Abstract
In addition to releasing preformed granular proteins, polymorphonuclear leukocytes (PMNs) synthesize chemokines and other factors under transcriptional control. Here we demonstrate that PMNs express an inducible transcriptional modulator by signal-dependent activation of specialized mechanisms that regulate messenger RNA (mRNA) translation. HL-60 myelocytic cells differentiated to surrogate PMNs respond to activation by platelet activating factor by initiating translation and with appearance of specific mRNA transcripts in polyribosomes. cDNA array analysis of the polyribosome fraction demonstrated that retinoic acid receptor (RAR)-α, a transcription factor that controls the expression of multiple genes, is one of the polyribosome-associated transcripts. Quiescent surrogate HL60 PMNs and primary human PMNs contain constitutive message for RAR-α but little or no protein. RAR-α protein is rapidly synthesized in response to platelet activating factor under the control of a specialized translational regulator, mammalian target of rapamycin, and is blocked by the therapeutic macrolide rapamycin, events consistent with features of the 5′ untranslated region of the transcript. Newly synthesized RAR-α modulates production of interleukin-8. Rapid expression of a transcription factor under translational control is a previously unrecognized mechanism in human PMNs that indicates unexpected diversity in gene regulation in this critical innate immune effector cell.
Keywords: inflammation, innate immunity, gene expression, translation, transcription
Introduction
The human polymorphonuclear leukocyte (PMN) is a crucial effector cell in the innate immune system. PMNs are the first myeloid leukocytes to respond to tissue damage or microbial infection and initiate bacterial phagocytosis and killing. Activated PMNs use a repertoire of rapid response mechanisms to accomplish wound surveillance and bacterial containment. These include inside-out signaling of β2 integrins with consequent increased cell adhesiveness and aggregation, polarization and directional migration, degranulation of preformed proteins, and generation of oxygen radicals (1).
Besides acute responses, there is now evidence that PMNs mediate inflammatory events over hours and can express new proteins and polypeptides, including chemokines (2). Soluble agonists, receptor-mediated phagocytosis, and integrin engagement dramatically alter levels of messenger RNA (mRNA) transcripts in human PMNs, and many of these gene products are under transcriptional control (2–5). New evidence also indicates that transcription is important in the regulation of PMN apoptosis and, thus, the duration and intensity of inflammation (1, 6). Although control of this process in activated PMNs is incompletely understood, key transcription factors are clearly central modulators. For example, constitutively expressed NF-κB translocates from the cytoplasm to the nucleus in agoinist-stimulated PMNs where it binds to promoter motifs and regulates transcription of specific genes in regulated and dysregulated inflammation (2, 7, 8).
Here, we report a previously unrecognized mechanism of gene regulation in activated PMNs that involves unique interplay at posttranscriptional and transcriptional checkpoints. Critical posttranscriptional pathways are currently recognized with increasing frequency as analysis of the human genome proceeds, and there is now clear evidence that posttranscriptional dysregulation is a mechanism of disease (9–11). Posttranscriptional control at translational checkpoints may be particularly important in biologic systems in which factors with potent activities are precisely synthesized with respect to timing and magnitude and in a cell-specific fashion (9–11). These features characterize synthesis of chemokines, cytokines, and other factors by activated PMNs (1–8). In the studies reported here, we found that, in response to cellular activation, PMNs rapidly translate constitutive mRNA encoding the retinoic acid receptor (RAR)-α, a nuclear hormone receptor and transcription factor that regulates the expression of multiple downstream genes in a variety of cells (12). In PMNs stimulated with platelet activating factor (PAF), translation of RAR-α mRNA is controlled by the intracellular signaling kinase mammalian target of rapamycin (mTOR), a specialized regulator of translation that we have found recently to respond to outside-in signals in human PMNs (13). The newly synthesized RAR-α protein then modulates expression of the chemokine IL-8. These observations identify a previously unrecognized mechanism in the inflammatory portfolio used by PMNs in which they modulate synthetic outcomes by rapid, regulated translation of a critical transcription factor.
Materials and Methods
Reagents.
PAF, AM-580 (an RAR-α–specific agonist), and the anti–human RAR-α monoclonal antibody were purchased from BioMol. Puromycin, propidium iodide, heparin, and cycloheximide were obtained from Sigma-Aldrich. Rapamycin was purchased from Calbiochem. Trizol and Trizol LS total RNA isolation reagents were purchased from Invitrogen. The 32P-labeled dATP (PB10204, 250 μCi/μL) was obtained from Amersham Biosciences. The anti–human CD71 (transferrin receptor) monoclonal antibody and goat anti–mouse HRP-conjugated secondary antibody were obtained from Biodesign. A second anti–human CD71 (transferrin receptor) monoclonal antibody was purchased from Immunotech. The anti–human β-actin monoclonal antibody was purchased from ICN Biomedicals, Inc. ProteaseArrest and EDTA were purchased from Genotechnology Inc. Fura-2 was obtained from Molecular Probes.
Polymorphonuclear Leukocyte Isolation.
PMNs were isolated from healthy adult donor heparinized whole blood (75 U heparin/10 ml) using techniques described previously by our laboratory (14). The procedures were approved by the University of Utah Institutional Review Board. Isolated PMNs were resuspended in HBSS (BioWhittaker, Walkersville) with 0.5% human serum albumin (Baxter Healthcare Inc.) to a concentration of 15–20 × 106 cells/ml. Purity of the human PMNs was >95%.
HL-60 Cell Culture and Differentiation.
Cells of the human promyelocytic leukemia HL-60 cell line were obtained from American Type Culture Collection and grown in Iscove's Modified Dulbecco's Medium (BioWhittaker) supplemented with 20% FBS (Hyclone), penicillin G, and Streptomycin (BioWhittaker). Amphotericin B (Sigma-Aldrich) was added to the culture medium (2.5 μg/ml). The cell cultures were passed biweekly to maintain logarithmic growth. For these experiments, 2 × 106 HL-60 cells were resuspended in fresh media and were then differentiated along the PMN (granulocytic) lineage with 1.3% DMSO (American Type Culture Collection) for 6 d (15) (also see Selection for Differentiated HL-60 Cells and Assessment of HL-60 Differentiation). Viability was assessed by exclusion of 0.2% Trypan blue (Sigma-Aldrich) and was routinely >80% after differentiation.
Selection for Differentiated HL-60 Cells.
The purity of differentiated HL-60 surrogate PMNs was increased by negative selection in which cells that expressed the transferrin receptor (CD71) were removed (16). HL-60 cells treated with DMSO as above were incubated with the mouse anti–human CD71 (transferrin receptor) monoclonal antibody (1 μg/ml) for 30 min. The CD71-positive cells were complexed to magnetic microbeads coated with goat anti–mouse IgG (Miltenyi Biotec) and subsequently removed by passage through a magnetized column.
Assessment of HL-60 Differentiation.
Differentiation of HL-60 cells to surrogate PMNs (HL-60 PMNs) (15) was assessed by several criteria including nuclear segmentation, loss of transferrin receptor expression (16), and gain of responsivity to PAF (17). Immunocytochemical and flow cytometric analyses were used to detect transferrin receptor expression. Immunocytochemical experiments were done according to our previously published protocols (18), and the cells were examined with a Bio-Rad Laboratories confocal microscope (Hercules). Flow cytometry was performed on a Becton Dickinson FACScanner using minor modifications of previously published protocols (16, 19). PAF-induced cytosolic calcium influxes were assayed using a Shimadzu spectrofluorophotometer after incubation with the calcium sensitive fluorescent dye Fura-2 (Molecular Probes) (20).
