Abstract
The NK cell–activating receptor NKG2D interacts with three different cellular ligands, all of which are regulated by mouse cytomegalovirus (MCMV). We set out to define the viral gene product regulating murine UL16-binding protein-like transcript (MULT)-1, a newly described NKG2D ligand. We show that MCMV infection strongly induces MULT-1 gene expression, but surface expression of this glycoprotein is nevertheless completely abolished by the virus. Screening a panel of MCMV deletion mutants defined the gene m145 as the viral regulator of MULT-1. The MCMV m145-encoded glycoprotein turned out to be necessary and sufficient to regulate MULT-1 by preventing plasma membrane residence of MULT-1. The importance of MULT-1 in NK cell regulation in vivo was confirmed by the attenuating effect of the m145 deletion that was lifted after NK cell depletion. Our findings underline the significance of escaping MULT-1/NKG2D signaling for viral survival and maintenance.
Mouse CMV (MCMV) shares many features with human CMV (HCMV) and serves as a model to study the immunobiology of CMV infections. Both innate and acquired immunity are important for host control of CMV (1, 2). In spite of this, CMV can establish life-long persistence characterized by alternate stages of latency and low-level virus productivity (3). It is generally assumed that viral proteins that counteract the host immune system are essential to tune the virus–host balance. For example, to escape CD8+ CTL control, three MCMV proteins efficiently down-regulate MHC class I expression (4). However, the altered MHC class I expression should predispose infected cells to lysis by NK cells, as suggested by the “missing self” hypothesis (5). In contrast, most laboratory mouse strains, as well as wild mice, fail to generate a significant NK cell response to MCMV (6), with the exception of strains in which the MCMV m157-encoded protein leads to NK cell activation (7–9).
The NKG2D receptor binds ligands that are poorly expressed on normal cells, but are up-regulated on infected, transformed, or stressed cells (10). These ligands are distantly related to MHC class I molecules. Known mouse NKG2D ligands comprised the retinoic acid early inducible (RAE)-1 family of proteins (11), the minor histocompatibility antigen H60 (12, 13), and the murine UL16-binding protein-like transcript (MULT)-1 glycoprotein (14, 15). NKG2D receptor triggering can override signals by MHC class I–specific inhibitory NK cell receptors (16, 17). This suggests, along with the fact that NKG2D is expressed on CD8+ T cells and γδ T cells, a pivotal position of NKG2D in both innate and adaptive immunity (18).
We have set out to define the viral modulators of NKG2D ligands. MCMV m152/gp40 has a double role and not only modulates the plasma membrane expression of MHC class I molecules but also of NKG2D ligands, namely RAE-1 (19–21). The protein encoded by MCMV m155 modulates the H60 ligand (22, 23). Considering the fact that MCMV-infected cells show a complete absence of all NKG2D ligands on the plasma membrane, we concluded that MCMV genes must also be involved in the down-regulation of MULT-1. Here, we identify the responsible MCMV gene m145 and describe its effect upon MULT-1 expression and its role during NK cell defense in vivo.
Results
Members of the MCMV m145 gene family down-modulate NKG2D ligands
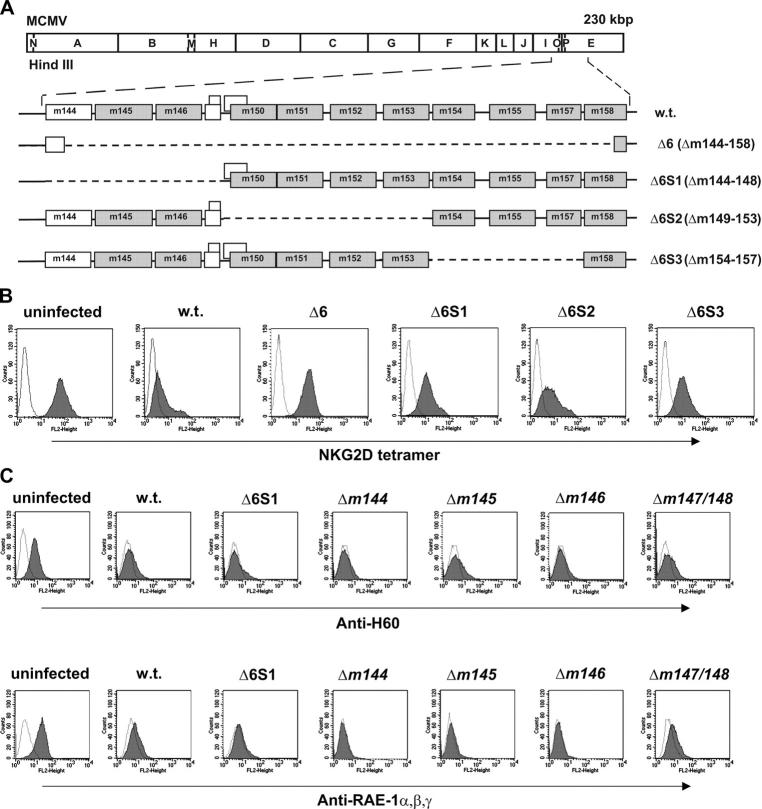
MCMV encodes proteins responsible for the down-regulation of all mouse NKG2D ligands (19). The m152 gene, responsible for the down-modulation of RAE-1 (20) and several other MCMV genes involved in the regulation of NK cell response, belongs to the m145 gene family (24), which shares properties with MHC class I proteins (7). Therefore, we postulated that other members of this gene family might be involved in the modulation of the expression of NKG2D ligands. To this aim, we assessed the expression of NKG2D ligands upon infection with several MCMV deletion mutants lacking genes of the m145 gene family (Fig. 1, A and B). NIH/3T3 cells were stained with NKG2D tetramer 12 h after infection with the deletion mutant Δ6 (Δm144–m158) and three additional mutants that dissect the genes deleted in Δ6 into three subgroups: Δ6S1 (Δm144–m148), Δ6S2 (Δm149–m153), and Δ6S3 (Δm154–m157). All the mutants, including the virus here referred to as WT MCMV, were constructed to express also GFP (25), to permit the selective analysis of infected cells. Cells infected with WT MCMV lost membrane staining with NKG2D tetramer. The same was observed upon infection with a WT virus that does not express GFP (unpublished data). In contrast, cells infected with the deletion mutant Δ6 displayed an expression level of NKG2D ligands on the cell surface comparable to that of uninfected control. The gene m152 is deleted in Δ6, and reexpression of RAE-1 should contribute to restoration of NKG2D ligands after infection with this mutant. Therefore, we tested the effect of mutants that lack subsets of genes of the m145 family. The Δ6S2 mutant also lacks m152 and revealed that the deletion of this genetic region only partially restores the NKG2D ligand expression. A similar phenotype was exhibited by mutant Δ6S3 in which the H60 regulator m155 is deleted (22). Because the mutant Δ6S1 also partially lifted NKG2D expression, we expected another unknown regulator in this genetic region. The staining with anti–RAE-1α,β,γ and anti-H60 mAbs upon infection with Δ6S1 and single gene deletion mutant viruses lacking defined open reading frames (ORFs) from the genomic region m144–m148 revealed down-regulation of RAE-1 and H60 to an extent identical with WT MCMV-infected cells (Fig. 1 C). We concluded that the target for the gene situated within this region must be a different NKG2D ligand, most likely MULT-1.
Figure 1.
