Abstract
Mycobacterium tuberculosis residing within pulmonary granulomas and cavities represents an important reservoir of persistent organisms during human latent tuberculosis infection. We present a novel in vivo model of tuberculosis involving the encapsulation of bacilli in semidiffusible hollow fibers that are implanted subcutaneously into mice. Granulomatous lesions develop around these hollow fibers, and in this microenvironment, the organisms demonstrate an altered physiologic state characterized by stationary-state colony-forming unit counts and decreased metabolic activity. Moreover, these organisms show an antimicrobial susceptibility pattern similar to persistent bacilli in current models of tuberculosis chemotherapy in that they are more susceptible to the sterilizing drug, rifampin, than to the bactericidal drug isoniazid. We used this model of extracellular persistence within host granulomas to study both gene expression patterns and mutant survival patterns. Our results demonstrate induction of dosR (Rv3133c) and 20 other members of the DosR regulon believed to mediate the transition into dormancy, and that rel Mtb is required for Mycobacterium tuberculosis survival during extracellular persistence within host granulomas. Interestingly, the dormancy phenotype of extracellular M. tuberculosis within host granulomas appears to be immune mediated and interferon-γ dependent.
Keywords: microarrays, gene expression, antibiotics, latency, persistence
Introduction
Mycobacterium tuberculosis infects approximately one third of the world's population, resulting in three million deaths annually (1). Soon after inhalation of tubercle bacilli, the organisms are phagocytosed by alveolar macrophages, resulting in potent cell-mediated immune responses and the formation of granulomas, which consist primarily of T cells and M. tuberculosis–infected macrophages (2, 3). 6–8 wk after infection in humans, and coincident with the development of a delayed-type hypersensitivity response manifested by tuberculin skin test positivity, these granulomas undergo caseous necrosis, resulting in the death of the majority of tubercle bacilli and destruction of surrounding host tissue (4). The small proportion of surviving bacilli are thought to exist in a nonreplicating hypometabolic state, as an adaptation to the unfavorable milieu in the solid caseous material (4). This altered physiologic state, termed latent tuberculosis infection, can endure for the lifetime of the infected individual, but in ∼10% of cases, through unknown mechanisms, these dormant bacilli reactivate many years to decades later to produce disease.
Efforts to gain insight into the adaptive mechanisms by which M. tuberculosis persists in the host have been impeded by the inability to recover sufficient quantities of M. tuberculosis RNA from host lesions consistent with contained latent tuberculosis infection (5). Consequently, several groups have turned to in vitro models that may reflect the persistent state, and have defined the gene expression profile of M. tuberculosis under conditions of hypoxia (6, 7), nutrient starvation (8), low pH (9), low concentrations of nitric oxide (10), and in the phagosomal compartment of murine macrophages (11). Current work has focused on the role of the two-component response regulator dormancy survival regulator (DosR), which initially was found to be the primary mediator of the hypoxic response in M. tuberculosis (6, 12). Bacilli exposed to low, nontoxic concentrations of nitric oxide in vitro enter a nonreplicating persistent state marked by the induction of a 48-gene regulon under the control of DosR, suggesting that the DosR regulon may mediate the transition of these bacilli into dormancy (10). Consistent with these findings, several DosR regulon genes, including acr (Rv2031c), Rv2623c, and Rv2626, are up-regulated in infected mouse tissues after the onset of Th1 immunity (10, 13).
We sought to mimic the caseous granuloma by physical containment of extracellular M. tuberculosis within mice as a means of comparing in vivo–cultivated persistent bacteria with in vitro latency models. Our model involves the encapsulation of tubercle bacilli in polyvinylidene fluoride (PVDF) hollow fibers (14, 15) and the implantation of these fibers into the subcutaneous space of mice (see Fig. 1). The PVDF fibers have a molecular mass cutoff of 500 kD, which allows for the diffusion of small soluble molecules, but prevents the entry of host immune cells and the exit of intact bacilli. The prevention of direct host–pathogen interactions imposed by the physical properties of the hollow fibers provides a unique opportunity to study the behavior of extracellular M. tuberculosis in the host.
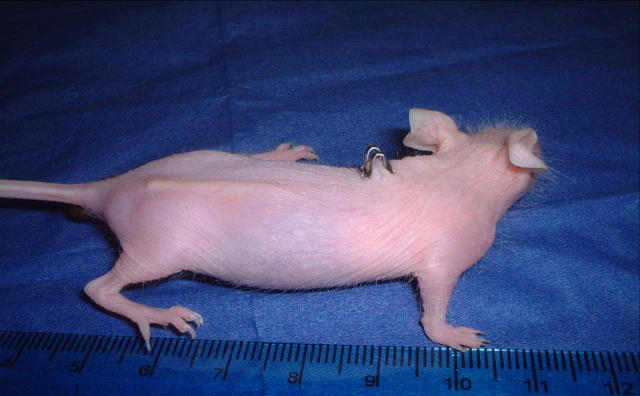
Figure 1.
SKH1 mouse with subcutaneously implanted hollow fiber containing M. tuberculosis.
Materials and Methods
Strains.
M. tuberculosis H37Rv-expressing firefly luciferase (H37Rv-lux) was passaged twice through mice and used for all infections. The organisms were grown in plastic roller bottles at 37°C in Middlebrook 7H9 liquid broth (Difco Laboratories) supplemented with 10% OADC (Becton Dickinson), 0.5% glycerol, and 0.05% Tween-80.
Hollow Fiber Assay.
Infection with M. tuberculosis was achieved in 6–8-wk-old female hairless, immunocompetent SKH1 mice (Charles River Laboratories) using the hollow fiber encapsulation/implantation technique as described previously (14, 15). In brief, liquid cultures of M. tuberculosis were inoculated into the lumen of PVDF hollow fibers (molecular mass cutoff 500 kD; Spectrum Laboratories) with a syringe and 20-gauge needle. The ends of the hollow fibers were heat sealed, and individual fibers were prepared by heat sealing at 2-cm intervals. Mice were anesthetized by intraperitoneal injection of 240 mg/kg of Avertin (2,2,2-tribromoethanol; Sigma-Aldrich), and the dorsal skin surface was sterilized with 70% ethanol. A small incision was made at the nape of the neck, and one fiber was deposited into the subcutaneous space of each flank with a tumor trocar (two fibers/mouse). Incisions were closed with a small surgical clip (Fig. 1). For immunology studies, BALB-C/J and IFNγ−/− (B6-IFNgtmITSIJ) mice were used (Jackson ImmunoResearch Laboratories). For experiments involving hollow fiber–encapsulated M. tuberculosis incubated in vitro, hollow fibers were incubated on a shaker at 37°C in 50-ml conical tubes containing 20 ml Middlebrook 7H9 liquid broth (Difco Laboratories) supplemented with 10% OADC (Becton Dickinson), 0.5% glycerol, and 0.05% Tween-80. For determination of CFU counts, hollow fibers were recovered from mice at the time of killing, and their contents were plated on 7H10 agar plates (Fischer Scientific). Log-transformed CFU values were used to calculate averages and standard errors for graphing purposes.
Luciferase Assay.
The luciferase reaction was initiated by the addition of 150 μl of luciferin (1 mM in 0.1 M Na citrate; Promega) to 50 μl of each undiluted sample. Luminescence was detected 20 s after the addition of substrate by a TD-20/20 luminometer (Turner Designs). Three successive measurements were made and the average relative light unit (RLU) values recorded. The log10 of average RLU values multiplied by 1,000 was represented graphically.
Microarrays.
