Abstract
Transgenic rice containing an antisense cDNA for the α subunit of rice heterotrimeric G protein produced little or no mRNA for the subunit and exhibited abnormal morphology, including dwarf traits and the setting of small seeds. In normal rice, the mRNA for the α subunit was abundant in the internodes and florets, the tissues closely related to abnormality in the dwarf transformants. The position of the α-subunit gene was mapped on rice chromosome 5 by mapping with the restriction fragment length polymorphism. The position was closely linked to the locus of a rice dwarf mutant, Daikoku dwarf (d-1), which is known to exhibit abnormal phenotypes similar to those of the transformants that suppressed the endogenous mRNA for the α subunit by antisense technology. Analysis of the cDNAs for the α subunits of five alleles of Daikoku dwarf (d-1), ID-1, DK22, DKT-1, DKT-2, and CM1361–1, showed that these dwarf mutants had mutated in the coding region of the α-subunit gene. These results show that the G protein functions in the formation of normal internodes and seeds in rice.
Heterotrimeric G proteins are known to function as mediators in the transduction of numerous signals that are perceived by receptors on cell surfaces in mammals and yeast (1, 2). In mammals, a set of genes has been identified for each subunit of the G proteins; more than 16 genes for the α subunits, 5 genes for the β subunit, and 6 genes for the γ subunit, most of which are expressed in a tissue-specific manner (3, 4). Thus mammals contain multiforms of the G proteins that are probably involved in separate systems of signal transduction. In contrast, yeast, a unicellular organism, carries only a few genes or a single gene for each subunit (5, 6). In higher plants, the genes for the putative α and β subunits of a heterotrimeric G protein have been isolated from various plant species (7–10). Studies with a recombinant protein produced in Escherichia coli by using a cDNA for the putative α subunit in rice (11) and Arabidopsis (12) have presented evidence that the cDNA encodes a true α subunit.
Although higher plants seem to contain a single molecular species of heterotrimeric G protein (13), the protein has been proposed to function in various systems of signal transduction in diverse tissues or cells; namely, in light-mediated gene expression (14), defense responses (15, 16), regulation of ion channels in guard cells (17), activation of phospholipase D (18), and secretion of α-amylase from aleurone cells (19). However, the evidence for involvement of the G protein in such systems has been based only on the effects of activators and inhibitors of mammalian heterotrimeric G proteins on intact higher-plant tissues or cells. Ma and coworkers (20) have suggested from studies using histological methods that the heterotrimeric G protein may participate in various biological processes such as cell growth, differentiation, and nutrient transport in Arabidopsis thaliana. Aharon et al. (21) reported that a recombinant α subunit of a tomato G protein (TGα1) activated a plant plasma membrane Ca2+ channel in vitro, which offers direct, though limited, evidence for the function of a heterotrimeric G protein in plants.
In the present work, we produced rice transformants in which the endogenous mRNA for the α subunit of rice heterotrimeric G protein was suppressed by antisense technology. These transformants exhibited several morphological abnormalities, suggesting that the G protein plays a role in the signal transduction involved in morphogenesis. We also studied the expression of the α-subunit mRNA in the tissue and cells of normal cultivars. In addition, we found that five alleles of the rice mutant Daikoku dwarf (d-1) contained different mutations in the coding region of the α-subunit gene.
MATERIALS AND METHODS
Plant Materials.
Four rice cultivars (Oryza sativa L. cv. Nipponbare, Shiokari, Kinmaze, and Taichung 65) and five alleles of a rice mutant, Daikoku dwarf (d-1) (DK22, ID-1, CM1361–1, DKT-1, and DKT-2), were used in this study. The recurrent parents of DK22, ID-1, and CM1361–1 are Nipponbare, Shiokari, and Kinmaze, respectively. DKT-1 and DKT-2 had been produced from a common parent, Taichung 65. All rice plants, normal cultivars, mutants, and transgenic plants, were grown under 14-hr light with cool-white fluorescent light at 30,000 lux at 30°C and 10-hr dark at 25°C cycles. Tissues were frozen in liquid N2 and stored at −80°C until extraction of nucleic acids.
Generation of Transgenic Rice Plants.
Binary vector, pBI121-Hm expressing β-glucuronidase (GUS) under the control of the cauliflower mosaic virus 35S promoter, was used as the control vector for rice transformation (22). The GUS gene in pBI121-Hm was replaced with a fragment of a full length cDNA for the α subunit of rice G protein (RGA1) (9), to generate pAntiRGA1, which expressed an antisense RNA of RGA1 under the control of the 35S promoter. Binary vectors, pBI121-Hm and pAntiRGA1, were introduced into Agrobacterium, EHA101.