NKG2D ligands are down-regulated by protein products of MCMV genes located between m144- m158 . (A) The HindIII cleavage map of the MCMV genome is shown (top) and the expanded region below indicates the positions of the m145 gene family members (shaded boxes) in the WT (w.t.) virus as well as the deletions (dashed lines) of the recombinant viruses. (B) NIH/3T3 cells were analyzed for expression of surface NKG2D ligands by staining with PE-NKG2D tetramer upon 12-h infection with indicated GFP-expressing viruses. Cells stained with streptavidin-PE were used as negative controls (dotted line). (C) NIH/3T3 and B12 cells were infected with indicated viruses and, after 12 h, stained with anti-H60 and anti–RAE-1α,β,γ mAbs, respectively. Cells stained with second antibody only were used as negative control (dotted line). (B and C) Each histogram represents 104 gated propidium iodide-negative, GFP-positive (infected) or GFP-negative (uninfected) cells.
Characterization of an anti–MULT-1 mAb and selection of MULT-1–expressing cell lines
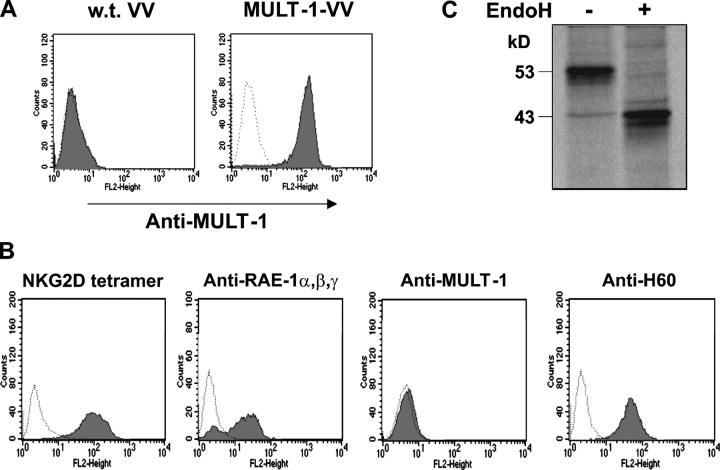
To focus on MULT-1, we generated a rat monoclonal antibody against the MULT-1 glycoprotein. The specificity of resulting antibodies was confirmed by the staining of cells infected with the recombinant vaccinia virus expressing HA-tagged MULT-1 protein (MULT-1-VV). As shown in Fig. 2 A, CV-1 cells infected with MULT-1-VV showed a bright staining with anti–MULT-1 mAb, whereas cells infected with WT VV were negative. Staining of BALB/3T3 cells with mAbs against H60, RAE-1, and MULT-1 clearly demonstrated that these cells constitutively express H60 and RAE-1 but not MULT-1 on their surface, and ruled out the possibility that anti–MULT-1 mAb cross-reacts with any of the other two NKG2D ligands (Fig. 2 B). In addition, immunoprecipitation of lysates of metabolically labeled cells, stably transfected with HA-tagged MULT-1 (MULT-1-3T3) using anti–MULT-1 antibody coupled to protein G–Sepharose, revealed the presence of an ∼53-kD band that shifts to a 43-kD–sized band upon EndoH digestion (Fig. 2 C). A band of the same size was seen upon immunoprecipitation with anti-HA mAb, confirming the specificity of the anti–MULT-1 antibody (unpublished data).
Figure 2.
Testing of anti–MULT-1 mAb. (A) CV-1 cells were infected with three MOI of WT VV or MULT-1-VV per cell and stained with anti–MULT-1 mAb 14 h after infection. (B) BALB/3T3 cells were stained with PE-NKG2D tetramer, anti–RAE-1α,β,γ or anti–MULT-1 antibodies. (A and B) Cells incubated with second antibody were used as a negative control (dotted line). Each histogram represents 104 gated propidium iodide–negative cells. (C) Upon metabolic labeling with [35S]methionine, lysates of MULT-1-3T3 cells were immunoprecipitated using anti–MULT-1 mAb coupled to protein G–Sepharose. Before separation by 11.5% SDS-PAGE, the samples were mock treated (−) or digested with EndoH (+).
Several cell lines that constitutively express MULT-1: SVEC4-10, B12, NIH/3T3, TpnT, IC21, and M2-10B4 were screened. Because SVEC4-10 cells expressed MULT-1 but neither RAE-1α,β,γ nor H60 (unpublished data), these cells were used to narrow down the MCMV gene region involved in the regulation of MULT-1.
The m145 gene product modulates cell membrane expression of MULT-1 protein
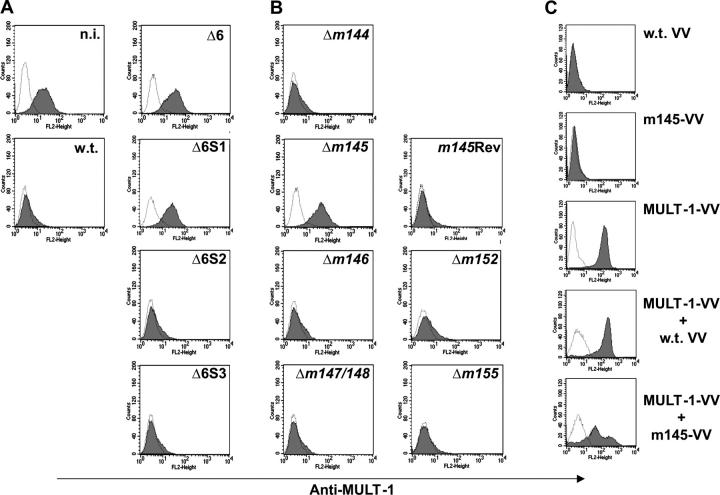
To assess which MCMV protein affects the surface expression of MULT-1, SVEC4-10 cells were infected with WT MCMV or specific deletion mutants and stained with the anti–MULT-1 mAb 12 h after infection. As suggested in Fig. 1 B and shown in Fig. 3 A, the Δ6 mutant lacked the MULT-1 down-regulating function, whereas the Δ6S2 or Δ6S3 mutants still maintained it. Cells infected with the Δ6S1 mutant lacking genes m144–m148 preserved the membrane expression of MULT-1 at a level comparable to or even higher than that of uninfected cells, pointing to five candidate genes that may be responsible for the down-regulation of surface expressed MULT-1. A panel of single deletion mutant viruses was tested that where lacking individual genes within the genomic region deleted in Δ6S1. Only the Δm145 mutant turned out to be unable to down-regulate MULT-1 (Fig. 3 B). The level of MULT-1 surface expression in Δm145 mutant-infected cells was similar to that observed upon infection with the Δ6 and the Δ6S1 mutants. To assess whether the expression of MULT-1 is affected by viral genes involved in regulation of two other NKG2D ligands, the cells were infected with Δm152 and Δm155 viruses and stained for the expression of surface MULT-1 (Fig. 3 B). The fact that both mutants caused a complete loss of membrane MULT-1 strongly argues against an additional involvement of m152 or m155 in the regulation of MULT-1 expression. The formal evidence that m145 expression is the crucial factor for down-modulation of surface MULT-1 came from the infection of cells with m145 revertant virus (m145Rev), which caused a down-modulation of MULT-1 comparable to that observed upon infection with WT virus (Fig. 3 B). Similar results were obtained on B12 and NIH/3T3 cells (unpublished data). We concluded that the m145 gene is essential for abrogating the surface expression of MULT-1. Led by the previously described capability of the m152 gene to regulate surface disposition of both NKG2D ligands and MHC class I molecules (19), we tested this possibility for m145. However, cells infected with Δm145 mutant down-modulated MHC class I to the same extent as the cells infected with WT virus (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20041617/DC1). This finding is in line with previously published results suggesting that besides m04, m06, and m152, there is no additional MCMV gene affecting MHC class I surface expression (4).
