Abstract
Recently, encouraging AIDS vaccine trials in macaques have implicated cytotoxic T lymphocytes (CTLs) in the control of the simian human immunodeficiency virus SHIV89.6P that induces acute CD4+ T cell depletion. However, none of these vaccine regimens have been successful in the containment of replication of the pathogenic simian immunodeficiency viruses (SIVs) that induce chronic disease progression. Indeed, it has remained unclear if vaccine-induced CTL can control SIV replication. Here, we show evidence suggesting that vaccine-induced CTLs control SIVmac239 replication in rhesus macaques. Eight macaques vaccinated with DNA-prime/Gag-expressing Sendai virus vector boost were challenged intravenously with SIVmac239. Five of the vaccinees controlled viral replication and had undetectable plasma viremia after 5 wk of infection. CTLs from all of these five macaques rapidly selected for escape mutations in Gag, indicating that vaccine-induced CTLs successfully contained replication of the challenge virus. Interestingly, analysis of the escape variant selected in three vaccinees that share a major histocompatibility complex class I haplotype revealed that the escape variant virus was at a replicative disadvantage compared with SIVmac239. These findings suggested that the vaccine-induced CTLs had “crippled” the challenge virus. Our results indicate that vaccine induction of highly effective CTLs can result in the containment of replication of a highly pathogenic immunodeficiency virus.
Keywords: CD8+ T lymphocytes, selection, MHC, SIV, Sendai virus
Introduction
Virus-specific CD8+ CTL responses are critical for the control of immunodeficiency virus infections. The importance of CTLs in the control has been indicated by several clinical correlations in HIV-1–infected humans (1–3) and CD8+ T cell depletion experiments in macaque AIDS models (4–6). Therefore, recent vaccine approaches have focused on eliciting CTL responses (7, 8). However, HIV-1–infected individuals often have high plasma virus concentrations despite the presence of high frequencies of CTLs (9) and it has remained unclear if HIV-1 replication can be contained by vaccine-elicited CTL responses.
DNA vaccines, recombinant viral vector–based vaccines, and their combinations are promising delivery methods for AIDS vaccine because of their potential for inducing CTL responses. Recently, encouraging trials of these vaccines in macaques have implicated vaccine-induced CTLs in the control of the simian HIV (SHIV)89.6P that induces acute CD4+ T cell depletion (10–14). However, most of these vaccine regimens used Env as an immunogen and it is likely that Env-specific antibodies played a role in control of this chimeric virus. Additionally, it has been suggested that SHIV89.6P may not be an appropriate challenge virus (15) and none of these vaccine regimens have been successful in the containment of the more realistic challenge of the pathogenic simian immunodeficiency viruses (SIVs) smE660, mac251, or mac239 (16–19). Thus, it is quite important to know if vaccine induction of CTL responses can lead to the containment of replication of these SIVs that induce chronic disease progression.
We previously developed a DNA-prime/Gag-expressing Sendai virus (SeV) vector boost vaccine system and showed its potential for efficiently inducing Gag-specific cellular immune responses (13, 20). In the preclinical trial, all the vaccinated macaques controlled viremia and were protected from acute AIDS progression after SHIV challenge (13, 21). In this study, we examined if CTL induction by our vaccine system can result in the containment of SIVmac239 replication.
Materials and Methods
Animals.
Male rhesus macaques (Macaca mulatta) originally from southeastern Asia (Myanmar) were maintained in accordance with the Guideline for Laboratory Animals of National Institute of Infectious Diseases. These macaques were tested negative for SeV, SIV, and simian retrovirus type D before use. Blood collection, vaccination, and virus challenge were performed under ketamine anesthesia.
Vaccination and Challenge.