Transgenic rice plants were generated by using the Agrobacterium-mediated transformation method described previously (23). Dehusked mature seeds (Oryza sativa L. cv. Nipponbare) were sterilized and inoculated into the callus-induction medium. After 3 weeks, the proliferated calli derived from the scutella were used for transformation. EHA101 containing the binary vector was cocultured with rice calli, and transgenic rice plants were selected in the presence of hygromycin.
Isolation of Nucleic Acids and Hybridization Analysis.
A 299-bp fragment between nucleotides 252 and 550 of RGA1 (9) was subcloned in pGEM3zf(+) vector (Promega) to obtain pGEM-RGA1. Digoxigenin (DIG)-11-dUTP was used for RNA labeling (Boehringer Mannheim). DIG-labeled sense and antisense strand RNAs were synthesized by in vitro transcription by using linearized pGEM-RGA1 and SP6 RNA polymerase for the sense RNA or T7 RNA polymerase for the antisense RNA. In Northern blots and in situ hybridization, these DIG-labeled RNAs were used as probes to detect of antisense and sense RNAs for the α subunit.
Total RNA from various tissues was extracted by using RNeasy Plant Mini Kits (Qiagen). Poly(A)+RNA-enriched fraction was isolated by using Straight A’s mRNA Isolation System (Novagen). Total RNA (10 μg) or poly(A)+RNA-enriched fractions (1 μg) were run on 1.2% (wt/vol) agarose gels containing 5% (vol/vol) formaldehyde in RH buffer (20 mM morpholinopropanesulfonic acid/5 mM sodium acetate/0.5 mM EDTA, pH 7.0), and transferred to a nylon membrane (Boehringer Mannheim). The conditions used for Northern blot were those recommended by the manufacturer of the membrane. The membrane was then incubated with anti-DIG antibody conjugated with alkaline phosphatase, and the blots were visualized by using CSPD chemiluminescent substrate (Tropix, Bedford, MA) as the substrate for alkaline phosphatase and exposed to x-ray film.
Genomic DNA from whole plants was isolated by an extraction method with cetyltrimethylammonium bromide (24). Genomic DNA (5 μg) was digested with HindIII, resolved on a 0.6% (wt/vol) agarose gel and transferred to a nylon membrane (Boehringer Mannheim). The conditions for Southern blot hybridization and the methods for DIG detection were those recommended by the manufacturers of the membranes. In Southern blots, a full length RGA1 cDNA was labeled with DIG-High Prime (Boehringer Mannheim) and used as a probe.
Mapping of the α-Subunit Gene on the Chromosome in Rice.
All procedures were carried out according to the methods used by the Rice Genome Research Program (25).
Reverse Transcription–PCR (RT-PCR) and Sequencing.
Poly(A)+RNA (0.5 μg) was used as a template in 50 μl of RT-PCR reaction buffer by using the Titan One Tube RT-PCR System (Boehringer Mannheim). Two oligonucleotides [(1) and (2)] for 5′ and 3′ noncoding regions were designed on the basis of the sequence of RGA1 (9) and used as primers for RT-PCR and sequencing: (1) 5′ GACGTCAACGTGCTTCCTGG 3′, corresponding to the sequence from the positions 1 to 20 of RGA1, and (2) 5′ AAAGATCAATCAATGGTCCACG 3′, corresponding to the sequence from the positions 1,361 to 1,340 of RGA1, in antisense orientation. PCR products were excised from the agarose gel and purified by QIAEX II Gel Extraction Kit and QIAquick PCR Purification Kit (Qiagen). The purified PCR products (100 ng) were sequenced with a THERMO sequence dye terminator cycle sequencing kit (Amersham Pharmacia) by using a DNA sequencer (model 377; Applied Biosystems/Perkin–Elmer).
In Situ Hybridization.
In situ hybridization with DIG-labeled RNA was conducted as described (26). Florets were collected 10 or 3 days before or 3 days after pollination and fixed at 4°C overnight with 4% (vol/vol) paraformaldehyde and 0.25% (vol/vol) glutaraldehyde in 10 mM sodium phosphate buffer, pH 7.2. Fixed florets were embedded in Paraplast (Oxford) and sectioned at 7–10 μm. Hybridization and immunological detection of the hybridized probe were performed by the method of Kouchi et al. (26).
RESULTS
Antisense Suppression of the α Subunit of a Heterotrimeric G Protein in Rice: Analysis of T1 Generation.