Hollow fibers each containing 106 bacilli were implanted into 15–20 SKH1 mice (two hollow fibers/mouse). Hollow fibers were retrieved from mice 10 d after hollow fiber encapsulation, and fiber contents were recovered and snap frozen. Pooled samples were suspended in TRIzol reagent (GIBCO BRL), and M. tuberculosis membranes were disrupted using zirconia/silica beads in a bead beater. M. tuberculosis RNA was recovered by centrifugation, chloroform extraction, and isopropyl alcohol precipitation, and purified using RNeasy column (QIAGEN) as described previously (6, 8). The same steps were followed to extract and purify RNA from mid-logarithmic phase (A 600, 0.600–0.850) M. tuberculosis H37Rv-lux cultures grown in plastic roller bottles at 37°C in supplemented liquid broth. Fluorescently labeled cDNA was generated using Powerscript (CLONTECH Laboratories, Inc.), using fluorescent dyes Cy3 and Cy5. These cDNA were competitively hybridized on microarray slides containing commercially available (Operon) M. tuberculosis 70-mer oligonucleotides representing all opening reading frames annotated in the H37Rv genome sequencing project (16), and fluorescence intensity data were collected with a GenePix 4000 scanner (Axon Instruments, Inc.) with Pro 4.0 software. Data were normalized based on total intensity of good-quality spots above background for each channel, and ratios of in vivo hollow fiber to in vitro cDNA were calculated based on normalized data. In this assay, the ratio of the signal from in vivo hollow fiber samples to that of in vitro control samples for a given open reading frame (ORF) should represent the relative abundance of the transcripts of that ORF under the two conditions. Three biological replicates were performed, and microarray samples were reverse labeled in one experiment. Significant differential regulation of genes was defined by greater than or equal to twofold change in gene expression as compared with control samples and P < 0.01.
Quantitative Real-Time RT-PCR.
RNA samples were treated with DNA-free kit (Ambion) according to the instructions of the manufacturer, and DNA contamination was excluded by PCR amplification using primers for Rv2031 (acr) and absence of PCR product on gel electrophoresis (unpublished data). Reverse transcription of RNA samples (∼0.5 μg RNA/sample) was accomplished using random hexamer primers (0.5 μg/reaction; Invitrogen) and Powerscript Reverse Transcriptase (CLONTECH Laboratories, Inc.). Real-time PCR was performed on cDNA using the SYBR green assay and Premix D (Epicentre) for all samples, and fluorescence was measured by iCycler (Bio-Rad Laboratories). Gene expression was compared with that of sigA, an M. tuberculosis housekeeping gene.
Transposon (Tn) Mutants.
Random insertion mutagenesis of M. tuberculosis CDC 1551 strain was performed using the Himar1 Tn as described previously (17, 18). Tn insertion sites were identified by sequencing the insertion junction as described previously (18). 100 different mutants, each with Tn insertion within the proximal 80% or proximal to the distal 100 base pairs of the ORF, were randomly selected from the library of mutants. Each of the selected mutants was separately grown in 37°C in supplemented Middlebrook 7H9 liquid broth to an A 600 of 0.8–1.0, and two master pools, each consisting of 50 mutants, were prepared by mixing an equal volume of culture of each mutant. This mixture was diluted to an A 600 of 0.1, and the latter was used to inoculate the hollow fibers. In separate experiments, the rel Mtb-disrupted Tn insertion mutant was grown separately in vitro using the aforementioned conditions to an A 600 of ∼1.0, and diluted 1:10. The diluted culture was added in a 1:1 ratio to a similarly grown and diluted culture of wild-type CDC 1551. The Δrel Mtb/wild-type culture suspension was encapsulated in hollow fibers as described before and either implanted subcutaneously into mice or incubated in vitro. Recovered hollow fiber samples were plated on Middlebrook 7H10 plates (Difco Laboratories) and on 7H10 plates containing 20 μg/ml kanamycin (the Tn insertion contains a kanamycin resistance gene; reference 18).
Viability Assay.
The LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes) was used to assay for mycobacterial viability after removal from hollow fibers. In brief, component A (3.34 mM SYTO 9 green fluorescent nucleic acid stain) and component B (20 mM propidium iodide) were mixed in equal volumes. An equivalent of 3 μl of this 1:1 nucleic acid stain mixture was added to each 1 ml of sample, and samples were incubated in the dark for ≥15 min. As a control, in vitro–grown M. tuberculosis H37Rv-lux was treated for 3 h with 70% ethanol to promote mycobacterial death and highlight differences in fluorescence staining between live and dead bacilli (see Fig. 3 a). Samples were treated with 20 μl of 4% paraformaldehyde (which does not alter cell permeability characteristics) for ≥15 min, and samples were observed using epifluorescence microscopy (Nikon Eclipse E800). Images were obtained using a Nikon digital camera DXM1200 and processed using Spot Version 3.4 software.
Figure 3.
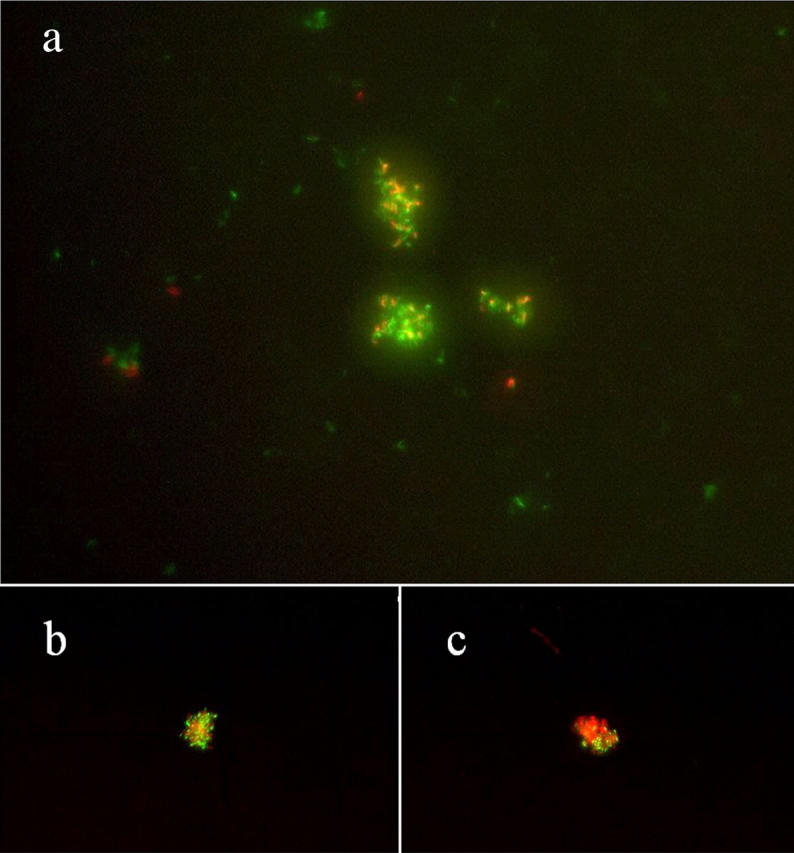
Hollow fiber–encapsulated bacilli in vivo remain viable. As a control, in vitro–grown cultures of M. tuberculosis H37Rv-lux were treated with 70% ethanol for 3 h to promote bacillary death (a). Live bacilli exhibit green fluorescence, whereas dead bacilli fluoresce red. Approximately half of all in vivo hollow fiber–encapsulated organisms on days 21 (b) and 28 (c) after fiber implantation were determined to be viable based on their staining properties.
Antibiotic Studies.
