Abstract
Mice deficient for the B cell–restricted transcription factor Pax5 show a defect in the VH to DJH rearrangement step of immunoglobulin heavy chain gene assembly even though the expression of the V(D)J recombinase is not diminished in Pax5 −/− pro–B cells. To investigate whether Pax5 is limiting for VH to DJH rearrangement, we generated transgenic mice which express Pax5 in developing thymocytes. We show that enforced expression of Pax5 in thymocytes results in a partial block in T cell development due to defective pre-TCR signaling in β-selection. Moreover, our results demonstrate that expression of Pax5 in early thymocytes is sufficient to induce VH to DJH rearrangements in CD4+CD8+ T cells and lead us to suggest that Pax5 may play a direct role in the lineage-specific regulation of immunoglobulin heavy chain gene rearrangement.
Keywords: lymphocyte development, V(D)J recombination, gene expression regulation, transgenesis, immunoglobulin
Introduction
V(D)J recombination is regulated in several ways (1). First, it is lineage specific, with Ig genes completely rearranging only in B cells and TCR genes completely rearranging only in T cells. Second, there is a preferred order to the rearrangement process with Ig heavy chain (IgHC) locus preceding Ig light chain (IgLC) locus rearrangement in B cells and TCRβ locus preceding TCRα locus rearrangement in T cells. Also, in both the IgHC and TCRβ loci D to J rearrangement precedes V to DJ rearrangement and direct V to D rearrangement occurs rarely if at all. Finally, the gene rearrangement process at most loci is subject to allelic exclusion. That is, an individual B or T cell makes only one productive (in frame) rearrangement at each locus, resulting in its ability to produce only a single Ig or TCR molecule.
Regulated changes in chromatin structure underlie most if not all aspects of substrate choice by the V(D)J recombinase (2). This idea, known as the “accessibility hypothesis,” arose from initial observations which correlated the transcription of unrearranged gene segments with their activation for rearrangement. It was shown subsequently that mutations which eliminate promoters or enhancers associated with rearranging loci greatly diminish rearrangement of the altered locus (1). Additional work has implicated specific transcription factors in the regulation of recombinase accessibility. For example, overexpression of either E47 or EBF can activate chromosomal Ig gene rearrangement in nonlymphoid cells transfected with RAG1 and RAG2 (3). Furthermore, mice with a null mutation in the Pax5 gene, which encodes the B cell–restricted transcription factor Pax5 (BSAP), show a profound block in B cell development at the pro–B cell stage (4). Most Pax5-null pro–B cells contain DJH but not VDJH rearrangements at their IgHC loci, leading to the suggestion that Pax5 is necessary to fully activate the VH to DJH step in IgHC gene assembly.
Two other aspects of the regulation of IgHC gene rearrangement focus particular interest on the V to DJ step. First, this is the step which is subject to allelic exclusion. Nearly all developing B cells undergo D to J rearrangement on both HC alleles, but only one of these alleles undergoes productive V to DJ rearrangement. Furthermore, IgHC transgenic mice show normal D to J rearrangement but greatly diminished V to DJ rearrangement at their endogenous IgHC loci (1). Finally, this is the step which shows strict lineage specificity. IgHC D to J rearrangement occurs frequently in developing T cells (5), but V to DJ rearrangement is limited to developing B cells.
Given the fact that the state of IgHC rearrangement in Pax5-null pro–B cells is similar to that in normal developing T cells and that developing T cells do not express Pax5, we hypothesized that T cells perform DH to JH but not VH to DJH rearrangement because Pax5 is absolutely required for the latter step in IgHC gene assembly. To test this idea, we have generated and analyzed Pax5 transgenic mice which express this B lineage transcription factor in developing thymocytes.
Materials and Methods
Antibodies and Flow Cytometry.
The following antibodies were purchased from either BD Biosciences or CALTAG Laboratories: FITC–anti-CD3ɛ (145–2C11), -CD4 (RM4–5), -CD8α (53–6.7), -Thy1.2 (CD90.2) (53–2.1); PE–anti-CD25 (PC61), -CD4 (RM4–5), -CD19 (1D3); CyChrome–anti-CD44 (IM7); and bi–anti-CD4 (RM4–5), -CD8α (CT-Cd8α). For analysis, live cells were gated based on forward and side scatter, and over 100,000 events were collected. CD4/CD8 double negative T cell (DN) and CD4/CD8 double positive T cell (DP) thymocytes were purified with an AutoMACS magnetic cell sorter (Miltenyi) as described (6) or using a MoFlo cell sorter (Cytometrix). Routinely, purity of all cell preparations was >95% unless otherwise specified in the text.