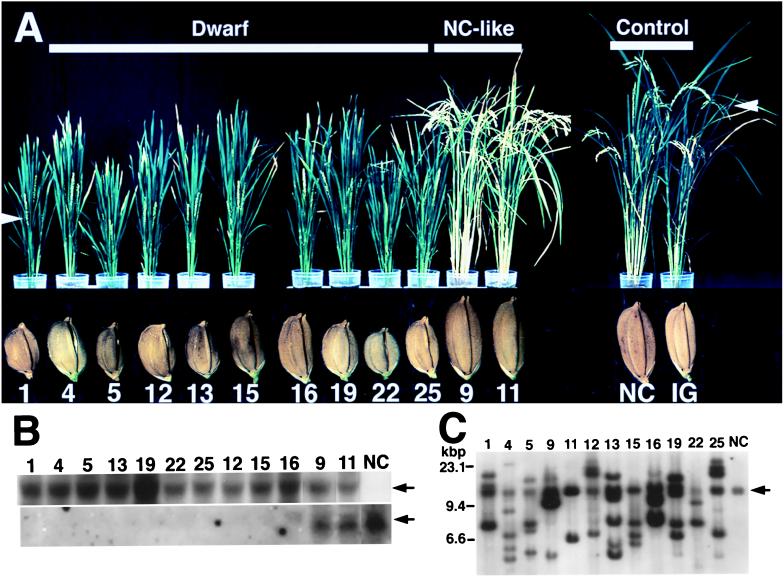
Transgenic rice plants carrying antisense RGA1 (cDNA for the rice α subunit) were generated by using an Agrobacterium-mediated transformation system (23). Ten transgenic rice plants, which expressed the antisense RNA, were analyzed. The plants at the first generation (T1 transgenic plants) were classified into two groups: group 1 (Anti α-1, Anti α-4, Anti α-5, Anti α-12, Anti α-13, Anti α-15, Anti α-16, Anti α-19, Anti α-22, and Anti α-25) contained no detectable endogenous mRNA for the α subunit, and group 2 (Anti α-9 and Anti α-11) accumulated the mRNA at levels similar to those for normal cultivar plants (Fig. 1B). The group 1 plants (T1 transformants) showed abnormal phenotypes such as shortening of internodes, leaves, and panicles, and the setting of small seeds (Fig. 1A), whereas the group 2 plants exhibited apparently normal phenotypes (Fig. 1A). The heads (ears) of mature group 1 plants remained erect, whereas those of mature group 2 plants and normal cultivars drooped. In addition, the leaves of mature group 1 plants were a darker green than normal cultivars and group 2 plants (Fig. 1A).
Figure 1.
Abnormal morphology of T1 transgenic rice plant carrying antisense RGA1 and their seeds (T2 seeds). (A) Morphology of the transgenic rice (4 mo old) and its seeds. Individual transgenic plants produced independently are designated as Anti α-1, Anti α-4, Anti α-5, and so on; Anti α, antisense RGA1 (cDNA for the α subunit of rice heterotrimeric G protein). Only the numbers of the individual transgenic plants are indicated below the plant pictures. Dwarf, dwarf plants; NC-like, apparently normal-cultivar-like plants; NC, normal cultivar (Nipponbare); IG, a transgenic plant expressing β-glucuronidase, a control for the transgenic plants with antisense RGA1. The white arrows indicate flowering (heads). T2 seeds from the transgenic plants are shown under the respective plants. (B) Northern blot analysis. See A for the numbers and NC. Total RNA (10 μg) from whole plants was loaded onto each lane. (Upper) The antisense RNA that is indicated by an arrow. (Lower) The sense RNA (endogenous mRNA) for the α subunit that is indicated by an arrow. (C) Southern blot analysis. See A for the numbers and NC (Upper). The arrow shows the α-subunit gene in the normal cultivar.
In the present work, we directed our attention to the abnormalities of dwarfism and the setting of small seeds. The dwarfism of our transformants was observed to be caused by shortening of all internodes, not only particular ones (data not shown). Microscopic observations showed no significant differences in the length of internode cells between the dwarf transformants and the control plants (data not shown), which suggests that the shortening of internodes may be attributable to a decrease in the number of cells per internode and so probably to reduction of cell division frequency. All seeds produced by every T1 transformant were normal in width but short in length compared with the seeds from the control plants, i.e., a normal cultivar plant, a transgenic plant expressing β-glucuronidase, and the group 2 plants (Fig. 1A).
In genomic Southern analysis with RGA1 as the probe, the different T1 transgenic plants showed different patterns of hybridization (Fig. 1C), indicating that they contain the antisense RGA1 gene at separate chromosome sites. The number of antisense RGA1 genes integrated for each T1 transgenic plant remains unconfirmed, but several antisense RGA1 genes seemed to have been inserted into the chromosomes for every plant.
Antisense Suppression of the α Subunit of a Heterotrimeric G Protein in Rice: Analysis of T2 Generation.