Figure 3.
The m145 protein down-regulates MULT-1. (A and B) SVEC4-10 cells were infected with 1 PFU of GFP-positive viruses per cell or left uninfected. 12 h after infection, cells were collected and stained with anti–MULT-1 mAb. Cells incubated with the second antibody upon binding to irrelevant primary antibody were used as a negative control (dotted line). Each histogram represents 104 gated propidium iodide-negative, GFP-positive cells (infected) or GFP-negative (uninfected) cells. n.i., noninfected cells. (C) CV-1 cells were infected with three MOI of WT VV, m145-VV, or MULT-1-VV or coinfected with MULT-1-VV/WT VV or MULT-1-VV/m145-VV per cell and, 14 h after infection, were analyzed for the expression of membrane-associated MULT-1 by staining with anti– MULT-1 mAb. Cells incubated with the second antibody in the absence of the primary antibody were used as a negative control (dotted line). Each histogram represents 104 gated propidium iodide-negative cells.
To assess whether the m145 protein requires the cooperation with other MCMV functions to down-modulate MULT-1, we coinfected CV-1 cells with MULT-1-VV and a recombinant vaccinia virus coding for the m145 protein (m145-VV). Coinfection with MULT-1-VV and WT VV served as a negative control. As shown in Fig. 3 C, the membrane expression of MULT-1 was significantly impaired upon coinfection with m145-VV, whereas WT VV had no effect. These results confirmed that m145 suffices for MULT-1 down-regulation.
MCMV induces MULT-1 expression
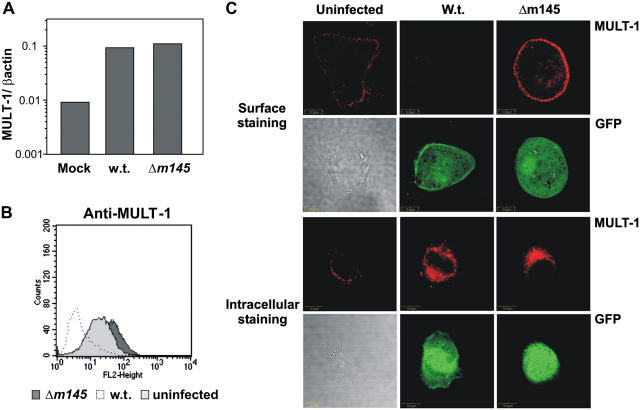
Cells can react to infection or stress by up-regulating the expression of ligands for the activating NK cell receptors (10). We tested the expression of MULT-1 mRNA in SVEC4-10 cells infected with WT or Δm145 MCMV and found an ∼10-fold induction of MULT-1 mRNA expression in comparison with uninfected cells (Fig. 4 A). Similar results were obtained on mouse embryonic fibroblasts (MEFs; unpublished data).
Figure 4.
MCMV infection stimulates MULT-1 expression. (A) SVEC4-10 cells were infected for 12 h with 1 PFU of the indicated GFP-expressing viruses per cell or were mock infected. Total RNA was isolated from the cells, and the expression of MULT-1 mRNA was quantified by real-time RT-PCR. MULT-1 cDNA copies in each sample were normalized by measuring the β-actin cDNA copy numbers. Bars represent the ratio of MULT-1 and β-actin cDNA copy numbers in each sample. (B) SVEC4-10 cells were infected with 1 PFU of the indicated GFP-expressing viruses per cell, or left untreated and, upon 12 h, harvested and stained with anti–MULT-1 mAb. Histogram represents 104 gated propidium iodide–negative, GFP-positive (infected) or GFP-negative (uninfected) cells. (C) SVEC4-10 cells were infected with 2 PFU/cell of the indicated GFP-expressing viruses and analyzed for the intracellular (bottom) or surface (top) expression of MULT-1. Infection was visualized by GFP expression.
We tested whether the mRNA induction would also lead to up-regulation of membrane-associated MULT-1. To this end, SVEC4-10 cells (Fig. 4 B) or MEFs (not depicted) were infected with WT or Δm145 viruses and stained with anti–MULT-1 mAb. Although WT infection of SVEC4-10 cells caused the down-modulation of surface MULT-1, infection with Δm145 virus resulted in an induction of surface-expressed MULT-1, as compared with uninfected cells (Fig. 4 B). Confocal analysis of SVEC4-10 cells confirmed the flow cytometry data (Fig. 4 C). Although WT MCMV infection caused a complete loss of surface MULT-1, the protein was clearly induced on the surface of the cells infected with the Δm145 virus. In contrast with surface staining, intracellular MULT-1 expression was equally induced in cells infected with WT and the Δm145 virus (Fig. 4 C). Similar results were obtained in MEFs (unpublished data). Altogether, these results demonstrated that MCMV infection induces the expression of the MULT-1 protein, but a viral function prevents cell membrane expression.
Intracellular processing and transport of newly synthesized MULT-1
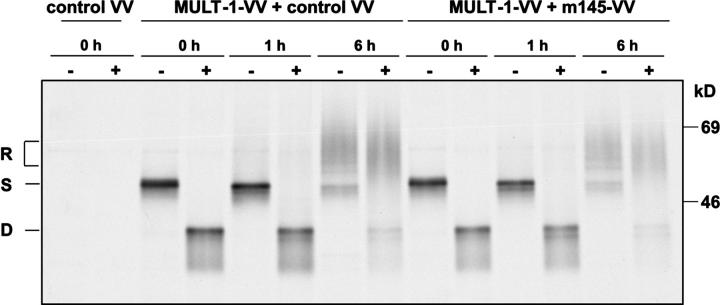
To study the effect of m145 on the fate of newly synthesized MULT-1, CV-1 cells were coinfected with MULT-1-VV and m145-VV. Coinfection of CV-1 cells with MULT-1-VV and an unrelated VV recombinant was performed as a negative control. Cells were metabolically labeled for 30 min, followed by chase periods of 1 and 6 h before immunoprecipitation of MULT-1 proteins was performed. Before separation on 10% SDS-PAGE, an aliquot of the precipitated material was subjected to EndoH digestion to determine the stage of maturation and intracellular transport of MULT-1. As shown in Fig. 5, the bulk of newly synthesized MULT-1 protein remained in an EndoH-sensitive state during the pulse period and 1 h of chase, suggesting a slow exit of MULT-1 from the ER and cis-Golgi compartment. Within 6 h of chase, a majority of MULT-1 molecules acquired EndoH resistance, indicative for the transit through the medial-Golgi compartment. Extended chase periods determined a half-life of MULT-1 of >16 h (unpublished data). Importantly, neither the glycosylation pattern, nor the rate of intracellular transport to EndoH-resistant forms nor the half-life of MULT-1 were affected by the m145 protein, even after 24 h of chase (unpublished data). Thus, the m145 glycoprotein alters the intracellular distribution of MULT-1 and its transport to the plasma membrane at a step after the passage through the ERGIC/cis-Golgi compartment. Because coinfection of CV-1 cells with MULT-1-VV and m145-VV resulted in an incomplete down-regulation of surface MULT-1, escaping MULT-1 molecules might have influenced the data obtained by immunoprecipitation. To exclude this possibility, endogenous MULT-1 was immunoprecipitated from MCMV-infected SVEC4-10 cells. The slow maturation pattern of endogenous MULT-1 was in agreement with the data obtained in MULT-1-VV–infected cells (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20041617/DC1). Importantly, there was no influence of m145 expression on MULT-1 maturation and stability in MCMV-infected cells. The presence of several differently glycosylated specific bands of endogenous MULT-1 might be a consequence of its slow exit from the ER/Golgi compartment. The infection conditions in the experiment allowed infection of the vast majority of cells (>95%) as confirmed by staining with anti-MCMV gp48 mAb in flow cytometry (unpublished data). Altogether, the results confirmed that m145 protein interferes with MULT-1 expression only beyond the ERGIC/cis-Golgi compartment.