Rearrangement PCR and Ligation-mediated–PCR.
100 ng of genomic DNA from DP or pre–B (CD43−B220+IgM−) cells was used to assay for IgHC gene rearrangements. PCR assays for DH to JH and VH to DJH rearrangements were performed as described (7). The locus-specific PCR primers and probes used for ligation-mediated (LM)–PCR were described previously (8, 9).
PCR Fragment Length Polymorphism.
Total thymocytes were first depleted of DP and SP populations by incubation of cell suspensions with biotinylated antibodies specific for CD4, CD8, and CD3ɛ followed by negative selection using streptavidin-conjugated microbeads (Miltenyi). DN cells were further stained with PE-conjugated anti-CD25 and CyChrome-conjugated anti-CD44 antibodies. CD44−CD25+ DN3 cells were purified using a MoFlo cell sorter (Cytometrix). PCR amplification of the TCRβ CDR3 region using specific V and J primers was performed essentially as described (10).
Online Supplemental Material.
Fig. S1 shows Western blot and RT-PCR analyses of transgene expression. Fig. S2 displays CD19 expression in Pax5 transgenic thymus. Fig. S3 shows RT-PCR analysis of gene expression in Pax5 transgenic thymus. Fig. S4 shows CDR3-length analysis of VDJβ rearrangements in Pax5 transgenic and control thymocytes. Fig. S5 shows VDJH rearrangement in Pax5 transgenic thymocytes from Igβ2/− mice. In addition, Supplemental Materials and Methods are provided regarding the generation of the transgenic construct, the performance of RT-PCR, and the analysis of transgene expression by Western blot. Figs. S1–S5 and Supplemental Materials and Methods are available at http://www.jem.org/cgi/content/full/jem.20032249/DC1.
Results and Discussion
Impaired T Cell Development in CD2EP-Pax5 Transgenic Mice.
A murine Pax5 cDNA, cloned in between the human CD2 promoter and its locus control region, was used to generate transgenic mice by pronuclear injection (11). Analysis by Western blot of whole cell thymic extracts from multiple founder lines showed that in two founders the expression level of Pax5 appeared comparable to that in the BM (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20032249/DC1).
In three independent lines of CD2EP-Pax5 transgenic mice, we observed an approximately threefold decrease in thymocyte numbers compared with those of control littermates (Fig. 1 B, data shown from only two founders). Analysis of CD4/CD8 expression revealed a fivefold increase in the percentage of DN thymocytes and a slight decrease in the percentage of DP thymocytes in the transgenic mice compared with littermate controls (Fig. 1 A, top). Interestingly, the absolute number of DN thymocytes in transgenic mice remained similar to that in control littermates, whereas the number of DP thymocytes was considerably decreased (Fig. 1 B). We went on to examine DN thymocyte populations for CD44 and CD25 expression and observed a small increase in the proportion of CD44−CD25+ (DN3) cells and a decrease in the proportion of CD44+CD25− (DN1) cells in the transgenic mice (Fig. 1 A). The decrease in DN1 cells may be due to expression of Pax5 in a fraction of early thymic progenitors leading to their inefficient entry into the pool of developing thymocytes (12). Collectively, these observations indicate that ectopic expression of Pax5 in early thymocytes leads to a partial developmental block at the DN to DP transition.
Figure 1.