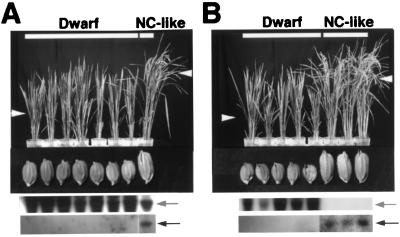
T2 seeds produced by four T1 transformants, Anti α-1, Anti α-15, Anti α-19, and Anti α-22, were grown to analyze their morphological phenotypes. Each T2 line (a set of T2 plants obtained with T2 seeds from a T1 transformant) had two types of plants, one exhibiting dwarf traits and the other, apparently normal cultivar phenotypes (Fig. 2 for Anti α-19 and Anti α-22). All T2 plants exhibiting dwarf traits contained the antisense RNA, but not the endogenous mRNA (sense RNA), for the α subunit. Every apparently normal cultivar plant contained the endogenous mRNA for the α subunit, whether the antisense RNA was present or not. The dwarf T2 plants set seeds of small sizes, whereas the apparently normal cultivar plants set normal seeds. The results show that the abnormal phenotypes are caused by the absence of the endogenous mRNA for the α subunit, and this trait is transmitted to some of the offspring.
Figure 2.
Morphology of T2 transgenic rice plants and their seeds (T3 seeds). Each eight T2 seeds from Anti α-19 (A) and Anti α-22 (B) were grown to analyze morphological phenotypes. Dwarf, dwarf plants; NC-like, apparently normal-cultivar-like plants. The white arrowheads indicate flowering (heads). (Upper and Lower) Under the plant photographs are Northern blots with the DIG-labeled sense and antisense RNAs, respectively, as probes. The arrows (Upper and Lower) indicate the antisense and sense (endogenous) RNAs, respectively, for the α subunit.
Expression of the Gene for the α Subunit in Normal Cultivar Plant.
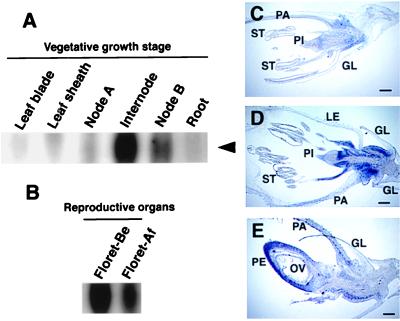
Northern blot analysis with control cultivars showed that the internodes of 1.5-mo-old plants carried significant amounts of the mRNA for the α subunit (Fig. 3A), but those of 3-mo-old plants contained only a little mRNA (data not shown), suggesting that the heterotrimeric G protein may play a role in young internodes. The leaves and roots of normal cultivar plants contained only small amounts of mRNA for the α subunit.
Figure 3.
Accumulation of endogenous mRNA for the α subunit of a heterotrimeric G protein in a normal rice cultivar. (A) Northern blot analysis of the mRNA in various tissues at the vegetative growth stage (1.5 mo old). Node A, upper node; Internode, internode between the upper and lower nodes; Node B, lower nodes and internodes. (B) Northern blot analysis of the mRNA from florets at reproductive growth stages (4 mo old). Floret-Be, florets 3 days before fertilization; Floret-Af, florets 3 days after fertilization. (C–E) In situ hybridization of florets, the longitudinal sections of florets on 10 days (C) and 3 days (D) before flowering and 3 days after fertilization (E). The DIG-labeled antisense RNAs were used as a probe. Positive hybridization signals are indicated in blue. Bars = 100 μm. GL, glume; LE, lemma; OV, ovary; PA, palea; PE, pericarp; PI, pistil; ST, stamen.
The florets accumulated a large quantity of the mRNA (Fig. 3B). In situ hybridization showed that the mRNA was present mainly in the bases of the stamen, pistil, lemma, and palea before flowering and in the pericarp of the ovary after fertilization (Fig. 3 C–E), which shows that the profile of tissue-specific expression of the gene for the α subunit changes during reproductive growth. The tissues, in which the mRNA for the α subunit is largely expressed in the control cultivars, correspond to those in which the T1 and T2 transformants show growth abnormalities.
Restriction Fragment Length Polymorphism (RFLP) Mapping of the α -Subunit Gene.
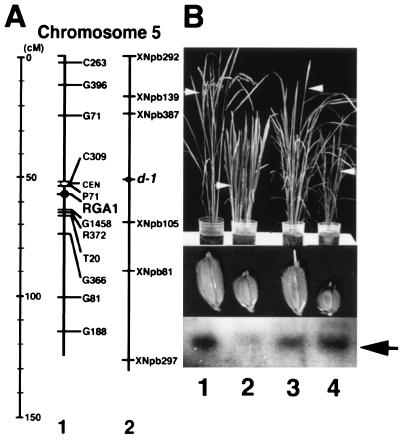
The α-subunit gene was located between RFLP markers P71 and G1458 on chromosome 5 in the rice high-density genetic linkage map (25) (Fig. 4A). The position was close to the mutated locus of Daikoku dwarf (d-1), which had been determined by integrated linkage mapping (Fig. 4A) (27). The Daikoku dwarf (d-1) mutant has pleiotropic phenotypes such as dwarfism and the setting of small seeds. This mutant was the first dwarf mutant to have been identified by genetic analysis in Japan (28). More than 10 alleles of Daikoku dwarf (d-1) had been isolated from mutagenic libraries by chemical mutagens and γ-ray irradiation. The morphological characteristics of Daikoku dwarf (d-1) closely resembled those of our transformants that lacked endogenous RGA1 mRNA after treatment by antisense technology. Thus, Daikoku dwarf (d-1) may have a mutation in the α- subunit gene.