Figure 5.
The m145 does not affect ER export of the MULT-1 protein. CV-1 cells were infected with control-VV or coinfected with MULT-1-VV and control VV or with MULT-1-VV and m145-VV and analyzed 12 h after infection. The cells were metabolically labeled for 30 min and chased for the indicated times, and the lysates were immunoprecipitated using anti-HA antibodies coupled to protein A–Sepharose. Before separation by 10% SDS-PAGE, the samples were mock treated (−) or digested by EndoH (+). R, EndoH resistant; S, EndoH sensitive; D, EndoH digested.
m145 compromises virus control by NK cells in vivo
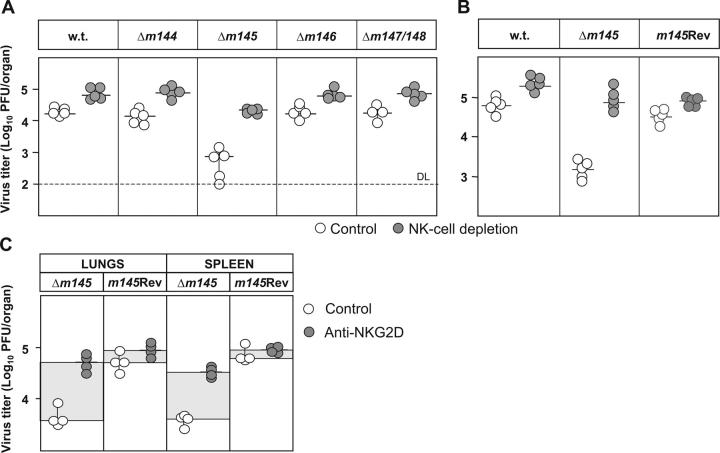
Provided that the data shown so far correctly reflect a major function of m145, the Δm145 mutant should be attenuated in vivo in an NK-dependent fashion. To assess this, BALB/c mice were infected with either WT MCMV, or Δm145, Δm146, Δm147/148 mutants or an m145 revertant virus (Fig. 6, A and B). Note that each of these mutants is indistinguishable from WT MCMV with respect to virus growth in fibroblasts (unpublished data). Half of the mouse groups were treated with anti-asialo GM1 serum to determine the contribution of NK cells to virus control in vivo. Virus titers in organs were determined by plaque assay 4 d after infection. As seen in Fig. 6 (A and B), the infection with Δm145 virus resulted in reduced virus titers as compared with WT and the other mutants, but the attenuation of the Δm145 mutant was found largely reversed after depletion of NK cells. These data confirmed our assumption that m145 has a relevant role in avoiding NK cell–mediated control of MCMV in vivo. Reconstitution of the m145 gene (m145Rev) resulted in virus resistance to NK cells and virus titers in tissues comparable with those found after WT MCMV infection (Fig. 6 B). More specific evidence for the involvement of NKG2D receptor in NK cell–mediated control of Δm145 mutant was obtained by injection of blocking anti-NKG2D mAb into the groups of infected mice. Similar to depletion of NK cells by cytolytic antibodies, injection of blocking anti-NKG2D mAb that does not affect the size of NK cell populations (unpublished data) led to a significant increase of Δm145 virus titers (Fig. 6 C), whereas the titer of m145 revertant virus was not affected. These data strongly support the interpretation that the interaction of MULT-1 with NKG2D is responsible for the NK cell–mediated control of Δm145 mutant virus. The Δm145 virus is also attenuated in BXD-8/Ty mice that possess the natural killer gene complex (NKC) from C57BL/6 mouse strain, but are susceptible to MCMV infection due to the mutation of Ly49h gene (references 26, 27 and Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20041617/DC1), pointing to a general role of m145-mediated inhibition of NK cell activity.
Figure 6.
Δm145 virus is attenuated in vivo in an NK-dependent fashion. (A and B) NK cell–depleted or undepleted BALB/c mice were injected i.v. with 2 × 105 PFU of indicated viruses. The mice were killed 4 d after infection, and viral titers in organs were determined. Titers in the spleen of individual mice (circles) and median values (horizontal bars) are shown. (C) BALB/c mice were injected i.p. with blocking anti-NKG2D mAb or left untreated. 4 d after i.v. infection with 2 × 105 PFU of indicated viruses, the mice were killed and viral titers in organs were determined. Titers in the spleen and lungs of individual mice (circles) and median values (horizontal bars) are shown.
Discussion
In this work, we describe the effects of MCMV infection on the expression of MULT-1, a recently described NKG2D ligand (14, 15). Despite the fact that MCMV infection strongly induces MULT-1 expression, the MULT-1 glycoprotein is down-regulated by an MCMV function from the surface of infected cells. The viral protein responsible for the down-modulation of MULT-1 is encoded by the m145 gene. In contrast with cells infected with WT MCMV, the Δm145-infected cells retain and even up-regulate MULT-1 on their plasma membrane. Consequently, the virus mutant lacking the m145 gene displays NK cell–dependent attenuation in vivo.
NKG2D ligands show either a constitutively low level or a complete lack of expression in normal tissues (13, 28). A tight regulation of cellular ligands for activating NK cell receptors may be essential for the prevention of autoaggressive NK cell activities. Inducibility by infection, as shown here for the MULT-1, points to their strong systemic role. To limit pathogen multiplication and dissemination at early stages of infection before activation of adaptive immunity, the host might alarm NK cells by up-regulating these ligands on the surface of infected cells.
Apparently, MCMV cannot avoid the induction of MULT-1 transcription in infected cells. However, to avoid the consequences of MULT-1 transcription, MCMV has developed a gene that encodes a protein that modulates the function of MULT-1. Rather than silencing the NKG2D receptor, evolution selected MCMV to control the signaling through the different ligands of NKG2D. This is why the MCMV genome harbors three individual genes to modulate the three known ligands of murine NKG2D. This evolutionary solution is probably very specific for large DNA viruses because only these viruses provide the genomic space to encompass multiple genes and gene families that seem to be selectively devoted to different aspects of virus host interaction because their absence has no effect on viral morphogenesis in cell culture.
Remarkably, the NK cell regulators of MCMV are comprised in the m145 gene family (Fig. 1 A) encoding glycoproteins that share some structural similarities with MHC class I proteins (7). Two other MCMV genes are involved in the control of NKG2D ligands: m152, responsible for the down-modulation of RAE-1 (19, 20), and m155, whose protein product regulates surface expression of H60 (22, 23). Another member of this gene family interferes with the function of NK cells. The m157 protein is a ligand for the activating NK cell receptor Ly49H, responsible for the resistance of Cmv1 r mouse strains to MCMV (7, 8). Deletion of the m157 gene results in higher virulence of the virus mutant in vivo (9). Under selective in vivo pressure of Ly49H, MCMV tends to inactivate the m157 gene by mutation (29, 30). In addition, MCMV homologue for MHC class I encoded by m144 gene inhibits NK function by serving as a ligand for an inhibitory NK cell receptor although the putative receptor remains elusive (31).