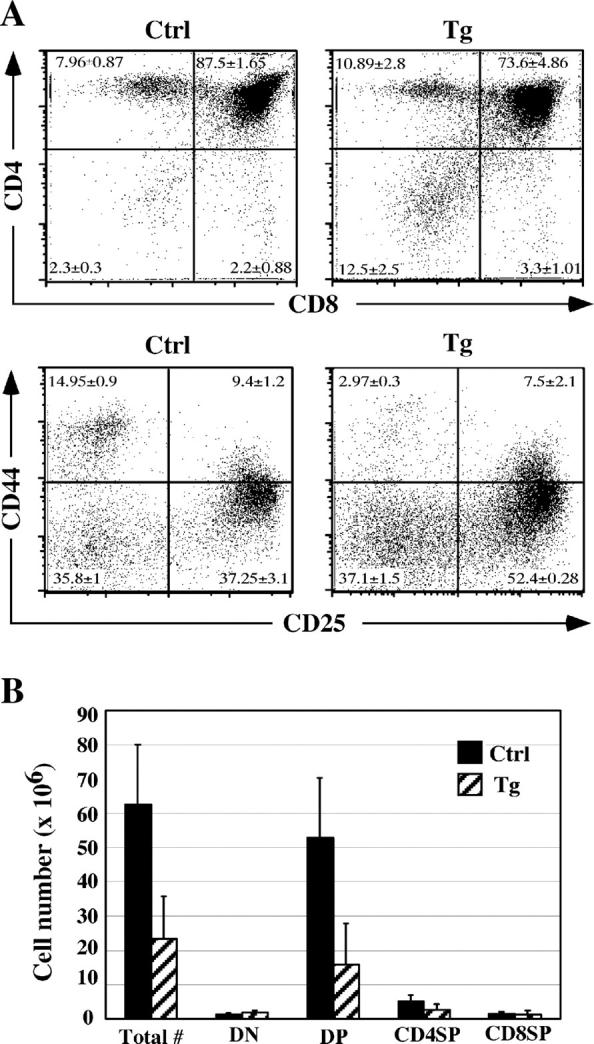
Impaired T cell development in CD2EP-Pax5 transgenic mice. (A) FACS® analysis of CD4 and CD8 expression on thymocytes from 4-wk-old littermate control or transgenic mice (top). Electronically gated CD3−CD4−CD8− DN thymocytes from 2-wk-old control or transgenic mice were stained with anti-CD25 and anti-CD44 antibodies (bottom). Percentages represent the means and SDs of live-gated subpopulations from six mice of each genotype. (B) Absolute cell numbers were calculated for total thymocytes and the thymocyte subsets described in A (top). The error bars represent mean ± SD values for control and transgenic mice (n = 12).
Appearance of Rare CD19+ Cells in the Thymi of CD2EP-Pax5 Transgenic Mice.
Given that ectopic Pax5 expression inhibits T cell development and B cell commitment depends on Pax5 (12), we asked whether overexpression of Pax5 in early thymocyte precursors promotes intrathymic B cell development. We found a small fraction (∼1–3%) of CD19+ cells in the thymus of transgenic mice but not in control littermates (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20032249/DC1). The majority of the CD19+ cells from founder #10 coexpress CD4 and CD8 or Thy1.2 (not depicted), whereas CD19 is not coexpressed with CD4 and CD8 on cells from founder #2. The level of CD19 expression on thymocytes from founder #10 is clearly greater than that expressed on littermate controls but far less than that expressed on wild-type splenic B cells (Fig. S2 C).
To test whether these T cells also express other B lineage markers, we purified DP thymocytes by flow cytometry from either transgenic or control mice and used RT-PCR to assay for expression of early B cell–related molecules. We found that Pax5 transgenic DP T cells express the direct Pax5 targets CD19 and BLNK (13) but do not express other early B cell–specific transcripts such as λ5, VpreB, and Igβ (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20032249/DC1). We conclude from these studies that Pax5 expression in thymocytes activates direct Pax5 targets but does not activate the full genetic program of early B cell development.
These and more detailed analyses failed to reveal a quantitative increase in B cell development in the BM or the induction of significant amounts of B cell development in the thymus (not depicted). These findings are in contrast to those from a recently reported study of knock-in mice in which Pax5 was placed under the control of the Ikaros locus (12). In these mice, Pax5 expression in hematopoietic stem cells resulted in an extreme skewing of lymphopoiesis toward the B cell lineage. T cell development was blocked, presumably due to the repression of Notch1 transcription (12). The discrepancy between our results and those of Souabini et al. could be due to a difference in the onset of ectopic Pax5 expression. It was shown previously that expression of a similar hCD2 promoter/locus control region–driven transgene is first detected at the DN1 stage of T cell development in a heterogeneous fashion but that transgene expression does not become uniform in all thymocytes until the DN3 to DN4 transition (11). Thus, it seems likely that commitment to the T cell lineage takes place before the expression of our CD2 promoter–driven Pax5 transgene. In distinction to the previous study, the relatively later onset of Pax5 expression in our transgenic system allows one to assess the effects of Pax5 expression after the onset of early T cell development.
Ectopic Pax5 Expression in the Thymus Inhibits β-Selection.
Transition from the DN to DP stage, often referred to as β-selection, is accompanied by various phenotypic changes, including cell cycle entry and selection for in-frame TCRβ rearrangement (14). To test whether the increased percentage of DN cells in the thymi of transgenic mice was a consequence of faulty β-selection, we first analyzed DN3 (CD25+CD44−) cells for forward scatter as an indirect marker of cell cycle activation. Strikingly, we found a significant reduction in the percentage of large, presumably cycling cells within the DN3 population of Pax5 transgenic mice (Fig. 2 A). To test whether this defect was due to deficient TCRβ locus rearrangement, we measured recombination reaction intermediates (broken signal ends) in sorted DN3 and DN4 cells from control and transgenic mice (Fig. 2 B). We found that dsDNA breaks at both the 3′ and 5′ of Dβ1 RSSs, which are associated with Dβ to Jβ and Vβ to DJβ rearrangements, respectively, appear slightly increased in DN3 cells from transgenic mice compared with littermate controls. Thus, the lower percentage of large cycling cells in transgenic thymocytes cannot be explained by deficient assembly of genes encoding the TCRβ chain.