Figure 4.
Map of the α-subunit gene on chromosome 5 in normal cultivar and accumulation of the endogenous mRNA for the α subunit in alleles of Daikoku dwarf (d-1). (A) Map on chromosome 5 on the basis of the result from the Rice Genome Research Program. cM, centimorgan. 1, RFLP markers used in this study are indicated on the right side of chromosome 5. The position of the α-subunit gene was mapped between RFLP markers P71 and G1458. The distances between P71 and RGA1 and between RGA1 and G1458 are approximately 3.5 cM and 6.4 cM, respectively. 2, The approximate locus of Daikoku dwarf (d-1) cited from the data by Yoshimura et al. (27). RFLP markers are indicated on the right side of chromosome 5. The RFLP marker XNpb81 is identical with RFLP marker G81 used in map 1. The distances between XNpb81 and d-1 and between G81 and RGA1 are approximately the same (36 cM and 40 cM, respectively). (B) Analysis of Daikoku dwarf (d-1). 1, Nipponbare (a normal cultivar); 2, Daikoku dwarf (d-1), DK22, from the recurrent parent, Nipponbare; 3, Shiokari (a normal cultivar); 4, Daikoku dwarf (d-1), ID-1, from the recurrent parent, Shiokari. (Top) Morphology of flowering plants. The white arrows indicate flowering (heads). (Middle) Morphology of their seeds. (Bottom) Northern blot analysis. Poly(A)+ RNA (2 μg) from whole plants was loaded on each lane and hybridized with the DIG-labeled antisense RNA (pGEM-RGA1) as a probe. The sense RNA (endogenous mRNA) for the α subunit is indicated by an arrow.
Analysis of the Rice Mutant Daikoku Dwarf (d-1).
Two alleles of Daikoku dwarf (d-1), DK22 and ID-1, showed dwarfism and set small seeds under our growth conditions, as compared with their recurrent parents, Nipponbare and Shiokari, respectively. In DK22, a low level of endogenous mRNA for the α subunit was detected (Fig. 4B Lower), which strongly suggests that DK22 is a mutant in the α-subunit gene. On the other hand, ID-1 contained the endogenous mRNA for the α subunit at a similar level to that in its recurrent parent (Fig. 4B Lower).
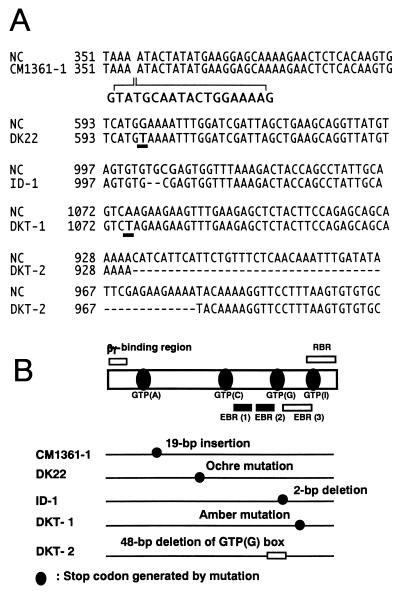
cDNAs for the α subunits in five alleles of Daikoku dwarf (d-1) (DK22, ID-1, CM1361–1, DKT-1, and DKT-2) and four normal cultivars (Nipponbare, Shiokari, Kinmaze, and Taichung 65) were synthesized by RT-PCR. DNA fragments at lengths of approximately 1,400 bp were synthesized for all plants with the primers described in Materials and Methods. The RGA1 sequences of the all normal cultivars (Nipponbare, Shiokari, Kinmaze, and Taichung 65) were identical (data not shown). Fig. 5 shows the aligned sequences of the mutated regions of Daikoku dwarf (d-1) (Fig. 5A) and a schematic diagram of the structure of the α subunit of rice heterotrimeric G protein and the positions of mutations in five alleles of Daikoku dwarf (d-1) (Fig. 5B).
Figure 5.