Isolation of Polyribosomal RNA from Differentiated HL-60 Surrogate PMNs for mRNA Expression Array Analysis.
For polyribosomal profiling experiments, we used differentiated HL-60 surrogate PMNs rather than primary PMNs to minimize difficulties in maintaining integrity of leukocyte RNA during cell fractionation and transcript analysis (21). Differentiated HL-60 PMNs (25 × 106 cells/sample) were left quiescent or stimulated with PAF (10−8 M) for 1 h at 37°C. The cells were lysed, and ribosomal profiling was conducted using minor modifications of procedures that we have described elsewhere (22). Total RNA was isolated from the polyribosomal fractions from each sample and used to generate 32P-dATP labeled cDNA, which was then employed as a template in expression analysis (Atlas Human Cancer cDNA Expression Array, 7742–1; CLONTECH Laboratories, Inc.).
RAR-α Protein Expression.
Differentiated HL-60 PMNs or freshly isolated primary human PMNs (2.5 × 106) were resuspended in M-199 media (37°C) and left quiescent or activated with PAF in indicated concentrations at 37°C in 5% CO2. In selected experiments, the cells were pretreated (30 min) before activation with specific translational inhibitors (puromycin, cycloheximide, and rapamycin). At the indicated time points, the cells were incubated with ProteaseArrest/EDTA (100×) for 5 min at room temperature to prevent protein degradation. The cells were pelleted and then lysed in 200 μl of 1× Laemmli buffer (0.025 M Tris, pH 6.8, 4% glycerol, 2% SDS, 1% β-mercaptoethanol, 0.25% bromophenyl blue). Samples were stored frozen at −20°C until SDS-PAGE and Western analysis was conducted as described previously (23).
RAR-α and IL-8 mRNA Expression.
Quiescent or PAF-stimulated human PMNs were lysed in Trizol reagent, and total RNA was isolated via a modification of previously described methods (24). SuperScript II or M-MLV reverse transcriptase (Invitrogen) was then used to generate cDNA. The cDNA was used for a PCR reaction designed to amplify a 233-bp product generated using primers against the RAR-α gene sequence (sense 5′-GGAGCTCATTGAGAAGGTGC-3′; antisense 5′-TTGAGGAGGGTGATCTGGTC-3′). Primers designed to amplify a 530-bp sequence of the GAPDH gene were used as an internal control. In selected experiments, primers specifically constructed based on the RAR-α1 (5′-CTGTCTGCCTCCCTTCTGA-3′) and RAR-α2 (5′-CCACCCCTAATCCCTTCCT-3′) isoforms (25) were used with a primer common to the coding region of RAR-α1 and RAR-α2 (see Characterization of the 5′-Untranslated Region of the RAR-α mRNA Using Rapid Amplification of cDNA Ends). For real time PCR measurements, we purchased premixed PCR primers against RAR-α (Hs00230907_m1) and IL-8 (Hs00174103) and separate primers for GAPDH (GAPDH Probe [designated “Joe”], GAPDH forward, GAPDH reverse). The Bio-Rad Laboratories iCycler IQ (Hercules), the MJ Research, Inc. Opticon 2, and Applied Biosystems 7900 HT real-time PCR analysis systems were used to generate quantitative measures of the expression of RAR-α, IL-8, and GAPDH mRNAs.
Characterization of the 5′-Untranslated Region of the RAR-α mRNA Using Rapid Amplification of cDNA Ends.
Total RNA was isolated from unstimulated human PMNs and used to generate first-strand rapid amplification of cDNA ends (RACE)–ready cDNA using reagents purchased from CLONTECH Laboratories, Inc. with the Smart RACE cDNA Amplification kit. The sequence for the 5′-untranslated region (UTR) of the RAR-α mRNA was amplified by PCR using the nested universal primer supplied by CLONTECH Laboratories, Inc. and a specific primer directed at the conserved regions of the RAR-α gene (5′-CCCCATAGTGGTAGCCTGAG-3′). The RAR-α 5′-UTR fragments were ligated into a pCR 2.1 vector and cloned into INF′ Escherichia coli bacteria supplied with the TA Cloning kit (pCR II, pCR 2.1) purchased from Invitrogen. Plasmid DNA was extracted and prepared for sequencing using a QIAprep Spin DNA Miniprep kit with supportive reagents purchased from QIAGEN. An Applied Biosystems ABI 3700 96-capillary sequencer operated by the DNA Sequencing Core Facility at the University of Utah was used to obtain sequences from the 5′-UTR containing plasmid DNA. DNASis software (MiraiBio) was used for analyzing the secondary structure of the 5′-UTR of the RAR-α mRNA.
Detection of IL-8 Protein.
Freshly isolated human PMNs were incubated with PAF (10−8 M), AM-580 (10−6 M), or the combination of both reagents at 37°C in 5% CO2 for various times. For other experiments, we used the same conditions with and without rapamycin pretreatment (10 nM) for 1 h. The supernatants were collected and frozen at −20°C until analysis by ELISA. IL-8 protein levels were quantitatively assessed using the human IL-8 DuoSet ELISA system from R&D Systems according to their protocols. Standard 96-well ELISA plates with samples were analyzed using SOFTmax for Macintosh Elisa software (Molecular Devices) and a Molecular Devices THERMOmax Microplate Reader.
Online Supplemental Material.
In Fig. S1, HL-60 cells were cultured, differentiated, and characterized as described in preceding sections. In Fig. S2, PMNs were isolated, stimulated with PAF, and IL-8 mRNA and protein were analyzed as outlined above. Figs. S1 and S2 are available at http://www.jem.org/cgi/content/full/jem.20040224/DC1.
Results
The mRNA for RAR-α Is Found in Polyribosomes of PAF-stimulated Surrogate PMNs.
PMNs release an array of basally stored granular proteins into the inflammatory milieu (1). In addition, they expand their proteomic repertoire by synthesizing new proteins, many of which are under transcriptional control, in response to cellular activation (2, 13, 26, 27). To date, only a limited number of newly synthesized proteins have been identified, and the molecular pathways that regulate their production are largely unknown (13, 27). To characterize additional genes under signal-dependent control, we first profiled mRNAs associated with polyribosomes (polysomes), an index of active translation (28), in resting and activated HL-60 surrogate PMNs. Polysome analysis provides an approach that links genomic and proteomic determinants of cellular phenotype (28). Differentiation of HL-60 cells along the PMN lineage was confirmed by segmented nuclear morphology, gain of responsiveness to PAF, and loss of transferrin receptor expression (15–17) (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20040224/DC1).