An env- and nef-deleted SHIV DNA, SIVGP1, was constructed from an infectious SHIVMD14YE clone DNA as described previously (13, 22). The DNA is deleted with a gene fragment encoding Env surface protein (SU; nucleotide [nt] 6211 to nt 7726 in HIV-1DH12; these sequence data are available from GenBank/EMBL/DDBJ under accession no. AF069140), the 3′ portion of the env gene (nt 8628 to nt 8764 in HIV-1DH12), and the 5′ quarter of the nef gene (nt 9333 to nt 9481 in SIVmac239; GenBank/EMBL/DDBJ accession no. M33262). From SIVGP1 DNA, the 5′ long terminal repeat region was replaced with a CMV promoter with immediate early enhancer and the 3′ portion containing the remaining nef and the 3′ long terminal repeat was replaced with Simian virus 40 poly A to obtain CMV-SHIVdEN DNA. Therefore, the CMV-SHIVdEN DNA has SIV-derived gag, pol, vif, vpx, and partial vpr sequences and HIV-1–derived partial vpr, tat, rev, and partial env (nt 7726 to nt 8628 containing the second exon of tat, the second exon of rev, and RRE) sequences. At DNA vaccination, animals received 5 mg CMV-SHIVdEN DNA intramuscularly. We used two kinds of SeV vectors, a transmissible one (SeV-Gag) and an F-deleted nontransmissible one (F[−]SeV-Gag), for the boost. Recombinant SeV-Gag and F(−)SeV-Gag were constructed and recovered as described previously (20, 23, 24). 6 wk after the DNA prime, animals received 108 cell-infectious units of SeV-Gag or 6 × 109 cell-infectious units of F(−)SeV-Gag intranasally as a boost. Four macaques (V1, V2, V3, and V4) were vaccinated with DNA-prime/SeV-Gag-boost, and the other four (V5, V6, V7, and V8) were vaccinated with DNA-prime/F(−)SeV-Gag-boost. 13 wk after the boost, animals were challenged intravenously with 1,000 TCID50 (50% tissue culture–infective dose) of SIVmac239 (25). An SIVmac239 molecular clone DNA, pBRmac239, was provided by T. Kodama (University of Pittsburg, Pittsburgh, PA) and R.C. Desrosiers (New England Primate Research Center, Southborough, MA), and the virus obtained from COS1 cells transfected with pBRmac239 was propagated on rhesus macaque PBMCs to prepare the SIVmac239 challenge stock.
Flow Cytometric Analysis of Virus-specific IFN-γ Induction.
We measured virus-specific T cell levels by flow cytometric analysis of IFN-γ induction after specific stimulation as described previously (13). In brief, PBMCs were cocultured with autologous herpesvirus papio-immortalized B lymphoblastoid cell lines (B-LCLs; reference 26) infected with a vaccinia virus (Vv) vector (27) for nonspecific Vv control stimulation and B-LCLs infected with a Vv vector expressing SIVmac239 Gag for Gag-specific Vv Gag stimulation, respectively. Intracellular IFN-γ staining was performed by using CytofixCytoperm kit (Becton Dickinson) according to the manufacturer's instructions. FITC-conjugated anti–human CD4, peridinin chlorophyll protein–conjugated anti–human CD8, allophycocyanin-conjugated anti–human CD3, and anti–human PE-conjugated IFN-γ antibodies (Becton Dickinson) were used. Gag-specific T cell levels were calculated by subtracting the IFN-γ+ T cell frequencies after nonspecific Vv control stimulation from those after Gag-specific Vv Gag stimulation. Alternatively, for measurement of SIV-specific T cell levels, lymphocytes were cocultured with B-LCLs infected with a vesicular stomatitis virus G (VSV-G)–pseudotyped murine leukemia virus for nonspecific stimulation and B-LCLs infected with a VSV-G–pseudotyped SIVGP1 for SIV-specific stimulation, respectively. In the case of examining peptide-specific T cell levels, B-LCLs were pulsed with each peptide (at a final concentration of 1 μM) or peptide mixture (final concentration of each peptide was 1–10 μM) for peptide-specific stimulation or incubated without peptide for nonspecific stimulation. The peptides, including a panel of 117 overlapping peptides (15–17 amino acids [aa] in length and overlapping by 10 to 12 aa) spanning the entire SIVmac239 Gag sequence, were purchased from Sigma Genosys Japan. Specific T cell levels <100 cells per million PBMCs were considered negative, those between 100 and 200 borderline, and those >200 positive. Gag-specific T cells were undetectable before the vaccination in all of the vaccinees and before the challenge in all of the naive controls.
Quantitation of Plasma Viral Loads.
Plasma RNA was extracted using High Pure Viral RNA kit (Roche Diagnostics). For quantitation of plasma SIV RNA levels, serial fivefold dilutions of RNA samples were amplified in quadruplicate by RT and nested PCR using SIV gag-specific primers to determine the end point as described previously (22). For preparing the RNA standard, we first set up the method for quantitation of SHIV RNA copy number by using HIV-1 vpu-specific primers and an HIV-1 standard quantitated by Amplicor HIV-1 Monitor (Roche Diagnostics). By using this method, we prepared an SHIV standard for the present assay. The lower limit of detection in this assay is ∼4 × 102 copies/ml. The plasma viral loads at several time points were confirmed by real time PCR (28).
Sequencing.