Figure 2.
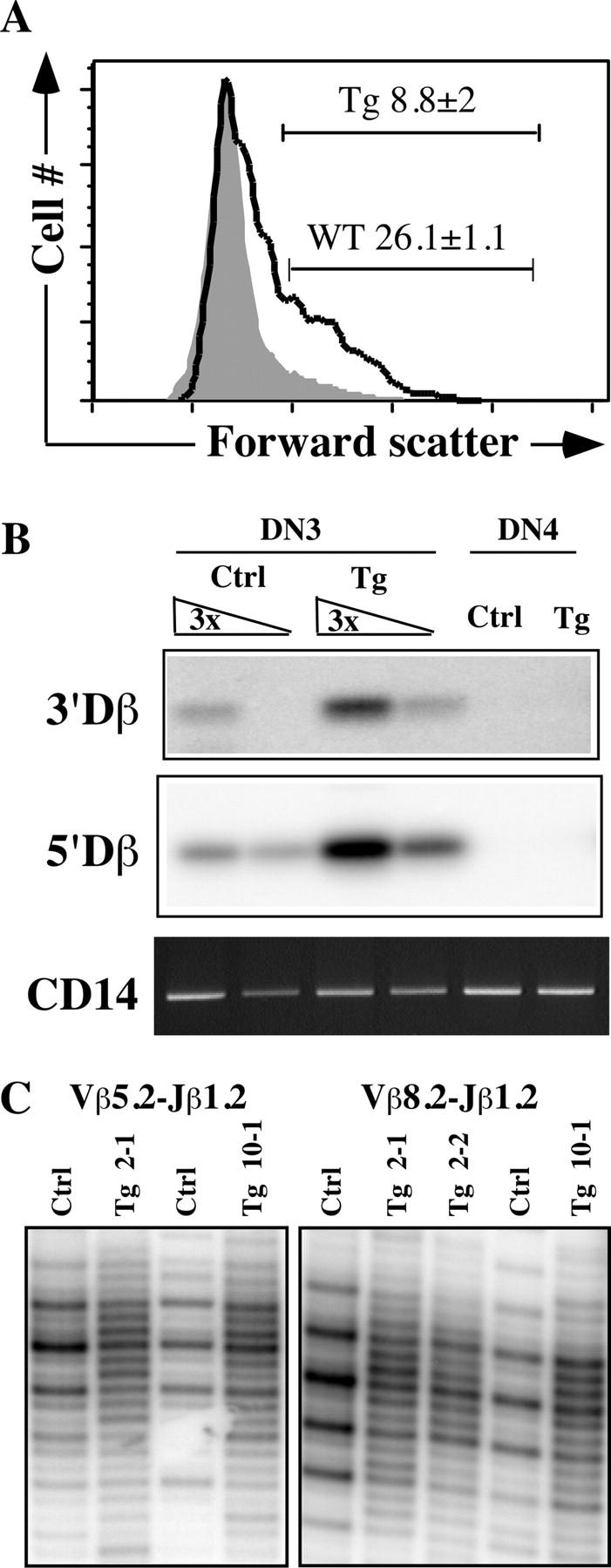
Pax5 expression in the thymus affects β-selection but not TCRβ gene segment rearrangement. (A) Gated DN3 (CD44−CD25+) cells were analyzed for forward scatter. The results are displayed as overlapping histograms of control (WT, unshaded) and transgenic (Tg, shaded) thymocytes. The percentage of large cycling cells is indicated, including the mean and SD (n = 3). (B) Linker-ligated DNAs from control and transgenic DN3 and DN4 T cells were subjected to LM-PCR with primers specific for the detection of double stranded breaks at the RSSs 3′ or 5′ of Dβ1 as indicated. CD14 is a nonrearranging locus control PCR. (C) PCR fragment length polymorphism analysis of the CDR3 region of fully rearranged TCRβ alleles using template DNA purified from sorted DN3 cells from control (Ctrl) or transgenic (Tg) mice (some assays used two independent samples of founder #2 DNA). Results of analyses focusing on Vβ5.2 or 8.2 to Jβ1.2 rearrangements are shown. Similar results were obtained using a downstream primer specific for Jβ2.1 (not depicted).