The molecular mechanisms by which the NKG2D ligand regulators act are not yet clear. The first regulator found, the gp40 encoded by m152, was identified by us as a modulator of MHC class I expression (21). Unlike certain MHC class I regulators of HCMV (32), the MCMV m152/gp40 does not act by retrograde translocation of the protein target from the ER into the cytosol and subsequent degradation by the cytosolic proteasome. Instead, the MHC class I protein binds antigenic peptides and matures to the stage of EndoH resistant glycoprotein modification, showing that the protein does not exit the ER in a retrograde fashion. Furthermore, the half-life of the target protein is not affected (21, 33). Thus, the m152 misdirects the MHC class I molecule into a compartment with ERGIC/cis-Golgi properties. Further studies are required to understand whether NKG2D ligands affected by m152 and other MCMV regulators follow similar patterns.
According to our understanding, the species-specific herpesviruses have coevolved with their hosts. Therefore, the presence of multiple NKG2D ligands and ligand modulators may simply reflect mutations and duplications of host defense principles and the necessary reciprocal viral counterstrategies. Alternatively, different ligands may have in fact diverse roles in different tissues due to tissue-specific ligand expression patterns. Because research on MCMV is based on only two isolates, Smith and K181, which, in fact, for unknown reasons differ in virulence, it is unknown whether wild-type strains of MCMV differ with respect to number and function of NKG2D ligand modulators that might affect virus distribution patterns in tissues. The fact that MULT-1 has a stronger affinity for NKG2D binding than that of RAE-1 and H60 (15), points to the relevance of MULT-1/NKG2D signaling as a target for the virus. Avoiding MULT-1/NKG2D signaling may be one of the first host checkpoints to be bypassed in the process of establishing MCMV infection.
NKG2D is an important costimulatory receptor for CD8+ T cells (18). The evasion function should also affect the triggering of virus-specific CTLs. Therefore, a virus lacking one or several NKG2D evasion genes should be more prone to activate CTLs. Thus, viral immunoevasins involved in the control of NKG2D ligands may not only contribute to the control of acute infection but also to viral immune surveillance during conditions of latent infection and reactivation episodes.
The results obtained in the MCMV system can be used to extrapolate the significance of HCMV-mediated down-regulation of NKG2D ligands. It has been shown that HCMV-encoded UL16 protein prevents membrane expression of ULBP1 and 2 as well as MICB and reduces sensitivity of infected cells to NK cell lysis (34–36). Although the precise mechanism by which UL16 down-regulates NKG2D ligands is still unknown, it seems that UL16 forms stable complexes with MICB in ER/cis-Golgi compartment of infected cells, thus preventing membrane expression of this NKG2D ligand (37, 38). Because, together with this paper, already three MCMV genes have been identified that interfere with the expression of NKG2D ligands, it is likely that HCMV also encodes additional, yet unidentified proteins that target NKG2D ligands that are not affected by UL16.
Materials and Methods
Cells
SVEC4-10 (endothelial cells derived from C3H/HeJ mouse strain, CRL-2181; American Type Culture Collection), CV-1 (fibroblasts derived from the kidney of a male adult African green monkey, CCL-70; American Type Culture Collection), B12 (subclone of an immortalized line of BALB/c fibroblast; reference 19), BALB/3T3 (fibroblasts derived from BALB/c mouse strain, CCL-163; American Type Culture Collection), and NIH/3T3 (fibroblasts derived from NIH Swiss mouse, CRL-1658; American Type Culture Collection) cells were cultivated in DMEM supplemented with 10% FCS. MEFs prepared from BALB/c mice were cultivated in MEM supplemented with 3% FCS or alternatively in DMEM supplemented with 10% FCS.
To obtain cell transfectants, the HA-tagged MULT-1 ORF was cloned from a p7.5K131-based plasmid (39) into the SalI restriction site of pB45-Neo, provided by E.R. Podack (University of Miami School of Medicine, Miami, FL; reference 40). The plasmid was transfected into NIH/3T3 fibroblasts using SuperFect Transfection Reagent (QIAGEN), according to the manufacturer's instructions. MULT-1–transfected 3T3 cells (MULT-1-3T3) were selected and cultured in DMEM supplemented with 10% FCS and 500 μg/ml G418 (GIBCO-BRL).
Production of anti–MULT-1 mAb 1D6
MULT-1 protein was generated by cloning the sequence of the extracellular domain (cDNA provided by W.M. Yokoyama, Washington University School of Medicine, St. Louis, MO) into a bacterial expression vector (pET3a; Novagen). Protein was expressed in inclusion bodies in BL21(DE3), subsequently purified, and in vitro refolded under identical conditions as described for the generation of soluble NKG2D (41).
DA rats were primed and boosted in 2-wk interval by intradermal injections of 50 μg MULT-1 protein diluted in PBS with addition of Freund's adjuvant (Sigma-Aldrich). Serum was screened after each injection by ELISA. 3 d preceding the fusion, animals were boosted i.p. with 50 μg MULT-1 protein diluted in PBS. Splenocytes from the boosted rats were fused with Sp2/0 murine myeloma cells. The hybridomas were selected by growing in RPMI 1640 medium (GIBCO-BRL) supplemented with HAT medium (GIBCO-BRL) and 10% FCS. Supernatants of hybridomas were collected and screened against the MULT-1 protein by ELISA. Positive hybridomas were tested in FACS on SVEC4-10 cell line. The hybridomas positive in both assays were subcloned to clone density, and supernatants were collected.
Viruses
The BAC-derived strain MCMV-GFP (25), here referred to as WT virus, is a derivative of the MCMV strain MW97.01 (42), which has previously been shown as biologically equivalent to the MCMV Smith strain (VR-194 recently reaccessed as VR-1399; American Type Culture Collection). To generate the deletion mutants listed in Table I, a PCR-based mutagenesis procedure was applied as described previously (43). In brief, a kanamycin resistance gene (kanR) was PCR amplified using primers containing 20–22 nucleotides (nt) at their 3′-ends specific for kanR and 50–60 nt at their 5′-ends homologous to the target region in the BAC-cloned genome. The PCR fragments were integrated into the BAC, thereby replacing the respective target gene. To construct a rescuant of the mutant strain MCMV-GFPΔm145, the kanR cassette in the respective BAC was excised by FLP-mediated recombination as described previously (4), resulting in BAC pSM3frΔm145-K. A 4.7 kbp KpnI fragment (nt 203952–208669 of the MCMV genome [24] was subcloned into the shuttle vector pST76KSR). Using the resulting shuttle plasmid, the DNA sequence with the m145 gene was reintroduced into the BAC pSM3frΔm145-K by a two-step replacement procedure as described previously (42). Recombinant MCMV BACs were characterized by restriction analysis and viruses were reconstituted by transfection of the MCMV-BACs into MEFs as described previously (4). For preparation of virus stocks, MCMV recombinants were propagated on MEFs and purified. Titers of virus stocks were determined by standard plaque assay on MEFs. Tissue culture-grown virus preparations were used for mouse inoculations.
Table I.