Mice received a powdered diet containing 1% sugar with either 0.02% (by weight) rifampin (Sigma-Aldrich) or 0.05% isoniazid (Sigma-Aldrich), beginning 14 d after implantation of hollow fibers. Untreated control mice received powdered diet containing 1% powdered sugar alone. In separate experiments, mice received diet containing 2.5% sugar alone (untreated controls) or with 0.25% moxifloxacin beginning on day 1 after hollow fiber implantation. Mouse dietary consumption was measured and recorded for all groups on a daily basis. At the time of killing, blood was obtained from antibiotic-treated mice by cardiac puncture, mouse serum was separated, and serum samples were stored at −70°C until the time of analysis. Mouse serum samples were mailed overnight on dry ice to C. Peloquin (National Jewish Medical Research Center, Denver, CO) for determination of serum antibiotic concentrations.
Online Supplemental Material.
A complete list of all Tn mutants used, including information on the gene mutated and the exact coordinate of Tn insertion, is presented as online supplemental material. DosR genes that fulfilled one, but not both, criteria for significant up-regulation (see Results) are presented in the online supplemental material. The complete lists of genes found to be significantly up-regulated or down-regulated by microarray analysis in hollow fiber–encapsulated bacilli in vivo, as well as all genes not found to be differentially regulated, are also presented. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20040646/DC1.
Results
Inhibition of Growth of Encapsulated Tubercle Bacilli In Vivo (Fig. 2)
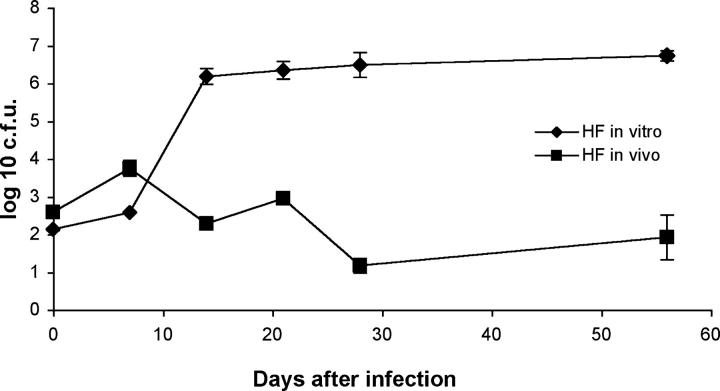
Figure 2.
Reduced growth of bacilli within hollow fibers in vivo. CFU counts per fiber of hollow fiber–encapsulated M. tuberculosis implanted into mice (HF in vivo) are compared with those of hollow fiber–encapsulated M. tuberculosis incubated in vitro (HF in vitro).
M. tuberculosis H37Rv-expressing firefly luciferase (H37Rv-lux) was encapsulated in hollow fibers and implanted into the subcutaneous space of mice or incubated at 37°C in supplemented Middlebrook 7H9 broth. Although hollow fiber–encapsulated bacilli incubated in vitro multiply exponentially for 14–21 d before reaching a plateau, bacilli encapsulated in hollow fibers and implanted into mice rapidly achieve stationary-state CFU counts (Fig. 2). The continued growth of in vitro hollow fiber–encapsulated bacilli implies that the failure of in vivo hollow fiber–encapsulated bacilli to multiply is not simply a result of physical containment imposed by the internal dimensions of the hollow fibers..
Assessment of Bacillary Viability and Metabolic Activity.
To further investigate the CFU equilibrium in hollow fiber–encapsulated bacilli in vivo, we examined the bacilli recovered from fibers using a bacterial viability assay in which live M. tuberculosis bacilli emit green fluorescence, whereas dead bacilli fluoresce red, based on differences in cell membrane permeability to the two nucleic acid stains used in the assay (Fig. 3). If the stationary-state CFU counts observed in hollow fiber–encapsulated bacilli in vivo represented an equilibrium between bacillary multiplication and death, one would expect that live-staining bacilli would comprise a very small proportion of all staining bacilli, due to the vast accumulation of dead bacilli over time. However, approximately half of all staining organisms on days 21 (Fig. 3 b) and 28 (Fig. 3 c) after hollow fiber implantation were determined to be alive by their staining properties, consistent with the hypothesis that these organisms are in a slowly replicating or nonreplicating persistent state. In addition, we detected a significant lag time in the appearance of colonies after plating the in vivo–cultivated bacilli, as compared with hollow fiber–encapsulated bacilli incubated in vitro. At the day 21 time point, microcolonies were detected 10–11 d after plating in the in vitro hollow fiber samples, whereas these were not detectable in the in vivo hollow fiber samples until 14 d after plating (unpublished data). Furthermore, average colony size for in vivo hollow fiber samples was significantly smaller than corresponding in vitro hollow fiber samples when examined 17 d after plating (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20040646/DC1). These data are consistent with the hypothesis that hollow fiber–encapsulated M. tuberculosis implanted in mice rapidly enter a state of decreased replication.
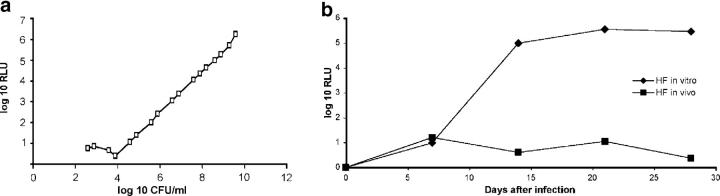
Because the oxidation of luciferin to oxyluciferin and photons (which can be detected by a luminometer at ∼560 nm) is an ATP-dependent process, the luciferase assay may be used as an indirect measure of the ATP content of cells. In log-phase in vitro cultures of M. tuberculosis (i.e., not encapsulated in fibers), the relationship between CFUs and RLUs is linear (Fig. 4 a). Hollow fiber-encapsulated M. tuberculosis grown in vitro maintains baseline metabolic activity, as reflected by RLU values that closely parallel CFU counts at the corresponding time points (Fig. 4 b). On the contrary, RLU values of in vivo–cultivated hollow fiber–encapsulated bacilli on days 14 and 21 after implantation (Fig. 4 b) were 0.8 log10 and 0.93 log10, respectively, less than predicted based on the corresponding CFU values (Fig. 2) and the proportionality of CFU:RLU (Fig. 4 a), suggesting reduced metabolic activity of these organisms as measured by the luciferase assay. These data suggest that in vivo hollow fiber–encapsulated M. tuberculosis bacilli rapidly enter a state of decreased metabolic activity.
Figure 4.
Reduced metabolic activity of encapsulated bacilli in vivo. (a) Relationship of relative light units (RLUs) to CFUs in mid-log phase M. tuberculosis H37Rv-lux grown in vitro. (b) Luciferase activity of hollow fiber–encapsulated M. tuberculosis implanted into mice (HF in vivo) versus hollow fiber–encapsulated M. tuberculosis incubated in vitro (HF in vitro).
Antibiotic Susceptibility of Hollow Fiber–encapsulated Bacilli.