Unstimulated HL-60 PMNs displayed typical ribosomal profiles (22, 28) consisting of 40S and 60S ribosomal subunit peaks and an intact 80S single ribosome peak, with smaller polysome peaks in the gradient (Fig. 1 A). When we stimulated these cells with PAF, a potent inflammatory phospholipid that rapidly activates PMNs (13, 29, 30), the heights of the 40S and 60S ribosomal subunit peaks decreased, and in parallel the height of the 80S monosome peak increased (Fig. 1 A) indicating translation initiation (10). We then examined patterns of mRNAs associated with polysomes in resting and activated surrogate PMNs by cDNA microarray analysis (22) to identify transcripts undergoing active translation (28). We found 28 mRNAs that were differentially associated with polysomes in PAF-stimulated cells compared with unstimulated HL-60 PMNs (Table I). One transcript of particular interest was RAR-α, a nuclear hormone receptor that binds retinoic acid (12). The RAR-α gene is best known as the target of chromosomal rearrangement in acute promyelocytic leukemia (31). Although RAR-α has not been identified in terminally differentiated mature PMNs, it modulates the commitment of pluripotent hematopoietic progenitors to the neutrophil lineage (32). Because the RARs directly and indirectly regulate transcription of multiple gene products (12), appearance of the mRNA encoding RAR-α in polysomes in response to activation of HL-60 surrogate PMNs suggested that it may represent a key checkpoint in PMN synthetic pathways.
Figure 1.
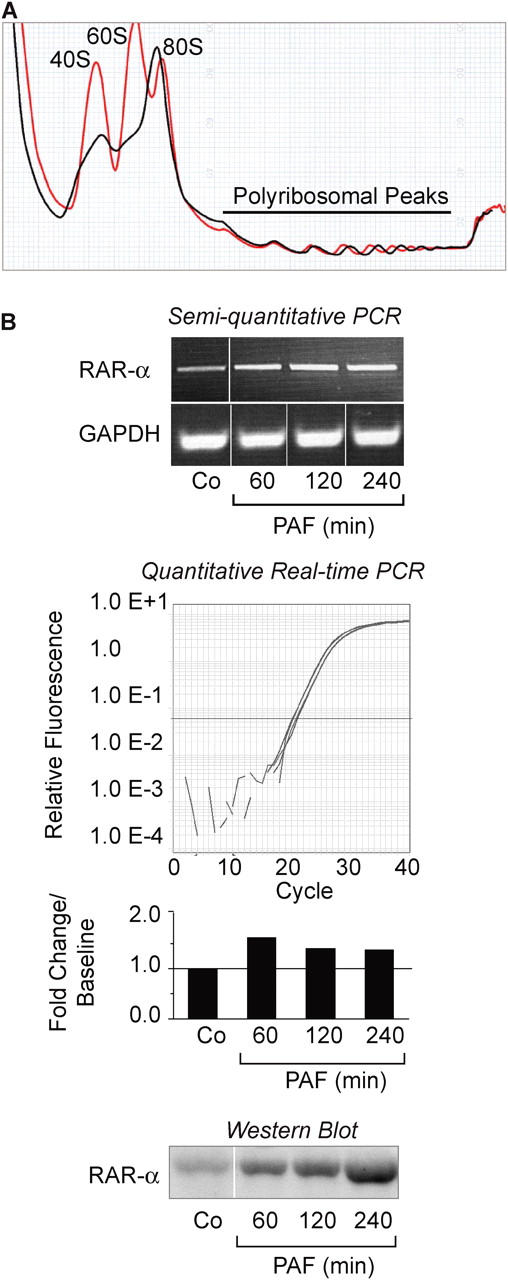
PAF regulates translational responses in differentiated HL-60 PMNs. (A) Ribosomal profile of HL-60 PMNs that were left quiescent (red tracing) or activated with 10 nM PAF for 1 h (black tracing). These profiles are representative of three independent experiments. Total RNA was isolated from the polyribosome peaks and used to generate cDNA for microarray analysis (as described in Materials and Methods and Table I). (B) The top panel illustrates RAR-α and GAPDH mRNA expression in PAF-stimulated HL-60 PMNs as measured by semiquantitative RT-PCR. (Dividing lines indicate elimination of additional time points and blank lanes between individual samples from the figure.) The middle panel displays RAR-α mRNA levels as measured by real-time PCR. The bars indicate the fold change in RAR-α mRNA copy numbers compared with control (Co) cells at baseline, which was arbitrarily set at 1. The tracings immediately above the bar graph indicate the amplification curves for each time point. The bottom panel demonstrates Western blots of RAR-α protein expression in differentiated HL-60 PMNs under control conditions or when stimulated with PAF. The results in B are representative of three independent experiments.
Table I.
mRNAs Expressed in Polysomes of Quiescent and PAF-stimulated HL-60 PMNs
| Messages expressed in polysomes of quiescent HL-60 PMNs only |
|---|
| GRB10 |
| BCL2-related protein A1 (BCL2A1) |
| Caspase 6 |
| CD27BP (Siva) |
| RXR-α |
| Messages expressed in polysomes of quiescent and PAF-stimulated HL-60 PMNs |
| Cell division protein kinase 6 (CDK6) |
| Cyclin-dependent kinase inhibitor 1A (CDKN1A) |
| Peptidyl-prolyl cis-transisomerase nima-interacting 1 (PIN1) |
| Prefoldin 5 (PFDN5) |
| Vimentin (VIM) |
| LPS-induced TNFα factor (LITAF) |
| Ras homolog gene family member A (RHOA) |
| Erb-3 proto-oncogene |
| TNF receptor 2 (TNFR2) |
| Villin 2 (VIL2) |
| Ninjurin 1 |
| Nucleoside diphosphate kinase β (NDKβ) |
| Rho-GAP hematopoietic protein C1 (RGC1) |
| Rho-GDP dissociation inhibitor 1 (RHO-GDI1) |
| Cadherin 5 (CDH5) |
| Early growth response protein 1 (EGR1) |
| Interferon γ antagonist |
| Interleukin 1 receptor antagonist protein (IL1RA) |
| Interleukin 1β |
| Messages expressed in polysomes of PAF-stimulated HL-60 PMNs only |
| Cyclin-dependent kinase 4 inhibitor 2D (CDKN2D) |
| Wee1Hu CDK tyrosine 15 kinase |
| CDC10 protein homolog |
| Ubiquitin-conjugating enzyme E2 |
| CDC37 homolog |
| Proliferating cyclic nuclear antigen (PCNA) |
| Growth factor receptor-bound protein 2 (GRB2) |
| B-raf proto-oncogene (RAFB1) |
| Myeloid cell leukemia protein 1 (MCL1) |
| Glutathione S-transferase–like protein |
| Chromatin assembly factor 1 p48 subunit |
| Growth arrest and DNA damage inducible protein 153 |
| Ubiquitin-conjugating enzyme E2 17-kD |
| UV excision repair protein RAD 23 homolog A |
| RAR-α |
| Aggrecan 1 (AGC1) |
| CD 59 glycoprotein |
| Integrin β8 |
| Malignant melanoma metastasis-suppressor gene |
| Interferon γ receptor β subunit |
mRNAs for key proteins are differentially associated with polysomes of quiescent and PAF-stimulated HL-60 PMNs. IL-8, which was examined elsewhere in these studies, was not represented in the array used for these experiments.
RAR-α Transcripts Are Constitutively Expressed in Resting HL-60 PMNs and Are Translated into Protein in Response to Cellular Activation.