MCMV mutants
| Deletion
|
||
|---|---|---|
| Mutants | Rangea | ORFs |
| MCMV-GFP | – | – |
| MCMV-GFPΔ6 | 203,002–217,799 | m144–m158 |
| MCMV-GFPΔ6S1 | 202,746–207,298 | m144–m148 |
| MCMV-GFPΔ6S2 | 207,354–212,803 | m149–m153 |
| MCMV-GFPΔ6S3 | 212,946–216,883 | m154–m157 |
| MCMV-GFPΔm144 | 202,746–203,892 | m144 |
| MCMV-GFPΔm145 | 204,032–205,492 | m145 |
| MCMV-GFPΔm146 | 205,646-206,774 | m146 |
| MCMV-GFPΔm147/m148 | 206,866–207,265 | m147, m148 |
| MCMV-GFPm145Revb | – | – |
Nucleotide positions refer to reference 24.
The genome of MCMV-GFPm145Rev was constructed by reinsertion of the m145 gene into the genome of MCMV-GFPΔm145.
Production of recombinant vaccinia viruses
For the generation of a recombinant vaccinia virus, the MULT-1 cDNA sequence (NCBI accession no. AK020784) was PCR amplified from pmX-IgMULT7 plasmid (obtained by courtesy of W.M. Yokoyama). Using forward primer 5′-CGCCCAAGCTTGGGATGGAGCTGACTGCCAGT3Ä and reverse primer 5′-CGAGGTACCCGCGGGTCGACCCGTCACGCGTAATCTGGAACATCGTATGGGTATGGGATCCCATCAAT-3′ the hemagglutinin (HA) epitope sequence was added COOH terminally. m145 DNA was PCR amplified from DNA of the MCMV Smith strain and COOH terminally flag-tagged using primers for 5′-CGAAGATCTTCCATGGACCGTCGGGTGGTC-3′ and rev 5′-CGGAATTCCTCACTTGTCGTCGTCGTCCTTGTAGTCCGCCTCTATCGTCTT-3′. The PCR products were cloned into 5′-HindIII and 3′-KpnI (MULT-1) or 5′-BglII and 3′-EcoRI (m145) restriction sites of plasmid p7.5K131 (39). The constructs were used for generation of recombinant vaccinia viruses expressing MULT-1 (MULT-1-VV) or m145 (m145-VV) by homologous recombination with the vaccinia strain Copenhagen. Vaccinia recombinants were selected by infection of 143 tk− cells in the presence of 100 μg/ml bromodeoxyuridine as described previously (44). As a VV control virus an ICP47 expression vaccinia recombinant was used (45).
Metabolic labeling of cells and immunoprecipitation
Subconfluent layers of cells were labeled with [35S]methionine (Amersham Biosciences) at a concentration of 500 μCi/ml at 37°C for 30 min and chased in the presence of 10 mM of nonlabeled methionine. After being washed with ice-cold PBS, cells were lysed in 1 ml of lysis buffer (140 mM NaCl, 5 mM MgCl2, 20 mM Tris, pH 7.6, 1 mM PMSF, 0.5 mM leupeptin, 1 μM pepstatin A), containing 1% (vol/vol) IGEPAL (Sigma-Aldrich) for 20 min and centrifuged at 13,000 g for 30 min.
The lysates were incubated for 1 h at 4°C with 0.5 μg of anti-HA (Sigma-Aldrich). Immunoprecipitation was performed as described previously (46, 47). In brief, immune complexes were retrieved with protein A-Sepharose (Amersham Biosciences) (60 μl of 1:1 buffer-Sepharose slurry, for 1 h at 4°C). The Sepharose beads were washed three times with (0.2% [vol/vol] IGEPAL, 10 mM Tris-HCl, pH 7.6, 140 mM NaCl, 2 mM EDTA), twice with (0.2% [vol/vol] IGEPAL, 10 mM Tris-HCl, pH 7.6, 500 mM NaCl, 2 mM EDTA), and once with (10 mM Tris-HCl, pH 7.6) washing buffers. The immune complexes bound to Sepharose beads were resuspended in 50 mM phosphate buffer, pH 5.5, containing 0.02% (wt/vol) SDS, 0.1% (vol/vol) IGEPAL, 0.2 mM PMSF, and 0.1 M 2-mercaptoethanol. Sepharose-bound immune complexes were mock treated or incubated with 2 mU of Endoglycosidase H (EndoH; Roche Diagnostics) at 37°C overnight. Digestion was stopped by addition of sample buffer and heating at 94°C for 5 min. The precipitates were analyzed by 10% SDS-PAGE. Dried gels were exposed to Kodak BioMaxMR films for 1–3 d.
Quantification of MULT-1 mRNA by real-time RT-PCR
SVEC4-10 and MEF cells (2 × 106) were mock-infected or infected with WT or Δm145 MCMV (1 and 0.5 PFU/cell, respectively). Upon 12 h of infection, the cells were collected and total RNA was isolated by TriPure reagent (Roche Diagnostics) according to the manufacturer's instruction. cDNA was synthetized from 1 μg of total RNA by using the 1st Strand cDNA Synthesis Kit for RT-PCR (Roche Diagnostics, containing: oligo (dT)15 primers (0.04 U A260), 1 mM dNTPs, 5 mM MgCl2, 2.5 U/μl RNase inhibitor, AMV RT buffer (×1), and 1 U/μl AMV reverse transcriptase; final volume of reaction, 20 μl) under the following conditions: 10 min at 25°C, 1 h at 42°C, 10 min at 95°C, and 10 min at 4°C. The cDNA was diluted 1:5 in water and 2 μl was used for each quantitative real-time PCR reaction designed to measure the number of MULT-1 and β-actin cDNA copies in each sample and performed on LightCycler (Roche Diagnostics). The MULT-1 cDNA PCR product (185 bp) was amplified by using exon spanning MULT-F (5′-CTCATAGGAACAGCATGA-3′) and MULT-R (5′-TCCTGTGAAATGTTTGTC-3′) primers and LightCycler-FastStart DNA Master SYBER green I Kit (Roche Diagnostics) according to manufacturer's instructions. Incorporation of Syber green I dye into the PCR products was automatically measured by the LightCycler after each of 45 cycles of amplification (0 s at 95°C, 10 s at 54°C, and 8 s at 72°C). The very same conditions and following primers: 5′-TTCTACAATGAGCTGCGT-3′ (βACT-LCF) and 5′-ATCACAATGCCTGTGGTA-3′ (βACT-LCR), spanning exons 2 and 3, were used to amplify the mouse β-actin cDNA PCR product (191 bp). Serial dilutions with known copy numbers (106, 105, 104, 103, 102, 101 in 2 μL) of plasmid pB45-Neo containing a complete cDNA sequence of MULT-1 were used to obtain a standard amplification curve (error = 0.0406, r = −1.00) to quantify MULT-1 and β-Actin cDNA copy numbers in each sample. The quantification was done automatically by LightCycler Software V3 and Second Derivative Maximum method.
Confocal microscopy
SVEC4-10 cells were grown on glass coverslips and infected with 1 PFU of WT or Δm145 MCMV per cell or mock infected. Upon 12 h of infection, the cells were washed in PBS, fixed with 2.5% (wt/vol) paraformaldehyde in PBS for 20 min, and immunostained. Unspecific binding of antibodies was blocked with 0.2% (wt/vol) fish skin gelatin. The staining was performed with anti–MULT-1 mAb in combination with goat anti–rat PE. Infected cells were visualized by GFP expression.