Comparison of the nucleotide sequences of the cDNA of the α subunit (RGA1) of the recurrent parents with those from five alleles of Daikoku dwarf (d-1). (A) Sequences of the mutated regions of Daikoku dwarf (d-1), CM1361–1, DK22, DKT-1, ID-1 and DKT-2 are aligned. The numbers of nucleotides correspond to those in RGA1 of Nipponbare. The RGA1 sequences of the all recurrent cultivars (Nipponbare, Shiokari, Kinmaze, and Taichung 65) were identical and these recurrent cultivars were indicated as NC (normal cultivar). (B) Schematic diagram of the structure of the α subunit of rice heterotrimeric G protein and the positions of mutations in five alleles of Daikoku dwarf (d-1). Highly conserved regions among the α subunits from various plants and animals, four GTP-binding sites (A, C, G, and I) and two of three effector-binding regions (EBR1 and 2), are indicated by closed boxes and bars, respectively. The regions, which show low homology among the plant and animal subunits, a βγ-binding region, one of three effector-binding regions (EBR 3) and a receptor-binding region (RBR), are indicated by open bars. The positions of stop codons generated by mutations in the cDNAs from four alleles of Daikoku dwarf (d-1), CM1361–1, DK22, ID-1, and DKT-1, are indicated by closed circles. A GTP-binding site (G), which is deleted in the cDNA of DKT-2, is indicated as an open box.
In CM1361–1, an insertion of 19 bp was present between the nucleotide positions of 354 and 355 in the cDNA (RGA1) for a normal cultivar. Thus the translation product would lack several functional domains: the GTP-binding regions (C), (G), and (I), the effector-binding regions (1), (2), and (3), and the receptor-binding region. In DKT-2, a region between the positions of 932 and 979 in RGA1 had been deleted, which would cause the loss of the GTP-binding region (G) in the translation product. In ID-1, a 2-bp-deletion was observed at the positions of 1,003 and 1,004 in RGA1, which results in a frameshift mutation. DK22 and DKT-1 were one-point mutants; the exchanges of G for T (positions 598 in RGA1) and A for T (positions 1,075 in RGA1) had occurred in the former and latter mutants, respectively, which result in the generation of ochre and amber mutants for the former and latter, respectively. The results show that Daikoku dwarf (d-1) is a mutant in the α-subunit gene of a heterotrimeric G protein in rice.
DISCUSSION
This study presents two lines of evidence for the involvement of rice heterotrimeric G protein in the development of normal internodes and seeds. First, the reduction of the endogenous mRNA for the α subunit by introduction of antisense cDNA into rice resulted in abnormal phenotypes such as short internodes and small seeds. Second, all alleles of a dwarf rice mutant, Daikoku dwarf (d-1), with similar abnormal phenotypes to those of the dwarf transformants had mutated in the coding region of the α-subunit gene. The mutated loci of other allele of Daikoku dwarf (d-1), HO541, was determined by map-based cloning to be located in the α-subunit gene (29). As the α-subunit gene is present as a single gene in rice (9) and Arabidopsis (7, 13), all alleles of Daikoku dwarf (d-1) did not contain the normal α-subunit gene. Because Daikoku dwarf (d-1) can survive and produce progeny, but not develop normally, the heterotrimeric G protein may function in a signal transduction pathway(s) required only for normal growth of specific, and not all, tissues in rice plants. The G protein also seems to participate in the normal development of rice leaves and panicles, but more precise observation may be needed to establish the abnormal traits of the leaves and panicles in the transformants and mutants.
We emphasize that the morphological abnormalities observed in the T1 transformants were caused by suppression of endogenous mRNA for the α subunit and not disruption of the α-subunit gene itself or of any particular gene(s) through insertion of the antisense RGA1 gene. Southern blot analysis of our transgenic plants supports this assessment. In addition, the abnormal phenotypes are inherited by the progeny, although the rate of segregation in the T2 transformants was not simple, for example, 3:1, because multiple antisense genes have been integrated into different chromosomes in the T1 transformants. Because some T2 transformants showed normal-like phenotypes, the possibility of artificial disruption of chromosomes during transformation may be ruled out.
The extent of accumulation of antisense RNA, the rate of suppression of the endogenous mRNA, and the level of the translation product varied among transgenic plants to which an antisense gene had been introduced (30), and some plants contain no or little endogenous mRNA (31, 32). We obtained two different types of transgenic rice plants expressing the antisense RNA in this study; one had no or little endogenous mRNA and showed abnormal phenotypes, and the other contained a significant amount of antisense RNA but endogenous mRNA at an amount nearly equal to that for the normal cultivar and showed a normal cultivar-like phenotype. Probably, the former transformants might contain so much antisense RNA that all endogenous mRNA might hybridize with the antisense RNA. We suspect that the hybridized RNA is broken down, and so no α subunit is synthesized. We also suppose that the latter may not contain the antisense RNA at the threshold level required to reduce the endogenous mRNA and thus to completely suppress the α subunit.