Translocation of RAR-α mRNA into polysomes of HL-60 PMNs was rapid, occurring within 1 h of activation with PAF. When we measured total RAR-α mRNA in resting and PAF-stimulated cells by standard and quantitative PCR, we found that it is present in unstimulated HL-60 PMNs and that its levels are only minimally altered by activation (Fig. 1 B). We then examined correlation of this constitutive RAR-α mRNA with the protein product in these cells. Only low levels of RAR-α protein were present in unstimulated HL-60 PMNs at baseline (Fig. 1 B, bottom) even though the transcript was present (Fig. 1 B, top). However, RAR-α protein dramatically accumulated within 60 min after cellular activation (Fig. 1 B, bottom) consistent with the observation that RAR-α mRNA associates with polysomes in PAF-activated HL-60 PMNs. These data indicate that constitutive RAR-α mRNA is mobilized to polysomes in HL-60 PMNs stimulated with PAF where it is then efficiently translated. This is consistent with a mechanism in which expression of the protein product is regulated at translational checkpoints (9, 10, 13, 28).
RAR-α mRNA Is Constitutively Expressed in Primary Human PMNs and Is Translated into Protein in a Signal-dependent Fashion.
Although differentiated HL-60 PMNs exhibit features of circulating human neutrophils (see The mRNA for RAR-α Is Found in Polyribosomes of PAF-stimulated Surrogate PMNs and Fig. S1) and are a useful surrogate model, the biological features of cell lines are often substantially different when compared with those observed in the corresponding primary human cell. Therefore, we characterized RAR-α mRNA and protein expression in freshly isolated human PMNs and in PMNs activated with PAF. Like HL-60 surrogate PMNs, quiescent primary PMNs express mRNA for RAR-α at baseline (Fig. 2 A). Levels of the RAR-α transcript did not increase and, in fact, decreased after stimulation with PAF (Fig. 2 A). Little or no RAR-α protein was present in resting PMNs, but in contrast it was dramatically induced after cellular activation with PAF (Fig. 2 A, bottom and not depicted). Recombinant PAF acetylhydrolase, an enzyme that selectively degrades PAF (30), blocked expression of RAR-α protein under these conditions indicating specificity of the agonist effect (not depicted). This same pattern of RAR-α expression was seen in an experiment using neonatal PMNs isolated from umbilical cord blood at the time of term delivery and stimulated with PAF under conditions similar to those used for PMNs from adult donors (not depicted). In addition to PAF, TNFα, lipopolysaccharide, and n-formyl-methionyl-leucyl-phenylalanine also triggered rapid synthesis of RAR-α (not depicted). Thus, outside-in signals delivered by other classes of receptors also induce this response, in addition to the PAF signaling pathway.
Figure 2.
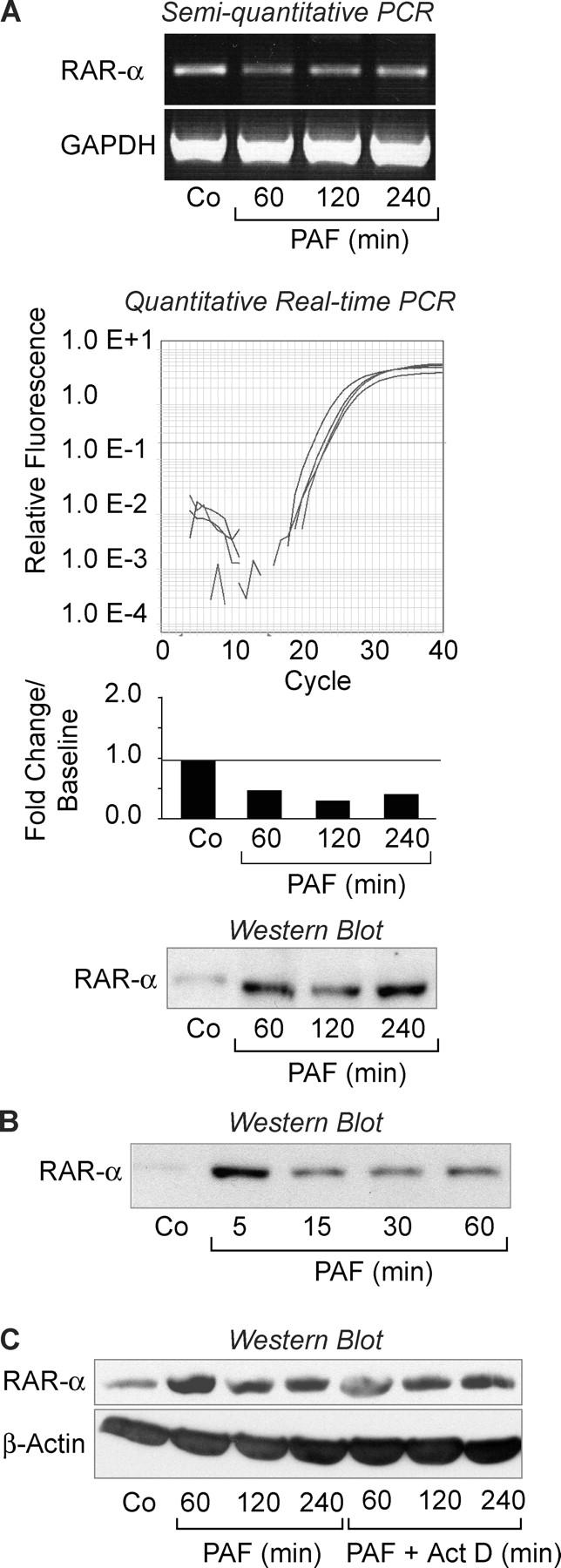
Cellular activation rapidly induces RAR-α protein expression from constitutive mRNA in human PMNs. (A) The top panel illustrates RAR-α and GAPDH mRNA expression in control (Co) PMNs or PMNs activated with PAF as assayed by semiquantitative RT-PCR. The middle panel illustrates RAR-α mRNA expression as measured by real-time PCR. The bars represent the fold change in RAR-α mRNA copy number compared with baseline (Co), which was arbitrarily set at 1. The tracings immediately above the bar graph show the amplification curves for each time point. The bottom panel illustrates RAR-α protein levels in control PMNs or PMNs stimulated with PAF over a 4-h time period. (B) A Western blot demonstrates RAR-α protein expression in PMNs stimulated with PAF over the course of 1 h. (c) RAR-α protein expression in PMNs activated with PAF after preincubation with control buffer or actinomycin D was examined by Western analysis. The same samples were also probed for β-actin. Experiments in this figure are representative of at least five independent studies.
Rapid de novo expression of RAR-α protein by activated PMNs suggests an alternate mode of transcriptional control that contrasts with NF-κB, which translocates to the nucleus and regulates transcription in response to cellular activation (2, 7). The experiments in Fig. 2 indicate that RAR-α is also controlled in a signal-dependent fashion, but its activity requires new synthesis rather than intracellular translocation of preformed protein from cytoplasmic to nuclear compartments as with NF-κB. Therefore, we further characterized features of this event in stimulated PMNs.
Synthesis of RAR-α Protein is Regulated at Translational Checkpoints in Activated PMNs.