Flow cytometry
SVEC4-10, MULT-1-3T3, NIH/3T3, BALB/3T3, and B12 cells were mock treated or infected with MCMV (1–2 PFU/cell) and trypsinized 12 h after infection. CV-1 cells were mock treated or infected with VV (3 multiplicity of infection [MOI]/cell) and harvested 14 h after infection. Cells were washed in PBS supplemented with 1% BSA and 0.1% NaN3 and stained with either PE-NKG2D tetramer (19), rat anti-H60 mAb clone 205303, provided by J.P. Houchins (R&D Systems, Minneapolis, MN), rat anti–RAE-1α,β,γ mAb CX1, provided by L.L. Lanier (University of California, San Francisco, CA; reference 20), or rat anti–MULT-1 mAb. After washing, bound antibodies were visualized by the addition of PE-labeled goat anti–rat IgG (Caltag Laboratories). Cells incubated with PE-streptavidin served as negative control for cells stained with PE-NKG2D tetramer, whereas cells incubated with the second antibody served as negative control for cells stained with anti-H60 and anti–RAE-1α, β, γ mAb. Cells stained with irrelevant rat mAb followed by PE-labeled goat anti–rat IgG were control for cells stained with anti–MULT-1 mAb. After staining, cells were analyzed with a Becton Dickinson FACScan and gated with propidium iodide-negative cells.
Animals, infection conditions, and detection of infectious MCMV in tissues
BALB/c (H-2d) and BXD-8/Ty (H-2b; The Jackson Laboratory) mice were housed and bred under specific pathogen-free conditions at the Central Animal Facility of the Medical Faculty, University of Rijeka, in accordance with the guidelines contained in the International Guiding Principles for Biomedical Research Involving Animals. The Ethical Committee at the University of Rijeka approved all animal experiments described here. 6–8-wk-old female mice were used in experiments.
Mice were injected i.v. with 2 × 105 PFU (BALB/c) or 4 × 105 PFU (BXD-8/Ty) of WT MCMV or recombinant viruses in a volume of 500 μl of diluents. Organs were collected 4 d after infection and viral titers were determined by a standard viral plaque-forming assay performed on MEFs.
Depletion of NK cell subsets in vivo
The depletion of NK cells was performed by i.p. injection of rabbit antibodies to asialo GM-1 (Wako Chemicals) at the dose of 25 μl, 6 h before infection. The efficacy of depletion was assessed by cytofluorometric analysis of spleen cells using biotin-labeled anti–mouse pan-NK cells mAb DX5 (BD Biosciences). The NKG2D was blocked by i.p. injection of blocking anti-NKG2D mAb (R&D Systems) at the dose of 100 μg/mouse, 12 h before infection.
Online supplemental material
Lack of influence of m145 gene on MHC class I expression is depicted in Fig. S1. The finding that MCMV infection does not alter maturation pattern of MULT-1 molecule before its passing through ERGIC/cis-Golgi compartment is shown in Fig. S2. In Fig. S3, the attenuation of Δm145 virus in BXD-8/Ty mice is presented. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20041617/DC1.
Acknowledgments
We thank J.P. Houchins and L.L. Lanier for generously providing anti-H60 mAb and anti–RAE-1α,β,γ mAb and W.M. Yokoyama for providing pmX-IgMULT7 plasmid. We are indebted to A. Zimmermann and W. Muranyi for technical advice. We also thank D. Rumora and E. Razic for technical assistance and M. Fritz-Boukhatem for organizing the experimental mouse facility.
This work was supported by Croatian Ministry of Science grant nos. 0062004 (to S. Jonjic) and 0062007 (to A. Krmpotic) and Deutsche Forschungsgemeinschaft grants SFB 455 (to U.H. Koszinowski), EU QLRT-2001-01112 and SFB421 A8 (to H. Hengel), ME1102/2-1 (to M. Messerle), and SFB576-A8 (to D.H. Busch).
The authors have no conflicting financial interests.
Abbreviations used: HCMV, human CMV; MCMV, mouse CMV; MEF, mouse embryonic fibroblast; MOI, multiplicity of infection; MULT, murine UL16-binding protein-like transcript; ORF, open reading frame; RAE, retinoic acid early inducible.
H. Hengel's present address is Institute for Virology, Heinrich Heine University Duesseldorf, 40225 Duesseldorf, Germany.
The online version of this article contains supplemental material.
References
- 1.Biron, C.A., K.B. Nguyen, G.C. Pien, L.P. Cousens, and T.P. Salazar-Mather. 1999. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 17:189–220. [DOI] [PubMed] [Google Scholar]
- 2.Reddehase, M.J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2:831–844. [DOI] [PubMed] [Google Scholar]
- 3.Polic, B., H. Hengel, A. Krmpotic, J. Trgovcich, I. Pavic, P. Luccaronin, S. Jonjic, and U.H. Koszinowski. 1998. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J. Exp. Med. 188:1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner, M., A. Gutermann, J. Podlech, M.J. Reddehase, and U.H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ljunggren, H.G., and K. Karre. 1990. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today. 11:237–244. [DOI] [PubMed] [Google Scholar]
- 6.Scalzo, A.A. 2002. Successful control of viruses by NK cells–a balance of opposing forces? Trends Microbiol. 10:470–474. [DOI] [PubMed] [Google Scholar]
- 7.Smith, H.R., J.W. Heusel, I.K. Mehta, S. Kim, B.G. Dorner, O.V. Naidenko, K. Iizuka, H. Furukawa, D.L. Beckman, J.T. Pingel, et al. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA. 99:8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arase, H., E.S. Mocarski, A.E. Campbell, A.B. Hill, and L.L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326. [DOI] [PubMed] [Google Scholar]
- 9.Bubic, I.W., M., Krmpotic, A., Saulig, T., Kim, S., Yokoyama, W.M., Jonjic, S., Koszinowski, U.H. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raulet, D.H. 2003. Roles of the NKG2D immunoreceptor and its ligands. Nat. Rev. Immunol. 3:781–790. [DOI] [PubMed] [Google Scholar]
- 11.Cerwenka, A., A.B. Bakker, T. McClanahan, J. Wagner, J. Wu, J.H. Phillips, and L.L. Lanier. 2000. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 12:721–727. [DOI] [PubMed] [Google Scholar]
- 12.Malarkannan, S., P.P. Shih, P.A. Eden, T. Horng, A.R. Zuberi, G. Christianson, D. Roopenian, and N. Shastri. 1998. The molecular and functional characterization of a dominant minor H antigen, H60. J. Immunol. 161:3501–3509. [PubMed] [Google Scholar]
- 13.Diefenbach, A., and D.H. Raulet. 2003. Innate immune recognition by stimulatory immunoreceptors. Curr. Opin. Immunol. 15:37–44. [DOI] [PubMed] [Google Scholar]
- 14.Diefenbach, A., J.K. Hsia, M.Y. Hsiung, and D.H. Raulet. 2003. A novel ligand for the NKG2D receptor activates NK cells and macrophages and induces tumor immunity. Eur. J. Immunol. 33:381–391. [DOI] [PubMed] [Google Scholar]
- 15.Carayannopoulos, L.N., O.V. Naidenko, D.H. Fremont, and W.M. Yokoyama. 2002. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J. Immunol. 169:4079–4083. [DOI] [PubMed] [Google Scholar]
- 16.Cerwenka, A., and L.L. Lanier. 2001. Natural killer cells, viruses and cancer. Nat. Rev. Immunol. 1:41–49. [DOI] [PubMed] [Google Scholar]