To determine whether the defect in β-selection might be due to a generalized disruption of the transcriptional program in progenitor T cells associated with the expression of this B lineage transcription factor, we used real-time RT-PCR to compare the levels of preTα, Notch1, and LEF-1 mRNA in DN thymocytes from transgenic and control mice (Fig. S3 C). No significant differences were observed, indicating that ectopic Pax5 expression does not affect expression of these critical genes.
Signals derived from the pre-TCR complex result in clonal expansion of thymocytes with functionally rearranged TCRβ chains (14). To determine whether ectopic Pax5 expression disrupts this aspect of β-selection, we used a PCR fragment–length polymorphism assay to assess both TCRβ CDR3 length heterogeneity and the distribution of in-frame rearrangements in sorted DN3 cells from control and transgenic mice. The imprecise nature of V(D)J joining results in a continuous distribution of CDR3 amplicon fragment lengths. Rapid cell division among cells with a productive TCRβ rearrangement results in the enrichment of PCR fragments whose lengths correspond to potential open reading frames within TCRβ. Using primers that amplify across the V(D)J junction within TCRβ alleles, PCR products generated using genomic DNA from control DN3 cells showed a length distribution with a periodicity of three nucleotides, which is indicative of enrichment for productive TCRβ rearrangements (Fig. 2 C and Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20032249/DC1). In contrast, CDR3 length distribution in DN3 cells from transgenic mice was relatively random, suggesting a defect in β-selection. We hypothesize that defects in pre-TCR signaling in Pax5 transgenic mice may result from up-regulation of BLNK and CD19 expression in early thymocytes. These components of the B cell signaling machinery may interfere with the function of the T cell–specific adaptors SLP-76 or LAT. Additional work will be required to test this hypothesis.
Pax5 Expression Is Sufficient to Induce VH to DJH Recombination at the IgHC Locus.
To test whether Pax5 plays an important role in controlling the B cell–specific VH to DJH rearrangement step of IgHC gene assembly, we examined the rearrangement status of the IgHC locus in sorted DP thymocytes from transgenic and control mice using a direct PCR assay (7). The purity of these sorted cells was >98%. Consistent with earlier work (5), frequent DH to JH rearrangements were readily detectable in both BM pre–B cells and DP thymocytes from either transgenic or control mice (Fig. 3 A). Interestingly, although the DP thymocytes from control mice lacked VH to DJH rearrangements, these rearrangements were readily detectable in DP thymocytes from transgenic mice (Fig. 3 B). By comparing PCR signals between transgenic DP thymocytes and sorted BM pre–B cells from wild-type mice, we estimate the frequency of VH to DJH rearrangements in DP Pax5 transgenic thymocytes to be ∼5–10% that of BM pre–B cells (Fig. 3 B).