Although there is differential expression of RARs in a variety of cells, most studies have focused on mechanisms by which RAR-α regulates the activity of target promoters (12, 31), and pathways that control synthesis of the protein are not well understood. Experiments in Fig. 2 A indicated that RAR-α protein is translated from constitutive mRNA in activated neutrophils. When we shortened the time course, we found that RAR-α protein could be expressed within intervals as short as 5 min in PAF-stimulated PMNs (Fig. 2 B). This, together with the discrepancy between RAR-α mRNA and protein levels in resting PMNs (Fig. 2, A and B), is a strong indication that RAR-α is under translational control (10, 13, 22, 23, 28). Actinomycin D, a transcriptional inhibitor, did not block synthesis of RAR-α protein under these conditions, indicating that its production is controlled post-transcriptionally (Fig. 2 C). In contrast, actinomycin D completely inhibited expression of the mRNA for IL-8 and synthesis of the IL-8 protein (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20040224/DC1), consistent with previous observations that transcriptional regulation is critical in its production by human PMNs (2). Using similar experimental protocols, actinomycin D also selectively inhibited production of other protein products of stimulated neutrophils (13). Studies with puromycin and cycloheximide, which are translational inhibitors, confirmed that accumulation of RAR-α protein requires new synthesis in neutrophils activated with PAF (Fig. 3 A).
Figure 3.
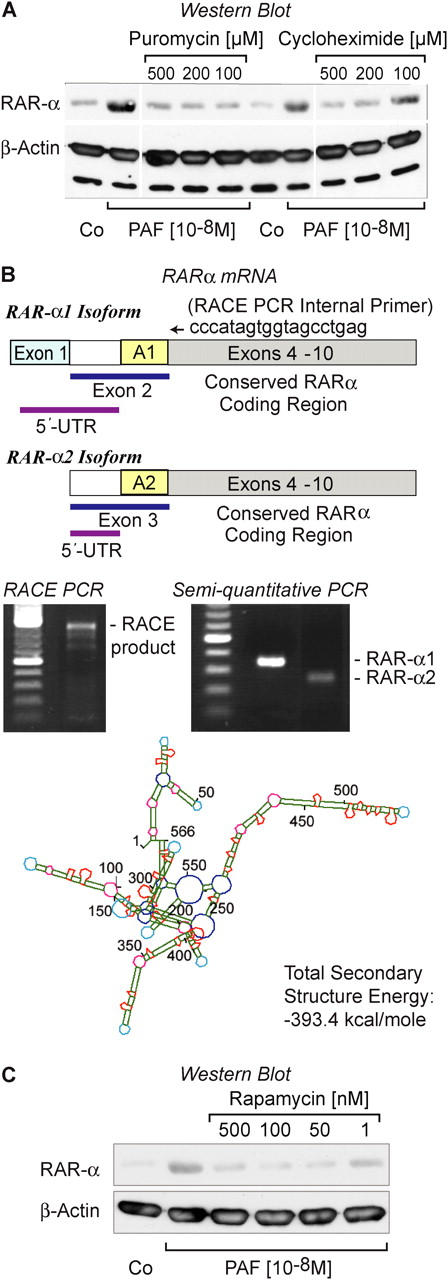
Signal-dependent synthesis of RAR-α is regulated at the translational level by mTOR in human PMNs stimulated by PAF. (A) PMNs were stimulated with PAF for 30 min in the presence of vehicle alone as a control (CO); puromycin, or cycloheximide and RAR-α protein levels were examined by Western analysis. The same samples were simultaneously probed for β-actin. (Dividing lines indicate elimination of additional concentrations of puromycin and cycloheximide from the figure.) These data are representative of four independent experiments. (B) Stick diagrams illustrating the structure of the transcripts for the RAR-α1 and RAR-α2 isoforms are shown. A 3′-flanking RACE primer (ccccatagtggtagcctgag) was directed at exon 4 of RAR-α (see Synthesis of RAR-α Protein is Regulated at Translational Checkpoints in Activated PMNs), which is conserved in the coding regions of the RAR-α1 and RAR-α2 isoforms. The 5′-UTR of RAR-α1 is derived from exon 1 and a portion of exon 2. The 5′-UTR of RAR-α2 is derived from a portion of exon 3 and is shorter than that of RAR-α1. See Zelent et al. (31) for details. The gels immediately below the stick figures illustrate results of experiments identifying the RAR-α isoform expressed by human PMNs. The left panel demonstrates the dominant 5′-UTR RACE product, which is RAR-α1 (see Results and Discussion for details). The right panel illustrates results of semiquantitative PCR analysis of RAR-α1 and RAR-α2 using primers specific for their respective 5′-UTRs (as described in Materials and Methods). These results are representative of two independent experiments. The bottom panel of B illustrates the predicted secondary structure of the 5′-UTR of RAR-α1 mRNA expressed in human PMNs. (C) PMNs were pretreated with rapamycin in increasing concentrations and then activated with PAF, followed by Western blot analysis for RAR-α protein. Expression of β-actin protein was determined in the same samples. This result is representative of three independent experiments.
Sequence and structure of the 5′-UTR of specific mRNA transcripts can regulate and suppress their translation (9–11, 13, 23, 24), although in many cases this has been examined in model systems and is not well documented in primary human cells. RAR-α transcripts are differentially expressed during development as two isoforms, α1 and α2. The most common isoform, RAR-α1, is found in many adult tissues (32). Exons 4–10, which cover the majority of the coding region, are conserved between RAR-α subtypes (31). However, RAR-α1 and RAR-α2 have different 5′-UTRs (31) (Fig. 3 B). Therefore, we characterized the 5′-UTR of the RAR-α mRNA in freshly isolated PMNs using sequence-specific primers and found that RAR-α1 is the predominant isoform (Fig. 3 B). In addition, RACE-PCR, using an internal primer that recognizes a sequence in the coding region common to both RAR-α1 and RAR-α2, yielded one dominant band (Fig. 3 B). Cloning and sequencing of this product demonstrated that the RAR-α1 isoform is the primary transcript in PMNs.
The 5′-UTR of the RAR-α1 mRNA in PMNs exceeds 500 nucleotides, is very GC rich, and computer folding indicates that it contains numerous hairpins and significant secondary structure (predicted ΔG = −393.4 kcal/mol) (Fig. 3 B). Transcripts with 5′-UTR secondary structures that require energy to unwind exceeding −60 kcal/mol are often repressed at the translational level (9, 10, 13, 23, 24, 33). Previously, it was noted that sequence features of the 5′-UTRs of mRNAs coding for RAR isoforms predict translational control (9, 11), but this mode of regulated expression of their proteins has not been reported in human leukocytes.
In other cell types, mTOR regulates events that unfold 5′-UTR secondary structure, relieving repression and initiating translation of a subset of key mRNAs that are under tight synthetic control (13, 23, 24, 33). In parallel studies, we found that mTOR and its downstream targets (p70S6 kinase and eukaryotic initiation factor-4E binding protein-1 [4E-BP1]) are present and active in stimulated PMNs (13), suggesting the possibility that mTOR controls RAR-α synthesis in response to PAF stimulation. Rapamycin, an immunosuppressant macrolide, specifically blocks mTOR-dependent signaling in myeloid leukocytes (13, 23) in addition to other cell types (33). We found that rapamycin potently inhibits the synthesis of RAR-α protein in PAF-stimulated PMNs (Fig. 3 C). In contrast, it had no effect on the level of actin (Fig. 3 C), a gene product that is independent of signal-dependent translational control (23).