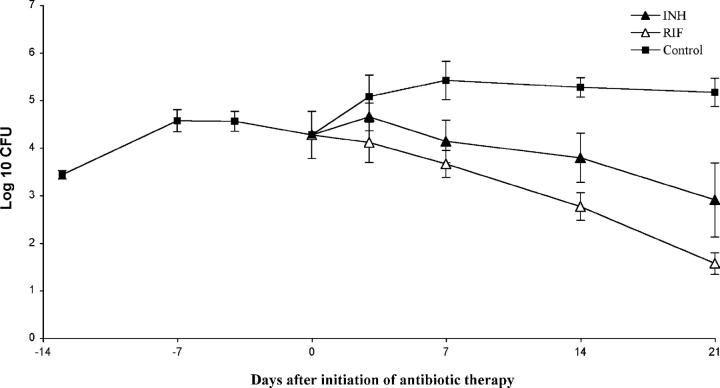
Animal and human chemotherapy studies have pointed to several populations of M. tuberculosis in the mammalian host, including rapidly multiplying and nonreplicating or sporadically replicating persistent bacilli (19, 20). In keeping with this mixed bacterial population model, drug treatment studies have demonstrated that the sterilizing drug rifampin is more active against sporadically multiplying and persistent bacilli than the bactericidal drug isoniazid; the former drug requires a significantly shorter duration of therapy to achieve acceptable relapse rates (21–25). We reasoned that rifampin likewise should be more effective than isoniazid against hollow fiber–encapsulated M. tuberculosis implanted in mice if these bacilli were truly in a slowly replicating, metabolically quiescent state. To allow bacilli to enter this altered physiologic state, antibiotics were not initiated until 14 d after hollow fiber implantation. Experimental groups of mice received either powdered diet containing isoniazid 0.05% or rifampin 0.02% (by weight), whereas untreated mice received antibiotic-free diet. Serum concentrations of each antibiotic were measured in killed mice at each time point, and were found to be at least 10 times greater than their respective MIC90 (Table I). Daily area under the serum concentration–time curve properties of each dietary regimen in mice were found to be similar to the area under the serum concentration–time curve in humans after standard daily doses of each drug (26, 27). Although isoniazid was somewhat effective against bacilli in the hollow fiber model in vivo, the activity of rifampin was more consistent and the magnitude of its effect was greater than that of isoniazid at each time point, resulting in a three-log kill by 21 d after initiation of therapy (Fig. 5). When mice were treated with the new 8-methoxyfluoroquinolone moxifloxacin, which has potent bactericidal activity against M. tuberculosis in mice (28), beginning on day 1 after hollow fiber implantation, fibers were culture negative by day 14 after daily therapy (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20040646/DC1). These data suggest that by 14 d after hollow fiber implantation, hollow fiber–encapsulated M. tuberculosis has entered an altered physiologic state in which bacilli are more resistant to the activity of antituberculosis drugs, but more susceptible to rifampin than to isoniazid, consistent with the antibiotic susceptibility of persistent bacilli in human latent tuberculosis infection (21–25).
Table I.
Serum Isoniazid and Rifampin Concentrations in Mice
| SKH1 mice
|
Humans
|
|||||
|---|---|---|---|---|---|---|
| Dose | Cmean | AUC | Dose | Cmean | AUC | |
| % diet | mg/L | mg · h/L | mg/kg | mg/L | mg · h/L | |
| Isoniazid | 0.02 | 0.77 ± 0.21 | 18.5 ± 5.0 | 5 | 5.4 ± 2.0a, 7.1 ± 1.9b | 19.9 ± 6.1a, 48.2 ± 1.5b |
| Rifampin | 0.05 | 3.09 ± 0.41 | 74.2 ± 9.8 | 10 | 14.91 | 117.93 |
Rapid acetylators.
Slow acetylators.
Figure 5.
Hollow fiber–encapsulated bacilli are more susceptible to rifampin than to isoniazid. The activities of isoniazid 0.05% in the diet (INH) and rifampin 0.02% in the diet (RIF) against hollow fiber–encapsulated bacilli in vivo are compared with no treatment (Control).
Granuloma Formation Surrounding Bacilli-containing Hollow Fibers in Mice.
Interestingly, we observed grossly the progressive formation of thick, granuloma-like lesions encasing hollow fibers containing M. tuberculosis H37Rv-lux (Fig. 6, d–f), but not those containing liquid broth alone (Fig. 6, a–c). By day 28 after hollow fiber implantation, histologic analysis revealed much greater cellular infiltration of macrophages, lymphocytes, and fibroblasts in the tissues surrounding hollow fibers containing tubercle bacilli (Fig. 6 l), as compared with those containing liquid broth (Fig. 6 i). Acid-fast staining of the surrounding tissue revealed no detectable bacilli, and inflammatory cells were not detected by microscopy within the hollow fibers (unpublished data). These results suggest the possibility that M. tuberculosis secretes soluble factors that diffuse from the fibers, leading to recruitment of host inflammatory cells. The formation of granulomatous lesions encasing hollow fibers may create a hostile microenvironment in which bacilli are forced to reduce their replication and metabolic activity, consequently becoming more susceptible to rifampin than to isoniazid.
Figure 6.
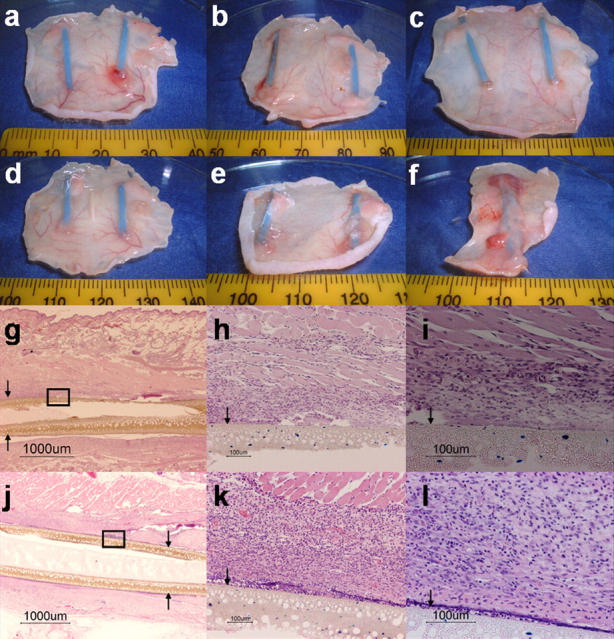
Formation of granuloma-like lesions surrounding M. tuberculosis–containing hollow fibers. Gross skin lesions surrounding hollow fibers containing liquid broth alone at days 1 (a), 14 (b), and 28 (c), and those containing M. tuberculosis H37Rv-lux at days 1 (d), 14 (e), and 28 (f) after hollow fiber implantation. Histopathology of tissues surrounding hollow fibers containing liquid broth (g–i) and those containing M. tuberculosis H37Rv-lux (j–l) 28 d after hollow fiber implantation (hematoxylin-eosin stain). Arrows indicate hollow fiber membrane.
Containment of Intrafiber Bacillary Growth In Vivo Is Immune Mediated and IFNγ Dependent.