Figure 3.
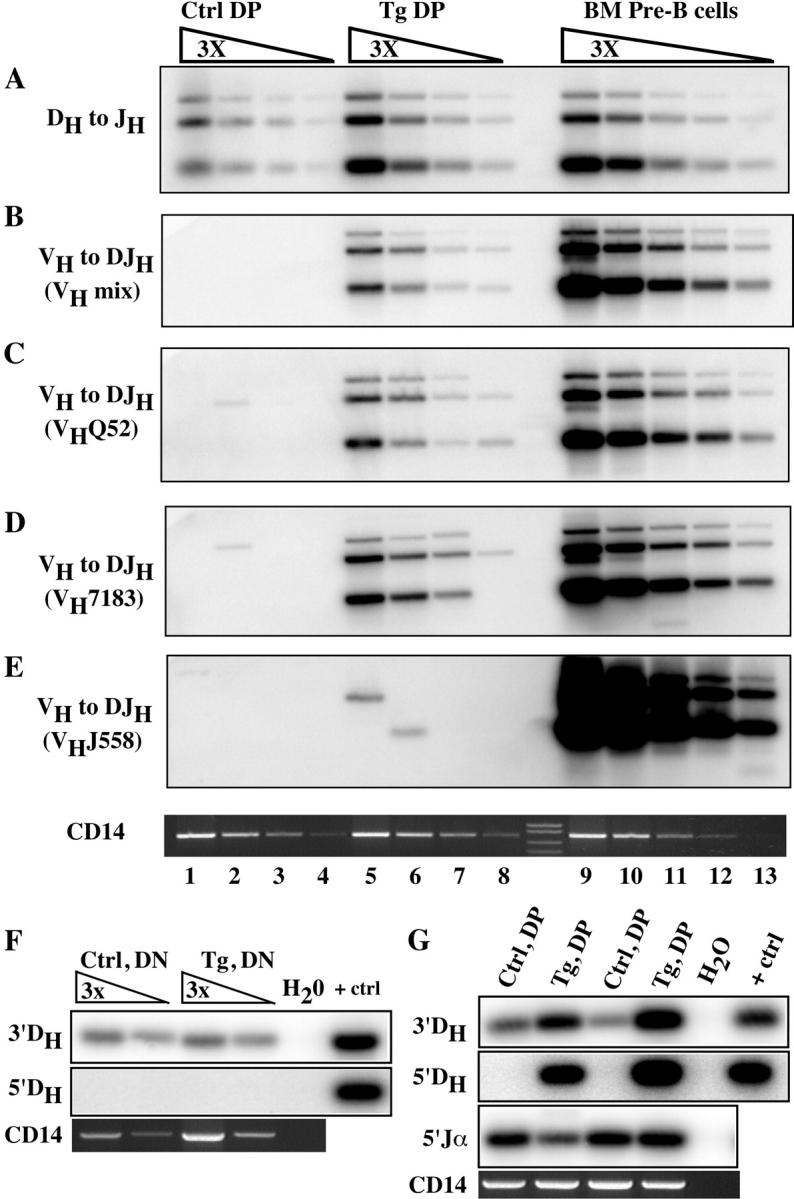
Ectopic Pax5 expression induces VH to DJH rearrangement in transgenic DP thymocytes. Threefold serially diluted genomic DNAs from FACS®-sorted control DP thymocytes (lanes 1–4), transgenic DP thymocytes (lanes 5–8), or wild-type BMpre–B cells (B220+CD43−IgM+) (lanes 9–13) were subjected to PCR analysis for DH to JH (A) and VH to DJH (B–E) gene rearrangements. An equimolar mixture of degenerate VHJ558, VH7183, and VHQ52 family-specific primers (VH mix) paired with a unique primer hybridizing 3′ of JH3 was used in B, whereas each degenerate VH primer was used in isolation in C–E as indicated. Genomic DNA samples purified from sorted DN (F) and DP (G) thymocytes from two independent samples of control and Pax5 transgenic mice were analyzed by LM-PCR to detect broken signal end recombination reaction intermediates associated with DH to JH rearrangement (3′DH, top), VH to DJH rearrangement (5′DH, middle), and Vα to Jα rearrangement (5′Jα, bottom). Amplification of a nonrearranging control locus (CD14) was used as a DNA recovery control. H2O denotes reactions lacking DNA template and + ctrl denotes control assays which contained BM DNA.
This observation was further supported by examination of V(D)J recombination reaction intermediates using a previously described LM-PCR approach. Double strand DNA breaks at RSSs 5′ of DH, which represent intermediates in the VH to DJH rearrangement step, were detected in DNA purified from the DP thymocytes of transgenic but not control mice, whereas DNA breaks associated with TCRα locus rearrangement were detected in all samples (Fig. 3 G). Interestingly, we failed to detect 5′ of DH broken ends in DN thymocytes from either control or transgenic mice (Fig. 3 F), leading us to conclude that Pax5-induced VH to DJH rearrangements occur mainly during the DP stage of T cell development. In contrast, 3′ of DH breaks were detected in DN and DP cells from both transgenic and control mice (Fig. 3, F and G). The level of these breaks was greater in DP samples from the transgenic samples, corresponding to greater levels of DJH joints observed by direct PCR (Fig. 3 A). We obtained similar results when we crossed the CD2EP-Pax5 transgene onto a Igβ2/− genetic background where B cell development is blocked at an early stage (15) (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20032249/DC1). This led us to conclude that contaminating B cells could not account for the VH to DJH rearrangements observed in transgenic thymocytes. In summary, our results indicate that expression of Pax5 alone in early thymocytes is sufficient to induce VH to DJH rearrangement in T lineage cells.