Rapamycin is used clinically to prevent rejection after organ transplantation and to reduce the incidence of vascular restenosis after interventional procedures in atherosclerotic vascular syndromes (34). It is generally believed that its therapeutic effects largely involve blocking proliferation of dividing cells (34, 35). Inhibition of expression of RAR-α (Fig. 3 C) and a subgroup of other key gene products in PMNs (13), which are terminally differentiated cells that do not divide, indicates that rapamycin also modulates other cellular responses and may have previously unrecognized effects on inflammation. Inhibition of RAR-α protein expression in activated PMNs by rapamycin, which is a critical molecular probe for the mTOR pathway (33, 36), also provides new insights into the interaction between specialized translation control pathways and transcriptional regulation (9) in this cell type. Although it has been observed that homologues of mTOR influence transcriptional activity in response to nutrient stress in yeast (36) and that mTOR regulates expression of Bcl-3, a member of the NF-κB family of transcriptional regulators, in other human cells (24, 37) selective translational control of a transcription factor in activated myeloid leukocytes has not been reported previously.
RAR-α Regulates the Expression of IL-8 by Activated PMNs.
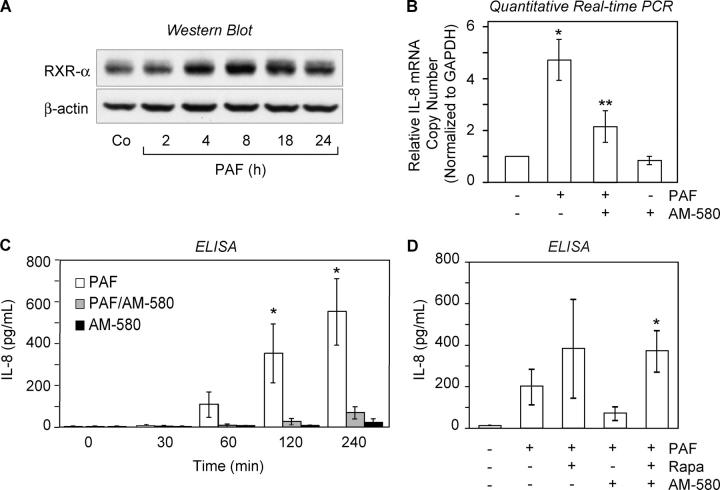
One mechanism by which RAR-α modulates transcription is by binding to promoter sequences that contain retinoic acid response elements (RAREs) (12). RAR-α binds consensus elements more efficiently in the presence of a second group of retinoic acid receptors, the retinoid X receptors (RXRs) (12). We found that RXR-α protein is constitutively present in resting PMNs (Fig. 4 A), indicating that it is available to form RXR-RAR heterodimers and that RXRα and RAR-α1 are differentially expressed in this cell type. IL-8 is one of a relatively small number of RAR-responsive products known to by synthesized by activated PMNs among the large group of genes whose expression is regulated by retinoic acid (12). We found that mRNA for IL-8 increased after stimulation of PMNs with PAF (Fig. S2 and Fig. 4 B), consistent with induced synthesis of this chemokine in response to stimulation (2). We then examined the effect of AM-580, a synthetic retinoid agonist with specificity for RAR-α (38), under conditions in which expression of RAR-α and IL-8 is induced. AM-580 significantly attenuated IL-8 transcript accumulation after 60 min of stimulation with PAF, whereas treatment of unactivated PMNs with AM-580 had no effect (Fig. 4 B). In parallel, we found that PAF-stimulated PMNs synthesize significant amounts of IL-8 protein. Treatment with AM-580 markedly attenuated PAF-induced IL-8 accumulation as measured by ELISA (Fig. 4 C). A similar effect on PAF-induced IL-8 protein synthesis was noted with all-trans retinoic acid treatment (not depicted). AM-580 (Fig. 4 C) or all-trans retinoic acid (not depicted) alone did not significantly alter IL-8 synthesis. We then pretreated PMNs with rapamycin before stimulation with PAF (Fig. 4 D) to inhibit the induction of RAR-α protein (Fig. 3 C). AM-580 failed to attenuate synthesis of IL-8 under these conditions (Fig. 4 D). Together these results indicate that engagement of newly expressed RAR-α1 by ligands can alter IL-8 transcription and synthesis of IL-8 protein in stimulated human PMNs and that mTOR regulates this sequence of events by controlling RAR-α translation. Furthermore, they indicate that RAR-α has activities beyond granulopoiesis (39) in this cell lineage. It is also possible that unliganded RAR-α1 regulates expression of IL-8 in PMNs activated with PAF (40). The PAF and retinoid systems have previously been reported to synergistically alter expression of cyclooxygenase 2 in neural cells and fibroblasts (41), but convergence of these signaling and regulatory mechanisms has not been observed in myeloid leukocytes.
Figure 4.
Retinoic acid receptor pathways regulate signal-dependent synthesis of IL-8 in human PMNs. (A) RXR-α and β-actin protein expression was determined by Western analysis in control PMNs and PMNs stimulated with PAF. (B) The relative mRNA copy number for IL-8, normalized to GAPDH, was analyzed by quantitative real-time PCR in PMNs treated for 60 min with PAF (10 nM), PAF together with AM-580, or AM-580 only. The bars represent fold changes compared with baseline (left bar), which was arbitrarily set to 1. The asterisk indicates statistical significance (P < 0.05) compared with control and the double asterisk significance (P < 0.05) comparing PAF and PAF/AM-580 treated PMNs. The data in this panel indicate the mean ± SEM of four separate experiments. (C) PMNs were activated with PAF or PAF in the presence of AM-580 or were treated with AM-580 alone. The bars indicate the mean ± SEM of five separate experiments. The asterisk identifies statistical significance (P < 0.05) between groups treated with PAF or PAF together with AM-580. (D) PMNs were pretreated with buffer or with rapamycin (10 nM) and then stimulated with PAF (10 nM) in buffer alone or together with AM-580 for 240 min. The bars indicate the mean ± SEM of three separate experiments. The asterisk identifies statistical significance (P < 0.05) between groups treated with PAF and AM-580 or PAF/AM-580 together with rapamycin.
Like many retinoid responsive genes the IL-8 promoter does not contain a classic RARE sequence, indicating that it may be modulated by RAR-α in an indirect fashion (12). Genes that contain consensus RAREs are less common and none of them have been identified in PMNs to date. Indirect regulation usually occurs via an intermediate transcription factor that is controlled by RAR-α or a nonclassical association of the receptor with another modulatory protein or transcription factor (12). These interactions can lead to protein sequestration and prevention of critical transactivating protein–protein interactions (12) with impaired synthesis of the corresponding protein product. These features remain to be explored in activated PMNs.