Plasma RNA was extracted using High Pure Viral RNA kit or RNA extraction system in Amplicor HIV-1 Monitor. The fragment spanning from nt 1231 to nt 2958 in SIVmac239 containing all of the gag region was amplified from plasma RNA by nested RT-PCR. In case of the plasma with low viral loads (<2,000 copies/ml), 8–16 tubes of nested RT-PCR amplifications were performed for each plasma to avoid obtaining only unrepresentative clones. The PCR products were sequenced using dye terminator chemistry and an automated DNA sequencer (Applied Biosystems). Alternatively, the PCR products were subcloned into a plasmid DNA by using the TOPO cloning system (Invitrogen) and sequenced.
Isolation of Mamu-A/B cDNA Clones.
Total cellular RNA was used to synthesize oligo(dT)-primed cDNA with reverse transcriptase (Superscript II; Invitrogen). Full-length cDNAs of Mamu-A and Mamu-B were amplified by PCR with locus-specific primer pairs (Mamu-A forward: 5′-ATGGCGCCCCGAACCCTCCTCCTG-3′, Mamu-A reverse: 5′-TCACACTTTACAAGCCGTGAGAGA-3′; Mamu-B forward: 5′-ATGGCGCCCCGAACCCTCCTCCTG-3′, Mamu-B reverse: 5′-TCAAGCC-GTGAGAGACACATC-3′) and cloned in pGEM-T Easy vector (Promega). The integrity of the clones was verified by reference strand–mediated conformation analysis (RSCA; 29) as the following and then sequenced.
Determination of Mamu MHC-I Haplotype.
Locus-specific RT-PCR products were subjected to second round PCR to obtain 725-bp-long DNA fragments encoding Mamu-A/B extracellular domains using Mamu-A/B universal forward (5A: 5′-ATGGCGCCCCGAACCCTC-3′) and reverse (4R: 5′-CCAGGTCAGTGTGATCTCCG-3′) primers. The product was analyzed by RSCA conformation analysis essentially as described previously (31). In brief, the second round PCR products and “a reference strand,” a fragment derived from the same PCR condition except for using 5′ Cy5-labeled forward primer and a certain cloned DNA template (its sequence is available upon request), were mixed together in a reaction tube, heat denatured, and then cooled down to form heteroduplex DNA. The mobility of heteroduplex DNA molecules in 6% nondenaturing Long Ranger gel (BioWhittaker Molecular Applications) was measured by ALF express II automated sequencing apparatus (Amersham Biosciences). Fluorescence electropherograms showed multiple peak patterns corresponding to multiple, different kinds of sequences expressed in individual macaques. The identity of each peak was determined by comparison of its mobility with those of heteroduplexes derived from parallel PCR using Mamu-A/B cDNA clones as templates. Alleles that were shared by a breeder macaque and subset of his sons were thought to be transmitted together and assigned to a single haplotype. The number of expressed alleles on one MHC-I haplotype ranged from one A and two B alleles to no less than three A and five B alleles.
Typing of MHC-II (Mamu-DRB and Mamu-DQA).
MHC-II alleles and haplotype compositions of macaques were analyzed by sequencing of cloned cDNA and denaturing gradient gel electrophoresis (DGGE; reference 30). Total RNA was extracted from B-LCLs and cDNA was generated by using SuperScript II reverse transcriptase. The entire DRB cDNA and the DQA exon 2 fragments were amplified by PCR using the following primer sets designed to hybridize with the conserved monomorphic regions: 5′-CGCGAATTCTCAGCTCAGGAGTCC-3′ and 5′-GCGGGATCCATGGTGTGTCTG-3′ for DRB, and 5′-CGCGAATTCGGTAGCAGCGGTAGAGT-3′ and 5′-GCGGGATCCGTGTAAACTTGTACCAGTT-3′ for DQA. The PCR products were subcloned into pUC19 and sequenced. When more than four clones with an identical sequence were obtained for an allele, the allele was considered to be expressed in the animal (see Table I).
Table I.