It was reported recently that pro–B cells from Pax5-deficient mice show a far more significant defect in rearrangement of the DH-distal VHJ558 gene family relative to that of the more DH-proximal VH gene families (16). Therefore, we used PCR assays to separately examine the use of VHJ558, VH7183, and VHQ52 gene segments. As shown in Fig. 3, C and D, the VH7183 and VHQ52 gene families accounted for the vast majority of VH to DJH rearrangements in DP thymocytes from transgenic mice. In striking contrast, rearrangements using the VHJ558 gene family were barely detectable in transgenic thymocytes (Fig. 3 E). In addition, we were able to detect VH7183 but not VHJ558 gene segment transcripts in transgenic DP thymocytes (not depicted). Thus, our findings suggest that the extent of VH to DJH rearrangement induced by ectopic Pax5 expression in the thymus progressively decreases as the distance of a VH gene family from the DH cluster increases. However, the results of Hesslein et al. (16) suggest just the opposite—that rearrangement of the distal VHJ558 gene family in pro–B cells is far more dependent on Pax5 than rearrangement of the more proximal VH gene families. These differences may be due to differential expression of any of a large number of other transacting factors in B and T cell progenitors.
Given the fact that pro–B cells from Pax5−/− mice display amounts of VHJ558 germline transcription and histone acetylation that are indistinguishable from wild-type pro–B cells, these workers (16) concluded that Pax5 regulates the accessibility of VHJ588 gene segments at a level distinct from germline transcription and histone acetylation. Although histone acetylation appears to correlate well with accessibility of the V(D)J recombinase in many cases (17–19), we were also unable to detect changes in H3 or H4 acetylation which correlated with the Pax5-dependent activation of VH to DJH rearrangement in DP transgenic thymocytes (not depicted). Surprisingly, we did find that the distal VHJ558 family of IgHC gene segments was relatively hyperacetylated (although not to the same level as seen in pro–B cells) in both control and Pax5 transgenic thymus even though they were not found to be accessible to the recombinase in either instance (not depicted). Thus, it appears that variations in histone acetylation may not in a simple fashion explain the selectivity of VH accessibility. Additional work will be required to determine whether other specific modifications more closely track with VH gene segment accessibility.
It is possible that the preferential rearrangement of DH-proximal VH gene segments observed in the DP transgenic thymocytes is a result of the IL-7 dependence of distal VH gene segment rearrangement. Several labs have shown that the IL-7 signaling pathway is involved in promoting local accessibility of VHJ558 genes (18, 20). Expression of the IL-7Rα chain is low at the DP stage of T cell development (21). Perhaps VHJ558 genes are inactive at the DP stage because low levels of IL-7Rα expression make these DP thymocytes poorly responsive to IL-7. Therefore, it is possible that IL-7R signaling and Pax5 cooperate in the control of distal VH gene segment recombination. However, given the phenotypic similarity between IL-7Rα−/− and Pax5 −/− mice and the observation that Pax5 transcript levels are decreased in IL-7Rα−/− BM cells (20), we cannot rule out the possibility that Pax5 acts as a downstream target of IL-7R signaling in developing B cells.
This work shows that Pax5 expression is sufficient to activate VH to DJH rearrangement in T cells and by inference may play a similar role in B cells. Evidence for the involvement of specific transcription factors in modulating the accessibility of specific gene segments to the recombination machinery is scarce. In one study, STAT5 was shown to bind to and promote recombinase accessibility of the TCRγ locus (22). Several studies have implicated E2A proteins (E12 and E47) in the regulation of both Ig and TCR gene segment recombination (3, 23, 24). These effects might depend on the known interaction between transcription factors and chromatin remodeling complexes. However, we have been unable to convincingly demonstrate direct Pax5 interaction with VH gene segments using a ChIP approach. Thus, further studies on the interactions of various chromatin-modifying complexes with VH region chromatin in transgenic mice should provide a better understanding of how Pax5 facilitates targeting of recombination machinery to specific VH gene segments.
Acknowledgments
We acknowledge Dr. Paul Love (National Institutes of Health, Bethesda, MD) for the pCD2EP transgene vector, Dr. Michel Nussenzweig (The Rockefeller University, New York, NY) for Igβ2/2 mice, and Ms. Dan Huang for performing the RT-PCR experiment in Fig. S1. We are especially grateful to Dr. Kathryn Calame for providing advice throughout and helpful criticism of this manuscript.
This work was funded by grants from the National Institutes of Health to M.S. Schlissel.
The online version of this article contains supplemental material.
Abbreviations used in this paper: BSAP, B cell–restricted transcription factor Pax5; DN, CD4/CD8 double negative T cells; DP, CD4/CD8 double positive T cells; IgHC, Ig heavy chain; IgLC, Ig light chain; RSS, recombination signal sequence; LM, ligation-mediated.