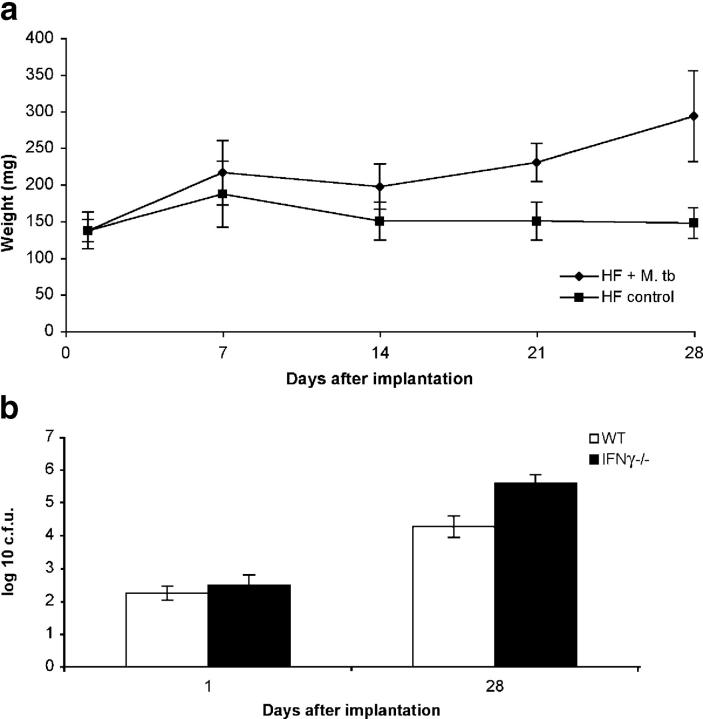
In addition to local granuloma formation, we detected significant spleen enlargement as early as 14 d after hollow fiber implantation in mice that had implanted fibers containing M. tuberculosis as compared with those that had implanted fibers containing media (Fig. 7 a), suggesting a systemic immune response in the former group of mice. To further investigate the phenomena of granuloma formation and splenomegaly and to exclude the possibility that intrafiber bacillary growth containment in vivo is mediated exclusively by nutrient starvation, we studied the growth of hollow fiber–encapsulated M. tuberculosis in IFNγ-deficient mice. In brief, ∼500 bacilli were encapsulated in each hollow fiber, and fibers were implanted subcutaneously either into wild-type BALB-C/J mice or BALB-C/J IFNγ-deficient mice, and colony counts were measured 28 d after implantation. Interestingly, although there was some growth of bacilli in wild-type mice over 28 d, CFU counts of hollow fiber–encapsulated M. tuberculosis in IFNγ-deficient mice were 1.3 log10 higher at 28 d than those in wild-type mice, suggesting that IFNγ plays a role in containment of intrafiber bacillary growth in vivo (Fig. 7 b).
Figure 7.
Containment of intrafiber bacillary growth in vivo is immune mediated and IFNγ dependent. Mice implanted with hollow fibers containing M. tuberculosis (HF + M. tb) developed enlarged spleens as measured by mean spleen weight (y axis) as early as 14 d after implantation as compared with mice implanted with fibers containing media (HF control; a). Wild-type BALB-C/J (WT) mice are able to contain the growth of hollow fiber–encapsulated M. tuberculosis to a greater extent than isogenic IFNγ-deficient (IFNγ−/−) mice 28 d after hollow fiber implantation (b).
Reduced Survival of relMtb-deficient M. tuberculosis in Hollow Fibers Implanted into Mice.
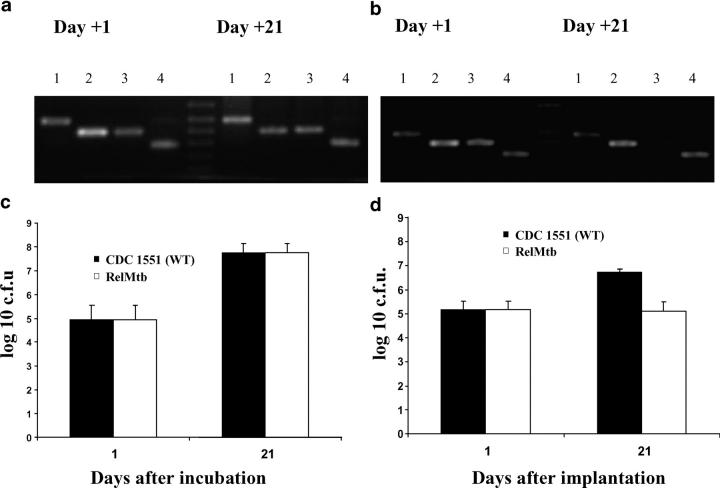
As proof-of-principle to determine whether this model could predict genes essential for extracellular persistence within granulomas, we generated a library of mutants by Tn mutagenesis of M. tuberculosis CDC 1551 strain using the Himar1 Tn as described previously (18). 100 genetically defined mutants, each deficient in a specific gene (as defined by Tn insertion within the proximal 80% or proximal to the distal 100 base pairs of the ORF of each particular gene; reference 18), were selected from the library and divided into two master pools, each consisting of 50 mutants (Tables S1 and S2, available at http://www.jem.org/cgi/content/full/jem.20040646/DC1). Each pool was generated by mixing an equal volume of pure culture from each mutant to ensure equal representation. Individual pools were encapsulated into separate hollow fibers and these were either implanted subcutaneously into mice or incubated in vitro. Hollow fiber contents were recovered on day 1 (input pool) and day 21 (output pool) after infection. PCR amplification of the junction sites between the Tn and the adjacent chromosomal DNA in the pooled genomic DNA from recovered mutants were performed at each time point to determine the presence or absence of each mutant. Of the 100 mutants tested, only those containing a Tn insertion in gene MT2660 (Rv2583c, rel Mtb) were present in the in vivo input pool, but absent in the in vivo output pool (Fig. 8 b). In contrast, this Tn mutant was readily detectable in both input and output pools in vitro (Fig. 8 a). Mice were also infected intravenously with pools representing the same 100 Tn mutants. In concordance with findings from the hollow fiber model in vivo, only the rel Mtb-deficient mutant was found to have significantly decreased survival 49 d after infection (unpublished data).
Figure 8.
Absence of rel Mtb-deficient mutant by PCR from a pool of mutants after 21 d of cultivation within mouse granulomas. PCR amplification of the Tn insertion junction (see Online Supplemental Material) reveals presence of the rel Mtb::Tn mutant in both input (day 1) and output (day 21) pools in hollow fibers incubated in vitro (a), but absence of the mutant in the output pool (day 21) in mouse-implanted hollow fibers (b), suggesting reduced survival of this mutant in vivo. 1, Rv1347 (hypothetical transcriptional regulator); 2, Rv0250 (miscellaneous oxidoreductase); 3, Rv2583c (rel Mtb); 4, Rv1069 (pra). Hollow fiber–encapsulated wild-type M. tuberculosis CDC 1551 (WT) and rel Mtb::Tn mutant (RelMtb) grow equally when incubated in hollow fibers in vitro (c), but the latter strain demonstrates significantly reduced survival when hollow fibers are implanted into mice (d).
M. tuberculosis deficient in RelMtb, an enzyme responsible for the synthesis and hydrolysis of hyperphosphorylated guanine nucleotides involved in the stringent response, has been shown to be significantly attenuated compared with wild type in the tissues of mice 38 wk after aerosol infection (29). We decided to specifically evaluate the growth phenotype of the rel Mtb Tn insertion mutant (rel Mtb::Tn), which carries a kanamycin resistance marker, in the hollow fiber model in vivo. To ensure that each strain was exposed to identical microenvironment conditions, a 1:1 mixture of rel Mtb::Tn and wild-type strain M. tuberculosis CDC 1551 was encapsulated in hollow fibers and either incubated in liquid broth in vitro or implanted subcutaneously in mice. 21 d later, hollow fibers were recovered and their contents were diluted and plated on antibiotic-free and kanamycin-containing plates to assess CFU counts for each hollow fiber–encapsulated strain. There was no growth defect of rel Mtb::Tn when incubated in vitro, as both hollow fiber-encapsulated wild-type and rel Mtb::Tn bacilli grew equally after 21 d of incubation (Fig. 8 c). However, although hollow fiber–encapsulated wild-type bacilli implanted in mice demonstrated reduced growth as compared with those incubated in vitro after 21 d (growth ∼1.5 log for in vivo wild type versus ∼3 log10 for in vitro wild type), hollow fiber–encapsulated rel Mtb::Tn showed markedly reduced survival as compared with wild-type bacilli implanted in vivo (Fig. 8 d). Significantly, the decreased survival of the rel Mtb-deficient mutant became apparent as early as 21 d after hollow fiber implantation, as compared with several months using the standard murine aerosol infection model (29). We observed no significant change in survival compared with wild type in an M. tuberculosis mutant containing a Tn insertion in an unrelated gene (MT2749, Rv2675), suggesting that the presence of the Tn insertion alone does not confer a survival disadvantage in the hollow fiber model in vivo.