Discussion
Synthesis of RAR-α under signal-dependent translational control is a new feature of gene expression in activated PMNs that may herald unrecognized regulatory mechanisms in acute inflammation. Using cDNA array analysis, we have found that PMNs also express multiple additional constitutive transcripts in the resting state and that in response to PAF and other agonists a subset of these messages is translated in an mTOR-dependent fashion. One protein product controlled in this manner is the IL-6 receptor α, a soluble factor that mediates signaling of endothelial cells and transition to the mononuclear phase of inflammation (13). Regulation of expression of the IL-6 receptor α protein by signal-dependent translation is consistent with the general prediction that specialized translational mechanisms frequently control synthesis of potent biologically active proteins (9). Rapid translation-dependent synthesis of a transcription factor that can then modulate expression of a critical chemokine, IL-8, and potentially other inflammatory factors—as we show here for RAR-α—is also consistent with this analysis and identifies a novel mechanism of regulatory interplay between transcriptional and translational systems in activated human PMNs. Such interplay between transcriptional and post-transcriptional mechanisms provides the potential for exquisite precision in timing and magnitude of expression of potent factors with key biologic activities (9–11) and may be particularly important in cells with critical and highly regulated functions such as those subserved by PMNs in inflammation. mTOR is one checkpoint at which specialized translational control can influence expression and activity of transcription factors and other regulatory proteins in response to outside-in signaling of PMNs. Additional evidence for differential expression of mRNAs and their corresponding proteins in lipopolysaccharide-stimulated PMNs indicates that regulated translational events occur under these conditions as well (27). Thus, signaling to posttranscriptional checkpoints and to translational control mechanisms appears to be an important determinant of the proteome of activated neutrophils. As in other specialized cell types, diverse posttranscriptional mechanisms are likely to operate (11, 22) in concert with transcriptional regulation to yield specific protein patterns and PMN phenotypes in the complex setting of the inflammatory milieu.
Acknowledgments
We thank our technical staff for help with cell isolation and culture, Michele Czerwinski and Mary Madsen for preparation of the article, our colleagues in the Program in Human Molecular Biology and Genetics for helpful discussions, and Diana Lim for preparation of the figures.
This work was supported in part by the Children's Health Research Center of the University of Utah and the Primary Children's Medical Center Foundation (awards to C.C. Yost and F.J. Rubner), a Physician-Scientist Training Award from the American Diabetes Association (American Diabetes Association no. PID2206052 to M.M. Denis), the National Institutes of Health (HL-44525 to G.A. Zimmerman, HL-66277 to A.S. Weyrich, HL-44513 to T.M. McIntyre, and P50 HL-50153 to T.M. McIntyre and G.A. Zimmerman), and Established Investigator Award from the American Heart Association (to A.S. Weyrich). C.C. Yost is a member of the Fellowship-to-Faculty Transition Program at the University of Utah, which is supported in part by the Howard Hughes Medical Institute Biomedical Research Program for Medical Schools.
The authors have no conflicting financial interests.
S. Lindemann's present address is I1 Medizinische Klinik der Johannes-Gutenberg Universitaet, 55101 Mainz, Germany.
Abbreviations used in this paper: mRNA, messenger RNA; mTOR, mammalian target of rapamycin; PAF, platelet activating factor; PMN, polymorphonuclear leukocyte; RACE, rapid amplification of cDNA ends; RAR, retinoic acid receptor; RARE, retinoic acid response element; RXR, retinoid X receptor; UTR, untranslated region.
References
- 1.Chilvers, E.R., K.A. Cadwallader, B.J. Reed, J.F. White, and A.M. Condliffe. 2000. The function and fate of neutrophils at the inflamed site: prospects for therapeutic intervention. J.R. Coll. Physicians Lond. 34:68–74. [PMC free article] [PubMed] [Google Scholar]
- 2.Scapini, P., J.A. Lapinet-Vera, S. Gasperini, F. Calzetti, F. Bazzoni, and M.A. Cassatella. 2000. The neutrophil as a cellular source of chemokines. Immunol. Rev. 177:195–203. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi, S.D., J.M. Voyich, C.L. Buhl, R.M. Stahl, and F.R. DeLeo. 2002. Global changes in gene expression by human polymorphonuclear leukocytes during receptor-mediated phagocytosis: cell fate is regulated at the level of gene expression. Proc. Natl. Acad. Sci. USA. 99:6901–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walzog, B., P. Weinmann, F. Jeblonski, K. Scharffetter-Kochanek, K. Bommert, and P. Gaehtgens. 1999. A role for beta(2) integrins (CD11/CD18) in the regulation of cytokine gene expression of polymorphonuclear neutrophils during the inflammatory response. FASEB J. 13:1855–1865. [DOI] [PubMed] [Google Scholar]
- 5.Sconocchia, G., L. Campagnano, D. Adorno, A. Iacona, N.Y. Cococcetta, V. Boffo, S. Amadori, and C.U. Casciani. 2001. CD44 ligation on peripheral blood polymorphonuclear cells induces interleukin-6 production. Blood. 97:3621–3627. [DOI] [PubMed] [Google Scholar]
- 6.Weinmann, P., P. Gaehtgens, and B. Walzog. 1999. Bcl-Xl- and Bax-alpha-mediated regulation of apoptosis of human neutrophils via caspase-3. Blood. 93:3106–3115. [PubMed] [Google Scholar]
- 7.McDonald, P.P., and M.A. Cassatella. 1997. Activation of transcription factor NF-kappa B by phagocytic stimuli in human neutrophils. FEBS Lett. 412:583–586. [DOI] [PubMed] [Google Scholar]
- 8.Abraham, E. 2003. Neutrophils and acute lung injury. Crit. Care Med. 31:S195–S199. [DOI] [PubMed] [Google Scholar]
- 9.Kozak, M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathews, M.B., N. Sonenberg, and J.W.B. Hershey. 2000. Origins and principles of translational control. In Translational Control of Gene Expression. N. Sonenberg and J.W.B. Hershey, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1–31.