References
- 1.Schlissel, M.S. 2003. Regulating antigen receptor gene assembly. Nat. Rev. Immunol. 3:890–899. [DOI] [PubMed] [Google Scholar]
- 2.Stanhope-Baker, P., K.M. Hudson, A.L. Shaffer, A. Constantinescu, and M.S. Schlissel. 1996. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 85:887–897. [DOI] [PubMed] [Google Scholar]
- 3.Romanow, W.J., A.W. Langerak, P. Goebel, I.L. Wolvers-Tettero, J.J. van Dongen, A.J. Feeney, and C. Murre. 2000. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell. 5:343–353. [DOI] [PubMed] [Google Scholar]
- 4.Nutt, S.L., P. Urbanek, A. Rolink, and M. Busslinger. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476–491. [DOI] [PubMed] [Google Scholar]
- 5.Kemp, D., A. Harris, S. Cory, and J. Adams. 1980. Expression of the immunoglobin Cμ gene in mouse T and B lymphoid and myeloid cell lines. Proc. Natl. Acad. Sci. USA. 77:2876–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu, L.Y., J. Lauring, H.E. Liang, S. Greenbaum, D. Cado, Y. Zhuang, and M.S. Schlissel. 2003. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 19:105–117. [DOI] [PubMed] [Google Scholar]
- 7.Schlissel, M.S., L.M. Corcoran, and D. Baltimore. 1991. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173:711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang, H.E., L.Y. Hsu, D. Cado, L.G. Cowell, G. Kelsoe, and M.S. Schlissel. 2002. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 17:639–651. [DOI] [PubMed] [Google Scholar]
- 9.Livak, F., and D.G. Schatz. 1996. T-cell receptor alpha locus V(D)J recombination by-products are abundant in thymocytes and mature T cells. Mol. Cell. Biol. 16:609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yassai, M., K. Ammon, J. Goverman, P. Marrack, Y. Naumov, and J. Gorski. 2002. A molecular marker for thymocyte-positive selection: selection of CD4 single-positive thymocytes with shorter TCRB CDR3 during T cell development. J. Immunol. 168:3801–3807. [DOI] [PubMed] [Google Scholar]
- 11.De Boer, J., A. Williams, G. Skavdis, N. Harker, M. Coles, M. Tolaini, T. Norton, K. Williams, K. Roderick, A.J. Potocnik, and D. Kioussis. 2003. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33:314–325. [DOI] [PubMed] [Google Scholar]
- 12.Souabni, A., C. Cobaleda, M. Schebesta, and M. Busslinger. 2002. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 17:781–793. [DOI] [PubMed] [Google Scholar]
- 13.Nutt, S.L., A.M. Morrison, P. Dorfler, A. Rolink, and M. Busslinger. 1998. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 17:2319–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borowski, C., C. Martin, F. Gounari, L. Haughn, I. Aifantis, F. Grassi, and H. von Boehmer. 2002. On the brink of becoming a T cell. Curr. Opin. Immunol. 14:200–206. [DOI] [PubMed] [Google Scholar]
- 15.Gong, S., and M.C. Nussenzweig. 1996. Regulation of an early developmental checkpoint in the B cell pathway by Ig beta. Science. 272:411–414. [DOI] [PubMed] [Google Scholar]
- 16.Hesslein, D.G., D.L. Pflugh, D. Chowdhury, A.L. Bothwell, R. Sen, and D.G. Schatz. 2003. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 17:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMurry, M.T., and M.S. Krangel. 2000. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 287:495–498. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhury, D., and R. Sen. 2001. Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J. 20:6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, K., C. Angelin-Duclos, S. Park, and K.L. Calame. 2003. Changes in histone acetylation are associated with differences in accessibility of V(H) gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell. Biol. 23:2438–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corcoran, A.E., A. Riddell, D. Krooshoop, and A.R. Venkitaraman. 1998. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 391:904–907. [DOI] [PubMed] [Google Scholar]
- 21.Fry, T.J., and C.L. Mackall. 2002. Interleukin-7: from bench to clinic. Blood. 99:3892–3904. [DOI] [PubMed] [Google Scholar]
- 22.Ye, S.K., Y. Agata, H.C. Lee, H. Kurooka, T. Kitamura, A. Shimizu, T. Honjo, and K. Ikuta. 2001. The IL-7 receptor controls the accessibility of the TCRgamma locus by Stat5 and histone acetylation. Immunity. 15:813–823. [DOI] [PubMed] [Google Scholar]
- 23.Fernex, C., M. Capone, and P. Ferrier. 1995. The V(D)J recombinational and transcriptional activities of the immunoglobulin heavy-chain intronic enhancer can be mediated through distinct protein-binding sites in a transgenic substrate. Mol. Cell. Biol. 15:3217–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlissel, M., A. Voronova, and D. Baltimore. 1991. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 5:1367–1376. [DOI] [PubMed] [Google Scholar]