Gene Expression of Extracellular M. tuberculosis within Granulomas in Mice.
Next, we sought to gain insight into the adaptive response of tubercle bacilli in granulomatous lesions in vivo by studying their global gene expression (for complete gene expression profile, Table S6, available at http://www.jem.org/cgi/content/full/jem.20040646/DC1). Significantly, the exclusion of host cells and containment of bacilli by the hollow fibers renders whole genome microarray analysis feasible. Hollow fibers containing M. tuberculosis were implanted subcutaneously into mice and retrieved 10 d later. Hollow fiber contents were recovered and immediately snap frozen, and bacilli from 30–40 implanted fibers were pooled to yield sufficient RNA for analysis. Hollow fiber–encapsulated M. tuberculosis gene expression was compared with that of log-phase in vitro–grown M. tuberculosis. Significant differential gene regulation was defined by both of the following: (a) a greater than or equal to twofold change in gene expression as compared with control samples, and (b) a p-value <0.01.
We found that in vivo hollow fiber–encapsulated M. tuberculosis demonstrates significant induction of ∼260 genes, including several key regulatory genes (Table S4, available at http://www.jem.org/cgi/content/full/jem.20040646/DC1). Using the aforementioned strict criteria, we found significant up-regulation of Rv3133c (dosR) and 20 other members of the previously described DosR regulon (10), including hspX, Rv2623c, and Rv2626 (Table II), suggesting that the adaptive response of hollow fiber–encapsulated M. tuberculosis in vivo is similar to that of bacilli exposed to low concentrations of nitric oxide in vitro. The M. tuberculosis hspX gene encodes α-crystallin, a member of the small heat shock protein family with chaperone activity (30), which is powerfully induced under hypoxic conditions (6) and in lung specimens obtained from patients with active tuberculosis disease (31). Significantly increased expression of hspX, Rv2623c, and Rv2626 has been shown by real-time RT-PCR after the onset of Th1-mediated immunity in the mouse model of tuberculosis (13) and in various models of latency in vitro (6, 8, 10, 13). In addition, we found up-regulation of 12 additional genes of the DosR regulon that fulfilled one, but not both, aforementioned criteria (Table S3, available at http://www.jem.org/cgi/content/full/jem.20040646/DC1). Hollow fiber–encapsulated M. tuberculosis in vivo also demonstrated significant up-regulation of sigB, sigC, and sigH, which belong to a family of alternative RNA polymerase σ factors shown to coordinate gene regulation in response to environmental conditions in M. tuberculosis and other bacterial species (32, 33). We detected significant up-regulation of dnaE2, which encodes a DNA polymerase in M. tuberculosis and in other organisms, and which has been shown to be up-regulated by several DNA-damaging agents and during infection of mice, contributing to in vivo survival and the emergence of drug resistance (34). In addition, hollow fiber–encapsulated M. tuberculosis in mice demonstrated significant induction of many other genes recently found to be up-regulated in the multibacillary model of murine tuberculosis (5), including Rv0967, Rv0970, Rv0978c (PE-PGRS 17), Rv0980c (PE-PGRS 18), Rv0982 (mprB), and Rv0988.
Table II.
Significantly Up-Regulated DosR Regulon Genes in the Hollow Fiber Model In Vivo by Microarray Analysis
| Rv no. | Gene | Protein function | Fold up-regulation | p-value |
|---|---|---|---|---|
| 80 | HP | 2.9 | 0.0009 | |
| 569 | CHP | 4.9 | 0.007 | |
| 570 | nrdZ | ribonucleotide reductase cl. II | 2 | 6.17E-07 |
| 572c | HP | 2.2 | 0.002 | |
| 1733c | CHP | 3 | 0.004 | |
| 1997 | ctpF | cation transport ATPase | 13.2 | 0.007 |
| 2005c | CHP-USPA motif | 2.2 | 0.003 | |
| 2030c | CHP | 7.8 | 3.30E-05 | |
| 2031c | hspX | α-crystallin | 25 | 0.009 |
| 2623 | CHP-USPA motif | 25.8 | 0.0002 | |
| 2626c | CHP | 14.6 | 0.0001 | |
| 2627c | CHP | 8 | 0.008 | |
| 2628 | HP | 8.4 | 0.004 | |
| 2631 | HP | 2.1 | 0.001 | |
| 3126c | HP | 2.2 | 0.0001 | |
| 3128c | CHP | 3.2 | 0.004 | |
| 3130c | CHP | 15.4 | 0.0003 | |
| 3131 | CHP | 11.4 | 0.0008 | |
| 3132c | sensor histidine kinase | 4.8 | 0.001 | |
| 3133c | dosR | 2-comp. response reg. | 2.1 | 0.007 |
| 3134c | CHP-USPA motif | 7.1 | 0.007 |
CHP, conserved hypothetical protein; HP, hypothetical protein.
Consistent with evidence that M. tuberculosis might switch to C2 carbon sources such as fatty acids during in vivo persistence (35), we detected significant up-regulation of several genes encoding enzymes involved in lipid degradation, including fadD10 (Rv0099), fadD19 (Rv3515c), fadD34 (Rv0035), fadE1 (Rv0131c), fadE10 (Rv0873), and fadE24 (Rv3139). In addition, we found significantly increased expression of pckA (Rv0211), suggesting limited availability of glucose within artificial granulomas in mice. De novo synthesis of glucose-6-phosphate in mycobacteria is achieved through the action of phosphoenolpyruvate carboxykinase (pckA), which converts oxaloacetate to phosphoenolpyruvate, thus diverting carbon derived from β-oxidation of fatty acids into gluconeogenesis (36). Increased gene expression of pckA has been reported in the mouse model of tuberculosis (31), and disruption of pckA in Mycobacterium bovis produced attenuated strains in animal models of infection (37, 38). In addition, we found a 1.5-fold induction of Rv0467 (aceA) (P = 0.004) that encodes isocitrate lyase, the first enzyme of the glyoxylate cycle, which is strongly induced in mouse lungs (31) and is required for long-term persistence of M. tuberculosis in mice (35).
Consistent with the hypothesis that hollow fiber–encapsulated bacilli in vivo display a reduced replication rate, we observed significantly decreased expression of certain genes encoding ribosomal RNA binding proteins, including Rv0701 (rplC), Rv0703 (rplW), Rv0704 (rplB), Rv0706 (rplV), Rv0708 (rplP), Rv0710 (rplQ), and Rv0716 (rplE). In agreement with our luciferase assay data suggesting that these bacilli have decreased intracellular energy stores (Fig. 4 b), we found significant down-regulation of several genes encoding components of ATP synthase, including Rv1304 (atpB), Rv1306 (atpF), Rv1308 (atpA), Rv1310 (atpD), and Rv1311 (atpC). A complete list of significantly down-regulated genes in hollow fiber–encapsulated M. tuberculosis implanted into mice can be found in Table S5 (available at http://www.jem.org/cgi/content/full/jem.20040646/DC1). Differential regulation of a subset of 13 genes identified by microarray analysis was confirmed using quantitative real-time RT-PCR (Table III).
Table III.