- 11.Brewer, G. 2001. Misregulated posttranscriptional checkpoints: inflammation and tumorigenesis. J. Exp. Med. 193:F1–F4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balmer, J.E., and R. Blomhoff. 2002. Gene expression regulation by retinoic acid. J. Lipid Res. 43:1773–1808. [DOI] [PubMed] [Google Scholar]
- 13.Lindemann, S.W., C.C. Yost, N.H. Riedel, T.M. McIntyre, A.S. Weyrich, and G.A. Zimmerman. 2004. Activated human neutrophils synthesize IL-6 receptor α by signal-dependent translational regulation: a new mechanism for rapid alteration of the acute inflammatory milieu. Proc. Natl. Acad. Sci. USA. 101:7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmerman, G.A., T.M. McIntyre, and S.M. Prescott. 1985. Thrombin stimulates the adherence of neutrophils to human endothelial cells in vitro. J. Clin. Invest. 76:2235–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi, T., T. Mukasa, E. Uchida, T. Kanayasu-Toyoda, and T. Hayakawa. 1999. The role of STAT3 in granulocyte colony-stimulating factor-induced enhancement of neutrophilic differentiation of Me2SO-treated HL-60 cells. GM-CSF inhibits the nuclear translocation of tyrosine-phosphorylated STAT3. J. Biol. Chem. 274:15575–15581. [DOI] [PubMed] [Google Scholar]
- 16.Kanayasu-Toyoda, T., T. Yamaguchi, T. Oshizawa, M. Kogi, E. Uchida, and T. Hayakawa. 2002. Role of the p70 S6 kinase cascade in neutrophilic differentiation and proliferation of HL-60 cells—a study of transferrin receptor-positive and -negative cells obtained from dimethyl sulfoxide- or retinoic acid-treated HL-60 cells. Arch. Biochem. Biophys. 405:21–31. [DOI] [PubMed] [Google Scholar]
- 17.Ye, R.D., E.R. Prossnitz, A.H. Zou, and C.G. Cochrane. 1991. Characterization of a human cDNA that encodes a functional receptor for platelet activating factor. Biochem. Biophys. Res. Commun. 180:105–111. [DOI] [PubMed] [Google Scholar]
- 18.Weyrich, A.S., M.R. Elstad, R.P. McEver, T.M. McIntyre, K.L. Moore, J.H. Morrissey, S.M. Prescott, and G.A. Zimmerman. 1996. Activated platelets signal chemokine synthesis by human monocytes. J. Clin. Invest. 97:1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos-Beneit, A.M., and F. Mollinedo. 2000. Expression of genes involved in initiation, regulation, and execution of apoptosis in human neutrophils and during neutrophil differentiation of HL-60 cells. J. Leukoc. Biol. 67:712–724. [DOI] [PubMed] [Google Scholar]
- 20.Marathe, G.K., G.A. Zimmerman, S.M. Prescott, and T.M. McIntyre. 2002. Activation of vascular cells by PAF-like lipids in oxidized LDL. Vascul. Pharmacol. 38:193–200. [DOI] [PubMed] [Google Scholar]
- 21.Kaspar, R.L., and L. Gehrke. 1994. Peripheral blood mononuclear cells stimulated with C5a or lipopolysaccharide to synthesize equivalent levels of IL-1 beta mRNA show unequal IL-1 beta protein accumulation but similar polyribosome profiles. J. Immunol. 153:277–286. [PubMed] [Google Scholar]
- 22.Lindemann, S., N.D. Tolley, D.A. Dixon, T.M. McIntyre, S.M. Prescott, G.A. Zimmerman, and A.S. Weyrich. 2001. Activated platelets mediate inflammatory signaling by regulated interleukin 1beta synthesis. J. Cell Biol. 154:485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahoney, T.S., A.S. Weyrich, D.A. Dixon, T. McIntyre, S.M. Prescott, and G.A. Zimmerman. 2001. Cell adhesion regulates gene expression at translational checkpoints in human myeloid leukocytes. Proc. Natl. Acad. Sci. USA. 98:10284–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weyrich, A.S., D.A. Dixon, R. Pabla, M.R. Elstad, T.M. McIntyre, S.M. Prescott, and G.A. Zimmerman. 1998. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc. Natl. Acad. Sci. USA. 95:5556–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leroy, P., A. Krust, A. Zelent, C. Mendelsohn, J.M. Garnier, P. Kastner, A. Dierich, and P. Chambon. 1991. Multiple isoforms of the mouse retinoic acid receptor alpha are generated by alternative splicing and differential induction by retinoic acid. EMBO J. 10:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashiro, S., H. Kamohara, J.M. Wang, D. Yang, W.H. Gong, and T. Yoshimura. 2001. Phenotypic and functional change of cytokine-activated neutrophils: inflammatory neutrophils are heterogeneous and enhance adaptive immune responses. J. Leukoc. Biol. 69:698–704. [PubMed] [Google Scholar]
- 27.Fessler, M.B., K.C. Malcolm, M.W. Duncan, and G.S. Worthen. 2002. A genomic and proteomic analysis of activation of the human neutrophil by lipopolysaccharide and its mediation by p38 mitogen-activated protein kinase. J. Biol. Chem. 277:31291–31302. [DOI] [PubMed] [Google Scholar]
- 28.Pradet-Balade, B., F. Boulme, H. Beug, E.W. Mullner, and J.A. Garcia-Sanz. 2001. Translation control: bridging the gap between genomics and proteomics? Trends Biochem. Sci. 26:225–229. 11295554 [Google Scholar]
- 29.Lorant, D.E., K.D. Patel, T.M. McIntyre, R.P. McEver, S.M. Prescott, and G.A. Zimmerman. 1991. Coexpression of GMP-140 and PAF by endothelium stimulated by histamine or thrombin: a juxtacrine system for adhesion and activation of neutrophils. J. Cell Biol. 115:223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prescott, S., G.A. Zimmerman, D.M. Stafforini, and T.M. McIntyre. 2000. Platelet-activating factor and related lipid mediators. Annu. Rev. Biochem. 69:419–445. [DOI] [PubMed] [Google Scholar]
- 31.Zelent, A., F. Guidez, A. Melnick, S. Waxman, and J.D. Licht. 2001. Translocations of the RARalpha gene in acute promyelocytic leukemia. Oncogene. 20:7186–7203. [DOI] [PubMed] [Google Scholar]
- 32.de The, H., A. Marchio, P. Tiollais, and A. Dejean. 1989. Differential expression and ligand regulation of the retinoic acid receptor alpha and beta genes. EMBO J. 8:429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gingras, A.C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807–826. [DOI] [PubMed] [Google Scholar]
- 34.Marx, S.O., and A.R. Marks. 2001. Bench to bedside: the development of rapamycin and its application to stent restenosis. Circulation. 104:852–855. [DOI] [PubMed] [Google Scholar]
- 35.Morice, W.G., G.J. Brunn, G. Wiederrecht, J.J. Siekierka, and R.T. Abraham. 1993. Rapamycin-induced inhibition of p34cdc2 kinase activation is associated with G1/S-phase growth arrest in T lymphocytes. J. Biol. Chem. 268:3734–3738. [PubMed] [Google Scholar]
- 36.Raught, B., A.C. Gingras, and N. Sonenberg. 2001. The target of rapamycin (TOR) proteins. Proc. Natl. Acad. Sci. USA. 98:7037–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraiss, L.W., A.S. Weyrich, N.M. Alto, D.A. Dixon, T.M. Ennis, V. Modur, T.M. McIntyre, S.M. Prescott, and G.A. Zimmerman. 2000. Fluid flow activates a regulator of translation, p70/p85 S6 kinase, in human endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 278:H1537–H1544. [DOI] [PubMed] [Google Scholar]
- 38.Teng, M., T.T. Duong, E.S. Klein, M.E. Pino, and R.A. Chandraratna. 1996. Identification of a retinoic acid receptor alpha subtype specific agonist. J. Med. Chem. 39:3035–3038. [DOI] [PubMed] [Google Scholar]
- 39.Kastner, P., H.J. Lawrence, C. Waltzinger, N.B. Ghyselinck, P. Chambon, and S. Chan. 2001. Positive and negative regulation of granulopoiesis by endogenour RARα. Blood. 97:1314–1320. [DOI] [PubMed] [Google Scholar]
- 40.Weston, A.D., B. Blumberg, and T.M. Underhill. 2003. Active repression by unliganded retinoid receptors in development: less is sometimes more. J. Cell Biol. 161:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bazan, N.G., B.S. Fletcher, H.R. Herschman, and P.K. Mukherjee. 1994. Platelet-activating factor and retinoic acid synergistically activate the inducible prostaglandin synthase gene. Proc. Natl. Acad. Sci. USA. 91:5252–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]