As mentioned in the Introduction, plant heterotrimeric G protein has been proposed to function in various signal transduction systems, including light-mediated gene expression (14), defense responses (15, 16), regulation of ion channels in guard cells (17), activation of phospholipase D (18), and secretion of α-amylase from aleurone cells (19). These proposals have been based on studies by using activators and inhibitors of the mammalian G proteins, but the reagents do not necessarily affect only the G proteins, as they also seem to display effects on calmodulin, phospholipase C, or small G proteins (1, 2, 33). These pharmacological studies have presented rather indirect evidence for the functions of the plant G protein. In contrast, our present work clearly shows that the G protein plays an important role in the normal development of rice internodes and seeds. Thus the most significant question raised by this study is how the G protein is involved in the normal development; in other words, how rice internodes and seeds become abnormal when the G protein does not work.
We suppose that cell division in the internodes may be reduced when the G protein does not function, because no significant difference was observed in the cell length between the cells in the corresponding internodes of normal cultivar and the dwarf transformant. This proposal is supported by the fact that, in normal cultivar, the mRNA for the α subunit exists at a high level in the young, but not in the mature, internodes. Kamijima supposed that the dwarf gene in Daikoku dwarf (d-1) might exert an inhibitory effect on cell division during the elongation of internodes (34). At present, the question remains of how the G protein functions in cell division.
Concerning seed development in rice plants, the lemma and palea are considered to play important roles in determining the size of rice seeds (35). The endogenous mRNA for the α subunit is not present at significant levels in the lemma and palea of the normal cultivar; it accumulates in the base of these tissues before pollination. We thus speculate that the base of the tissues needs the G protein to develop normal tissues. The pericarp of the ovary may also play a part in the development of normal-sized seeds.
Like heterotrimeric G proteins in mammals and yeast, the plant G protein must mediate an extracellular signal(s) to effectors. The other important question raised in this study is which signal(s) is mediated by the G protein to result in normal development of rice internodes and seeds. An allele of Daikoku dwarf (d-1), a 2-bp deletion mutant (see Fig. 5), has been proposed to be a gibberellin-insensitive mutant (36), because it produces little α-amylase even when gibberellin (GA3) is applied. The gibberellin-induced expression of the α-amylase gene in the oat aleurone layer has been shown to be affected by an activator of the mammalian G proteins, mastoparan 7 (19). In deepwater rice, gibberellin seems to play roles in the G2/M phase transition and subsequent enhancement of DNA synthesis during cell division (37). Thus we surmise that gibberellin may be the most probable candidate for an exogenous molecule(s), of which the signal(s) is transmitted through the G protein to the genes that conduct normal development of rice internodes and seeds. Because gibberellin is known to play a role in controlling chlorophyll content (38, 39), the abnormal phenotype in which the leaves of our transformants and Daikoku dwarf (d-1) are dark green may also be explained in connection with the plant hormone. If the exogenous molecule for the G protein is truly gibberellin, the Daikoku dwarf (d-1) may be a useful material for studying the signal transduction from gibberellin to genes.
Acknowledgments
We thank Dr. Kenzo Nakamura for the gift of binary vector pBI101-Hm, Drs. Seiichi Toki, Kinya Toriyama, Junko Kyozuka, and Ko Shimamoto for conducting the rice transformation technique, Drs. Makoto Matsuoka and Taku Demura for conducting the in situ hybridization technique, Drs. Itshuro Takamure, Yoshio Sano, and Toshiro Kinoshita for the gift of ID-1, Dr. Hikaru Satoh for the gift of CM1361–1, DK22, DKT-1, and DKT-2, and Drs. Koichiro Tsunewaki and Christopher J. Leaver for critically reviewing this manuscript. Part of the work was carried out at the Biological Resource Research and Development Center, Fukui Prefectural University. We acknowledge funding from three sources: a Grant-in-Aid for Scientific Research on Priority Areas (no. 06278102), a Grant-in-Aid for Scientific Research C (no. 10660318) from the Ministry of Education, Science and Culture, Japan, and a grant from the Science Promotion Foundation for Fukui Prefectural University.