Confirmation of Differential Regulation of a Subset of Genes by Quantitative Real-Time RT-PCR
| Rv. no. | Gene | Protein function | Fold induction |
|---|---|---|---|
| 1997 | HP | 32 | |
| 2031c | acr | α-crystallin | 294.1 |
| 2623 | CHP-USPA motif | 512 | |
| 2626c | CHP | 68.6 | |
| 2710 | sigB | RNA polymerase σ factor | 42.2 |
| 2744 | 35 kD Ag | 4.3 | |
| 3130c | CHP | 168.9 | |
| 3131 | CHP | 274.3 | |
| 3133c | dosR | 2-comp. response regulator | 78.8 |
| 3223c | sigH | RNA polymerase σ factor | 17.1 |
| 3370c | dnaE2 | DNA polymerase III | 9.8 |
| 3477 | PE family protein | −4.3 | |
| 3648c | cspA | prob. cold shock protein | −7 |
CHP, conserved hypothetical protein; HP, hypothetical protein.
Discussion
The hollow fiber encapsulation/implantation technique provides a means to establish a paucibacillary infection with M. tuberculosis in which the bacilli are readily recoverable from infected animals for further analysis. In this model of infection, the tubercle bacilli rapidly enter an altered physiologic state characterized by stationary-state CFU counts and decreased metabolic activity. These bacilli are more susceptible to the antituberculous drug rifampin than they are to isoniazid, consistent with the antibiotic susceptibility profile of persistent bacilli in animal chemotherapy models (21, 23) and in human latent tuberculosis infection (22, 24, 25). In addition, expression of rel Mtb, a gene that is essential for long-term persistence in the mouse model of chronic tuberculosis (29), is also necessary for short-term bacillary persistence in the hollow fiber granuloma model in vivo, suggesting a common adaptive strategy of M. tuberculosis in these two models of infection.
In this work, we present for the first time the whole-genome transcriptional profile of extracellular M. tuberculosis within granulomas in mice. In vivo hollow fiber–encapsulated M. tuberculosis demonstrates significant induction of several key regulatory genes, including Rv3133c (dosR), as well as 20 other genes of the recently described DosR regulon (10); the σ factor genes sigB, sigC, and sigH; the DNA polymerase-encoding dnaE2; and many other genes that were found to be significantly up-regulated in the mouse model of pulmonary tuberculosis (5). Increased expression of genes involved in lipid metabolism suggests depletion of glucose and the importance of fatty acids as a primary carbon source for extracellular M. tuberculosis in artificial granulomas in mice, which corroborates previous data in the mouse model of chronic tuberculosis (35). Consistent with the hypothesis that these organisms demonstrate an altered physiologic state characterized by reduced replication and metabolism, we detected significant down-regulation of genes encoding key ribosomal RNA binding proteins and ATP synthase, respectively. A subset of 13 genes identified by microarray analysis was confirmed to be significantly differentially regulated by quantitative real-time RT-PCR. The common transcriptional profile we observed lends further support to the hypothesis that the altered physiologic state of hollow fiber–encapsulated bacilli in vivo is similar to that of persistent bacilli in other models of latency.
It is important to note that, although the artificial subcutaneous granulomas surrounding hollow fibers containing M. tuberculosis resemble those formed in mouse lungs after aerosol infection with the same pathogen, granulomas in mice differ significantly from those in humans. In human infection, the granuloma is composed of a central core of macrophages, including multinucleated giant cells, surrounded by macrophages and lymphocytes, which include CD4 and CD8 T cells, and B cells (39). Although individual components of mouse granulomas are similar to those in humans, the architecture of the mouse granuloma is better characterized as a loose collection of activated and epithelioid macrophages and lymphocytic clusters (40). Unlike in human granulomas, multinucleated giant cells are absent in mouse granulomas, and necrosis and caseation are rarely observed (40). However, despite structural differences between granulomas in mice and humans, their function is likely to be similar, with respect to containment of infection and creation of a localized environment for the immune response to kill organisms.
Animal models of tuberculosis have shown that M. tuberculosis is an intracellular pathogen, residing within host macrophages (2). However, the precise location of persistent bacilli during human latent tuberculosis infection remains elusive. Specifically, autopsy data of persons who died of nontuberculosis-related causes have demonstrated that tubercle bacilli may be found outside lung granulomas, in normal-appearing lung tissue (41), and in nonmacrophage cell types, including alveolar epithelial cells (42). In addition, bacilli residing extracellularly in the caseous material of lung granulomas and cavities may represent an important reservoir of persistent organisms during human latent tuberculosis infection (4). Mouse-implanted hollow fibers containing M. tuberculosis induce splenomegaly and the accumulation of host inflammatory cells, including macrophages and lymphocytes, leading to the formation of artificial granulomas around the fibers. The precise microenvironment conditions prevailing within subcutaneously implanted hollow fibers are unclear, but may include hypoxia, nutrient starvation, and soluble immunologic factors (e.g., diffusible nitric oxide) secreted by surrounding host immune cells. Diminished ability to contain the growth of hollow fiber–encapsulated M. tuberculosis by IFNγ-deficient mice, as well as significant induction of the DosR regulon by these bacilli suggests that diffusible nitric oxide and/or hypoxia may contribute to the microenvironment experienced by intrafiber bacilli in vivo. The basis for mouse spleen enlargement and recruitment of host immune cells to the tissues surrounding the hollow fibers is unknown, but may involve diffusion of M. tuberculosis–secreted soluble factors across the hollow fiber membranes. Experiments designed to address these questions are currently underway.
Acknowledgments
We thank L. Einck for suggesting use of the hollow fiber encapsulation/implantation technique, and M. Hollingshead for generous technical advice and assistance.
The support of National Institutes of Health grants AI 37856, AI 36973, AI 43846, and AI 51668, and contract NOI 30036, as well as a contract from the Global Alliance for Tuberculosis Drug Development, is gratefully acknowledged. P.C. Karakousis received support from T32 AI 07608 and from a grant from the Potts Foundation.
The authors have no conflicting financial interests.
Abbreviations used in this paper: ORF, open reading frame; PVDF, polyvinylidene fluoride; RLU, relative light unit; Tn, transposon.
