218:6187–6196. [PubMed] [Google Scholar]
- 26.Wang CL, Lum AM, Ozuna SC, Clark DS, Keasling JD. Aerobic sulfide production and cadmium precipitation by Escherichia coli expressing the Treponema denticola cysteine desulfhydrase gene. Appl Microbiol Biotechnol. 2001;56:425–430. doi: 10.1007/s002530100660. [DOI] [PubMed] [Google Scholar]
- 27.Pagès D, Sanchez L, Conrod S, Gidrol X, Fekete A, et al. Exploration of intraclonal strategies of Pseudomonas brassicacearum facing Cd toxicity. Environ Microbiol. 2007;9:2820–35. doi: 10.1111/j.1462-2920.2007.01394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rijstenbil JW, Wijnholds JA. HPLC analysis of nonprotein thiols in planktonic diatoms: pool size, redox state and response to copper and cadmium exposure. Mar Biol. 1996;127:45–54. [Google Scholar]
- 29.Michalowicz A. Logiciels pour la Chimie. Paris: Société Francaise de Chimie; 1991. p. 102. [Google Scholar]
- 30.Teo BK. Berlin: Springer-Verlag; 1986. Inorganic Chemistry Concepts. [Google Scholar]
- 31.Rehr JJ, Albers RC. Scattering-matrix formulation of curved-wave multiple-scattering theory: Application to x-ray-absorption fine structure. Phys Rev B Condens Matter. 1990;41:8139–8149. doi: 10.1103/physrevb.41.8139. [DOI] [PubMed] [Google Scholar]
- 32.Zabinsky SI, Rehr JJ, Ankudinov A, Albers RC, Eller MJ. Multiple-scattering calculations of x-ray-absorption spectra. Phys Rev B. 1995;52:2995–3009. doi: 10.1103/physrevb.52.2995. [DOI] [PubMed] [Google Scholar]