ABBREVIATIONS
- DIG
digoxigenin
- RGA1
α subunit of rice heterotrimeric G protein
- RFLP
restriction fragment length polymorphism
- RT-PCR
reverse transcription–PCR
Footnotes
References
- 1.Gilman A G. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 2.Kaziro Y, Itoh H, Kozasa T, Nakafuku M, Satoh T. Annu Rev Biochem. 1991;60:349–400. doi: 10.1146/annurev.bi.60.070191.002025. [DOI] [PubMed] [Google Scholar]
- 3.Simon M I, Strathmann M P, Gautam N. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 4.Neer E J. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 5.Miyajima I, Nakafuku M, Nakayama N, Brenner C, Miyajima A, Kaibuchi K, Arai K, Kaziro Y, Matsumoto K. Cell. 1987;50:1011–1019. doi: 10.1016/0092-8674(87)90167-x. [DOI] [PubMed] [Google Scholar]
- 6.Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Genes Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- 7.Ma H, Yanofsky M F, Meyerowitz E M. Proc Natl Acad Sci USA. 1990;87:3821–3825. doi: 10.1073/pnas.87.10.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo H S, Kim H Y, Jeong J Y, Lee S Y, Cho M J, Bahk J D. Plant Mol Biol. 1995;27:1119–1131. doi: 10.1007/BF00020885. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa A, Tsubouchi H, Iwasaki Y, Asahi T. Plant Cell Physiol. 1995;36:353–359. doi: 10.1093/oxfordjournals.pcp.a078767. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa A, Iwasaki Y, Asahi T. Plant Cell Physiol. 1996;37:223–228. doi: 10.1093/oxfordjournals.pcp.a028935. [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki Y, Kato T, Kaidoh T, Ishikawa A, Asahi T. Plant Mol Biol. 1997;34:563–572. doi: 10.1023/a:1005807010811. [DOI] [PubMed] [Google Scholar]
- 12.Wise A, Thomas P G, Carr T H, Murphy G A, Millner P A. Plant Mol Biol. 1997;33:723–728. doi: 10.1023/a:1005732423622. [DOI] [PubMed] [Google Scholar]
- 13.Ma H. Plant Mol Biol. 1994;26:1611–1636. doi: 10.1007/BF00016493. [DOI] [PubMed] [Google Scholar]
- 14.Barnes S A, McGrath R B, Chua N-H. Trends Cell Biol. 1997;7:21–26. doi: 10.1016/S0962-8924(97)10043-5. [DOI] [PubMed] [Google Scholar]
- 15.Beffa R, Szell M, Meuwly P, Pay A, Vogeli-Lange R, Metraux J-P, Neuhaus G, Meins F J, Nagy F. EMBO J. 1995;14:5753–5761. doi: 10.1002/j.1460-2075.1995.tb00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blumward E, Aharon G S, Lam B C-H. Trends Plant Sci. 1998;3:342–346. [Google Scholar]
- 17.Assmann S M. Trends Plant Sci. 1996;1:73–74. [Google Scholar]
- 18.Munnik T, Arisz S A, Vrije T d, Musgrave A. Plant Cell. 1995;7:2197–2210. doi: 10.1105/tpc.7.12.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones H D, Smith S J, Desikan R, Plakidou-Dymock S, Lovegrove A, Hooley R. Plant Cell. 1998;10:245–254. doi: 10.1105/tpc.10.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H, Weiss C A, Ma H. Int J Plant Sci. 1994;155:3–14. [Google Scholar]
- 21.Aharon G S, Gelli A, Snedden W A, Blumwald E. FEBS Lett. 1998;424:17–21. doi: 10.1016/s0014-5793(98)00129-x. [DOI] [PubMed] [Google Scholar]
- 22.Hiei Y, Ohta S, Komari T, Kumashiro T. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 23.Toki S. Plant Mol Biol Rep. 1997;15:16–21. [Google Scholar]
- 24.Murray M G, Thompson W F. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin S Y, Antonio B A, Parco A, et al. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouchi H, Hata S. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimura A, Ideta O, Iwata N. Plant Mol Biol. 1997;35:49–60. [PubMed] [Google Scholar]
- 28.Akemine M. Rep Jpn Sci Assoc (Nihon gakujutsu kyokai hokoku) 1925;1:108–114. [Google Scholar]
- 29.Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. Breed. Sci. Vol. 48. abstr.; 1998. p. 70. [Google Scholar]
- 30.Bourque J E. Plant Sci. 1995;105:125–149. [Google Scholar]
- 31.Fan L, Zheng S, Wang X. Plant Cell. 1997;9:2183–2196. doi: 10.1105/tpc.9.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shintani D, Roesler K, Shorrosh B, Savage L, Ohlrogge J. Plant Physiol. 1997;114:881–886. doi: 10.1104/pp.114.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas A, Conradt B, Wickner W. J Cell Biol. 1994;126:87–97. doi: 10.1083/jcb.126.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamijima O. Jpn J Breed. 1981;31:302–315. [Google Scholar]
- 35.Matsuzaki A. In: Science of the Rice Plant II (Physiology) Matsuo T, Kumazawa K, Ishii R, Ishihara K, Hirata H, editors. Nobunkyo Tokyo; 1995. pp. 156–166. [Google Scholar]
- 36.Mitsunaga S, Tashiro T, Yamaguchi J. Theor Appl Genet. 1994;87:705–712. doi: 10.1007/BF00222896. [DOI] [PubMed] [Google Scholar]
- 37.Sauter M, Mekhedov S L, Kende H. Plant J. 1995;7:623–632. doi: 10.1046/j.1365-313x.1995.7040623.x. [DOI] [PubMed] [Google Scholar]
- 38.Jordan E T, Hatfield P M, Hondred D, Talon M, Zeevaart J A D, Vierstra R D. Plant Physiol. 1995;107:797–805. doi: 10.1104/pp.107.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor C B. Plant Cell. 1998;10:131–133. doi: 10.1105/tpc.10.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]