References
- 1.Cegielski, J.P., D.P. Chin, M.A. Espinal, T.R. Frieden, R. Rodriquez Cruz, E.A. Talbot, D.E. Weil, R. Zaleskis, and M.C. Raviglione. 2002. The global tuberculosis situation. Progress and problems in the 20th century, prospects for the 21st century. Infect. Dis. Clin. North Am. 16:1–58. [DOI] [PubMed] [Google Scholar]
- 2.Flynn, J.L., and J. Chan. 2001. Tuberculosis: latency and reactivation. Infect. Immun. 69:4195–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaplan, G., F.A. Post, A.L. Moreira, H. Wainwright, B.N. Kreiswirth, M. Tanverdi, B. Mathema, S.V. Ramaswamy, G. Walther, L.M. Steyn, et al. 2003. Mycobacterium tuberculosis growth at the cavity surface: a microenvironment with failed immunity. Infect. Immun. 71:7099–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grosset, J. 2003. Mycobacterium tuberculosis in the extracellular compartment: an underestimated adversary. Antimicrob. Agents Chemother. 47:833–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talaat, A.M., R. Lyons, S.T. Howard, and S.A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA. 101:4602–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherman, D.R., M. Voskuil, D. Schnappinger, R. Liao, M.I. Harrell, and G.K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA. 98:7534–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenkrands, I., R.A. Slayden, J. Crawford, C. Aagaard, C.E. Barry III, and P. Andersen. 2002. Hypoxic response of Mycobacterium tuberculosis studied by metabolic labeling and proteosome analysis of cellular and extracellular proteins. J. Bacteriol. 184:3485–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betts, J.C., P.T. Lukey, L.C. Robb, R.A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, M.A., B.B. Plikaytis, and T.M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voskuil, M.I., D. Schnappinger, K.C. Visconti, M.I. Harrell, G.M. Dolganov, D.R. Sherman, and G.K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnappinger, D., S. Ehrt, M.I. Voskuil, Y. Liu, J.A. Mangan, I.M. Monahan, G. Dolganov, B. Efron, P.D. Butcher, C. Nathan, and G.K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, H.D., K.M. Guinn, M.I. Harrell, R. Liao, M.I. Voskuil, M. Tompa, G.K. Schoolnik, and D.R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi, L., Y.J. Jung, S. Tyagi, M.L. Gennaro, and R.J. North. 2003. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. USA. 100:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollingshead, M.G., M.C. Alley, R.F. Camalier, B.J. Abbott, J.G. Mayo, L. Malspeis, and M.R. Grever. 1995. In vivo cultivation of tumor cells in hollow fibers. Life Sci. 57:131–141. [DOI] [PubMed] [Google Scholar]
- 15.Xu, Z.Q., M.G. Hollingshead, S. Borgel, C. Elder, A. Khilevich, and M.T. Flavin. 1999. In vivo anti-HIV activity of (+)-calanolide A in the hollow fiber mouse model. Bioorg. Med. Chem. Lett. 9:133–138. [DOI] [PubMed] [Google Scholar]
- 16.Cole, S.T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S.V. Gordon, K. Eiglmeier, S. Gas, C.E. Barry III, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 393:537–544. [DOI] [PubMed] [Google Scholar]
- 17.Rubin, E.J., B.J. Akerley, V.N. Novik, D.J. Lampe, R.N. Husson, and J.J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA. 96:1645–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamichhane, G., M. Zignol, N.J. Blades, D.E. Geiman, A. Dougherty, J. Grosset, K.W. Broman, and W.R. Bishai. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 100:7213–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchison, D.A. 1992. The Garrod Lecture. Understanding the chemotherapy of tuberculosis–current problems. J. Antimicrob. Chemother. 29:477–493. [DOI] [PubMed] [Google Scholar]
- 20.McKinney, J.D. 2000. In vivo veritas: the search for TB drug targets goes live. Nat. Med. 6:1330–1333. [DOI] [PubMed] [Google Scholar]
- 21.Lecoeur, H.F., C. Truffot-Pernot, and J.H. Grosset. 1989. Experimental short-course preventive therapy of tuberculosis with rifampin and pyrazinamide. Am. Rev. Respir. Dis. 140:1189–1193. [DOI] [PubMed] [Google Scholar]
- 22.1992. A double-blind placebo-controlled clinical trial of three antituberculosis chemoprophylaxis regimens in patients with silicosis in Hong Kong. Hong Kong Chest Service/Tuberculosis Research Centre, Madras/British Medical Research Council. Am. Rev. Respir. Dis. 145:36–41. [DOI] [PubMed]
- 23.Dickinson, J.M., and D.A. Mitchison. 1981. Experimental models to explain the high sterilizing activity of rifampin in the chemotherapy of tuberculosis. Am. Rev. Respir. Dis. 123:367–371. [DOI] [PubMed] [Google Scholar]
- 24.Jasmer, R.M., P. Nahid, and P.C. Hopewell. 2002. Clinical practice. Latent tuberculosis infection. N. Engl. J. Med. 347:1860–1866. [DOI] [PubMed] [Google Scholar]
- 25.Fox, W., and D.A. Mitchison. 1975. Short-course chemotherapy for pulmonary tuberculosis. Am. Rev. Respir. Dis. 111:325–353. [DOI] [PubMed] [Google Scholar]
- 26.Kim, Y.G., J.G. Shin, S.G. Shin, I.J. Jang, S. Kim, J.S. Lee, J.S. Han, and Y.N. Cha. 1993. Decreased acetylation of isoniazid in chronic renal failure. Clin. Pharmacol. Ther. 54:612–620. [DOI] [PubMed] [Google Scholar]
- 27.Kenny, M.T., and B. Strates. 1981. Metabolism and pharmacokinetics of the antibiotic rifampin. Drug Metab. Rev. 12:159–218. [DOI] [PubMed] [Google Scholar]
- 28.Nuermberger, E.L., T. Yoshimatsu, S. Tyagi, R.J. O'Brien, A.N. Vernon, R.E. Chaisson, W.R. Bishai, and J.H. Grosset. 2004. Moxifloxacin-containing regimen greatly reduces time to culture conversion in murine tuberculosis. Am. J. Respir. Crit. Care Med. 169:421–426. [DOI] [PubMed] [Google Scholar]
- 29.Dahl, J.L., C.N. Kraus, H.I. Boshoff, B. Doan, K. Foley, D. Avarbock, G. Kaplan, V. Mizrahi, H. Rubin, and C.E. Barry III. 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. USA. 100:10026–10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan, Y., D.D. Crane, and C.E. Barry III. 1996. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial alpha-crystallin homolog. J. Bacteriol. 178:4484–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timm, J., F.A. Post, L.G. Bekker, G.B. Walther, H.C. Wainwright, R. Manganelli, W.T. Chan, L. Tsenova, B. Gold, I. Smith, et al. 2003. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. USA. 100:14321–14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haldenwang, W.G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez, J.E., J.M. Chen, and W.R. Bishai. 1997. Sigma factors of Mycobacterium tuberculosis. Tuber. Lung Dis. 78:175–183. [DOI] [PubMed] [Google Scholar]
- 34.Boshoff, H.I., M.B. Reed, C.E. Barry III, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 113:183–193. [DOI] [PubMed] [Google Scholar]
- 35.McKinney, J.D., K. Honer zu Bentrup, E.J. Munoz-Elias, A. Miczak, B. Chen, W.T. Chan, D. Swenson, J.C. Sacchettini, W.R. Jacobs, Jr., and D.G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 406:735–738. [DOI] [PubMed] [Google Scholar]
- 36.Tuckman, D., R.J. Donnelly, F.X. Zhao, W.R. Jacobs, Jr., and N.D. Connell. 1997. Interruption of the phosphoglucose isomerase gene results in glucose auxotrophy in Mycobacterium smegmatis. J. Bacteriol. 179:2724–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, K., J. Yu, and D.G. Russell. 2003. pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and in macrophages. Microbiol. 149:1829–1835. [DOI] [PubMed] [Google Scholar]
- 38.Collins, D.M., T. Wilson, S. Campbell, B.M. Buddle, B.J. Wards, G. Hotter, and G.W. De Lisle. 2002. Production of avirulent mutants of Mycobacterium bovis with vaccine properties by the use of illegitimate recombination and screening of stationary-phase cultures. Microbiol. 148:3019–3027. [DOI] [PubMed] [Google Scholar]
- 39.Randhawa, P.S. 1990. Lymphocyte subsets in granulomas of human tuberculosis: an in situ immunofluorescence study using monoclonal antibodies. Pathology. 22:153–155. [DOI] [PubMed] [Google Scholar]
- 40.Flynn, J.L., and J. Chan. 2004. Animal models of tuberculosis. Tuberculosis. W.N. Rom and S.M. Garay, editors. Lippicott Williams & Wilkins, Philadelphia. 237–250.
- 41.Opie EL, A.J. 1927. Tubercle bacilli in latent tuberculous lesions and in lung tissue without tuberculous lesions. Arch. Pathol. Lab. Med. 4:1–21. [Google Scholar]
- 42.Hernandez-Pando, R., M. Jeyanathan, G. Mengistu, D. Aguilar, H. Orozco, M. Harboe, G.A. Rook, and G. Bjune. 2000. Persistence of DNA from Mycobacterium tuberculosis in superficially normal lung tissue during latent infection. Lancet. 356:2133–2138. [DOI] [PubMed] [Google Scholar]