- 17.Diefenbach, A., E.R. Jensen, A.M. Jamieson, and D.H. Raulet. 2001. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 413:165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamieson, A.M., A. Diefenbach, C.W. McMahon, N. Xiong, J.R. Carlyle, and D.H. Raulet. 2002. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 17:19–29. [DOI] [PubMed] [Google Scholar]
- 19.Krmpotic, A., D.H. Busch, I. Bubic, F. Gebhardt, H. Hengel, M. Hasan, A.A. Scalzo, U.H. Koszinowski, and S. Jonjic. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat. Immunol. 3:529–535. [DOI] [PubMed] [Google Scholar]
- 20.Lodoen, M., K. Ogasawara, J.A. Hamerman, H. Arase, J.P. Houchins, E.S. Mocarski, and L.L. Lanier. 2003. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J. Exp. Med. 197:1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziegler, H., R. Thäle, P. Lucin, W. Muranyi, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U.H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity. 6:57–66. [DOI] [PubMed] [Google Scholar]
- 22.Hasan, M., A. Krmpotic, Z. Ruzsics, I. Bubic, T. Lenac, A. Halenius, A. Loewendorf, M. Messerle, H. Hengel, S. Jonjic, and U.H. Koszinowski. 2005. Selective down-regulation of the NKG2D ligand H60 by mouse cytomegalovirus m155 glycoprotein. J. Virol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodoen, M.B., G. Abenes, S. Umamoto, J.P. Houchins, F. Liu, and L.L. Lanier. 2004. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J. Exp. Med. 200:1075–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawlinson, W.D., H.E. Farrell, and B.G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathys, S., T. Schroeder, J. Ellwart, U.H. Koszinowski, M. Messerle, and U. Just. 2003. Dendritic cells under influence of mouse cytomegalovirus have a physiologic dual role: to initiate and to restrict T cell activation. J. Infect. Dis. 187:988–999. [DOI] [PubMed] [Google Scholar]
- 26.Brown, M.G., A.O. Dokun, J.W. Heusel, H.R. Smith, D.L. Beckman, E.A. Blattenberger, C.E. Dubbelde, L.R. Stone, A.A. Scalzo, and W.M. Yokoyama. 2001. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 292:934–937. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S.H., S. Girard, D. Macina, M. Busa, A. Zafer, A. Belouchi, P. Gros, and S.M. Vidal. 2001. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat. Genet. 28:42–45. [DOI] [PubMed] [Google Scholar]
- 28.Cerwenka, A., and L.L. Lanier. 2003. NKG2D ligands: unconventional MHC class I-like molecules exploited by viruses and cancer. Tissue Antigens. 61:335–343. [DOI] [PubMed] [Google Scholar]
- 29.Voigt, V., C.A. Forbes, J.N. Tonkin, M.A. Degli-Esposti, H.R. Smith, W.M. Yokoyama, and A.A. Scalzo. 2003. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc. Natl. Acad. Sci. USA. 100:13483–13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.French, A.R., J.T. Pingel, M. Wagner, I. Bubic, L. Yang, S. Kim, U. Koszinowski, S. Jonjic, and W.M. Yokoyama. 2004. Escape of mutant double-stranded DNA virus from innate immune control. Immunity. 20:747–756. [DOI] [PubMed] [Google Scholar]
- 31.Farrell, H.E., H. Vally, D.M. Lynch, P. Fleming, G.R. Shellam, A.A. Scalzo, and N.J. Davis-Poynter. 1997. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature. 386:510–514. [DOI] [PubMed] [Google Scholar]
- 32.Loenen, W.A., C.A. Bruggeman, and E.J. Wiertz. 2001. Immune evasion by human cytomegalovirus: lessons in immunology and cell biology. Semin. Immunol. 13:41–49. [DOI] [PubMed] [Google Scholar]
- 33.Ziegler, H., W. Muranyi, H.G. Burgert, E. Kremmer, and U.H. Koszinowski. 2000. The luminal part of the murine cytomegalovirus glycoprotein gp40 catalyzes the retention of MHC class I molecules. EMBO J. 19:870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubin, M., L. Cassiano, J. Chalupny, W. Chin, D. Cosman, W. Fanslow, J. Mullberg, A.M. Rousseau, D. Ulrich, and R. Armitage. 2001. ULBP1, 2, 3: novel MHC class I-related molecules that bind to human cytomegalovirus glycoprotein UL16, activate NK cells. Eur. J. Immunol. 31:1428–1437. [DOI] [PubMed] [Google Scholar]
- 35.Cosman, D., J. Mullberg, C.L. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and N.J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 14:123–133. [DOI] [PubMed] [Google Scholar]
- 36.Vales-Gomez, M., H. Browne, and H.T. Reyburn. 2003. Expression of the UL16 glycoprotein of human cytomegalovirus protects the virus-infected cell from attack by natural killer cells. BMC Immunol. 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunn, C., N.J. Chalupny, C.L. Sutherland, S. Dosch, P.V. Sivakumar, D.C. Johnson, and D. Cosman. 2003. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J. Exp. Med. 197:1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, J., N.J. Chalupny, T.J. Manley, S.R. Riddell, D. Cosman, and T. Spies. 2003. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J. Immunol. 170:4196–4200. [DOI] [PubMed] [Google Scholar]
- 39.Schlicht, H.J., and H. Schaller. 1989. The secretory core protein of human hepatitis B virus is expressed on the cell surface. J. Virol. 63:5399–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohe, Y., D. Zhao, N. Saijo, and E.R. Podack. 1995. Construction of a novel bovine papillomavirus vector without detectable transforming activity suitable for gene transfer. Hum. Gene Ther. 6:325–333. [DOI] [PubMed] [Google Scholar]
- 41.Wolan, D.W., L. Teyton, M.G. Rudolph, B. Villmow, S. Bauer, D.H. Busch, and I.A. Wilson. 2001. Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A. Nat. Immunol. 2:248–254. [DOI] [PubMed] [Google Scholar]
- 42.Wagner, M., S. Jonjic, U.H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, M., Z. Ruzsics, and U.H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318–324. [DOI] [PubMed] [Google Scholar]
- 44.Volkmer, H., C. Bertholet, S. Jonjic, R. Wittek, and U.H. Koszinowski. 1987. Cytolytic T lymphocyte recognition of the murine cytomegalovirus nonstructural immediate-early protein pp89 expressed by recombinant vaccinia virus. J. Exp. Med. 166:668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banks, T.A., F.J. Jenkins, S. Kanangat, S. Nair, S. Dasgupta, C.M. Foster, and B.T. Rouse. 1994. Vaccination with the immediate-early protein ICP47 of herpes simplex virus-type 1 (HSV-1) induces virus-specific lymphoproliferation, but fails to protect against lethal challenge. Virology. 200:236–245. [DOI] [PubMed] [Google Scholar]
- 46.del Val, M., H. Hengel, H. Hacker, U. Hartlaub, T. Ruppert, P. Lucin, and U.H. Koszinowski. 1992. Cytomegalovirus prevents antigen presentation by blocking the transport of peptide-loaded major histocompatibility complex class I molecules into the medial-Golgi compartment. J. Exp. Med. 176:729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hengel, H., C. Esslinger, J. Pool, E. Goulmy, and U.H. Koszinowski. 1995. Cytokines restore MHC class I complex formation and control antigen presentation in human cytomegalovirus-infected cells. J. Gen. Virol. 76:2987–2997. [DOI] [PubMed] [Google Scholar]