MHC-I and MHC-II Alleles of Macaques Used in This Study
| Animala | Father | MHC-I Mamu-A & B RSCA patternb |
MHC-II Mamu-DRB & DQA allelesc |
|---|---|---|---|
| Naive control | |||
| N1 | R90-088 | *1 | DRB1(Z26148), DRB*W502, DQA1*03(M76230) DRB(AB112040), DRB*W2603, DRB*W402, DQA1*0502 |
| N2 | R90-120 | 90-120-Ia | DRB1*1007, DRB1(Z26137), DQA1*03(M76228) DRB(Z26165), DRB(AB112039), DRB(AB112043), DQA1*06(MM76195) |
| N3 | R94-027d | *2 | DRB1*0316, DRB*W2507, DQA1*01(M76202) DRB*W2104, DRB*W2603, DRB*W606, DQA1*0502 |
| N4 | R90-010 | ND | DRB*W2104, DRB*W2603, DQA1*0502 DRB*0321, DRB*0323, DRB*W606, DQA1*05(M76227) |
| Vaccinee | |||
| V1 | R90-088 | *1 | DRB1(Z26148), DRB*W502, DQA1*03(M76230) DRB*W2505, DRB(AB112046), DRB(AB124813), DQA(AB124814) |
| V2 | R20-120 | 90-120-Ib | DRB*W2002, DRB*W2501, DQA1*0502 DRB1(Z26148), DRB*W502, DQA1*03(M76230) |
| V3 | R90-120 | 90-120-Ia | DRB1*1007, DRB1(Z26137), DQA1*03(M76228) DRB1(Z26148), DRB*W502, DQA1*03(M76230) |
| V4 | R94-027 | 90-120-Ia | DRB1*1007, DRB1(Z26137), DQA1*03(M76228) DRB*W2505, DRB(AB112046), DRB(AB124813), DQA(AB124814) |
| V5 | R94-027 | 90-120-Ia | DRB1*1007, DRB1(Z26137), DQA1*03(M76228) DRB(AB112043), DRB(AB112047) |
| V6 | R94-027 | *2 | DRB1*0316, DRB*W2507, DQA1*01(M76202) DRB1(Z26148), DRB*W502, DQA1*03(M76230) |
| V7 | R94-027 | *2 | DRB1*0316, DRB*W2507, DQA1*01(M76202) DRB*W2104, DRB*W2603, DRB*W606, DQA1*0502 |
| V8 | R90-010 | ND | DRB*W2104, DRB*W2603, DQA1*0502 DRB1*0316, DRB*W2507, DQA1*09(M76200) |
| Breeder | |||
| R90-088 | unknown | *1 | DRB1(Z26148), DRB*W502, DQA1*03(M76230) |
| R90-120 | unknown | 90-120-Ia | DRB1*1007, DRB1(Z26137), DQA1*03(M76228) |
| 90-120-Ib | DRB*W2002, DRB*W2501, DQA1*0502 | ||
| R90-010 | unknown | ND | DRB*W2104, DRB*W2603, DQA1*0502 DRB1*0316, DRB*2507, DQA1*01(M76202) |
The underlined macaques showed control of SIV replication.
MHC-I Mamu-A and Mamu-B alleles and haplotype compositions of macaques were examined by RSCA and sequencing of cloned cDNA. The haplotype 90-120-Ia derived from macaque R90-120 consists of three Mamu-A alleles (Mamu-A120-1, Mamu-A120-4, and Mamu-A120-5) and four Mamu-B alleles (Mamu-B120-1, Mamu-B120-6, Mamu-B120-8, and Mamu-B120-9). The haplotype 90-120-Ib derived from macaque R90-120 consists of two Mamu-A alleles (Mamu-A120-2 and Mamu-A120-3 [= Mamu-A*05]) and five Mamu-B alleles (Mamu-B120-2, Mamu-B120-3, Mamu-B120-4, Mamu-B120-5 [= Mamu-B*36], and Mamu-B120-7). Macaques N1 and V1 shared an RSCA pattern of a haplotype derived from R90-088 (*1). Macaques N3, V6, and V7 shared an RSCA pattern of a haplotype not derived from R90-120 (*2).
MHC-II Mamu-DRB and Mamu-DQA alleles were analyzed by DGGE and sequencing of cDNA. The determined alleles are shown. Each number in parentheses indicates the corresponding accession number for the nt sequence of the allele that has not yet been designated.
The father of macaque R94-027 is macaque R90-120.
The number of DRB alleles expressed in macaques and their haplotype relationships were analyzed by comparing the patterns of DGGE and by cloning and sequencing of the DNA extracted from each band in the gel. For DGGE analyses, the DRB exon 2 fragment was amplified by PCR using the forward (5′-CACTGGCTTTGGCTGGGGACAC-3′) and the GC-clamped reverse (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCGCCCCCGCCCGCAGGATACACAGTCACCTTAG-3′) primers. DGGE was performed in 6% polyacrylamide gel containing a gradient of 36–50% of the denaturant mixture (7 M urea and 40% formamide) at 100 V at 60°C for 2.5 h in a DCode system (Bio-Rad Laboratories). DNA eluted from each separate band was subcloned into a plasmid by using the TOPO cloning system and sequenced.
Results
Gag-specific T Cell Induction after SeV-Gag-Boost.
Our extremely simple vaccine protocol consisted of a single prime with DNA followed by a single boost with a recombinant SeV vector expressing SIVmac239 Gag 6 wk after the prime. Eight rhesus macaques (V1, V2, V3, V4, V5, V6, V7, and V8) were vaccinated with the prime/boost, and four naive controls (N1, N2, N3, and N4) received no vaccination before an intravenous SIVmac239 challenge.
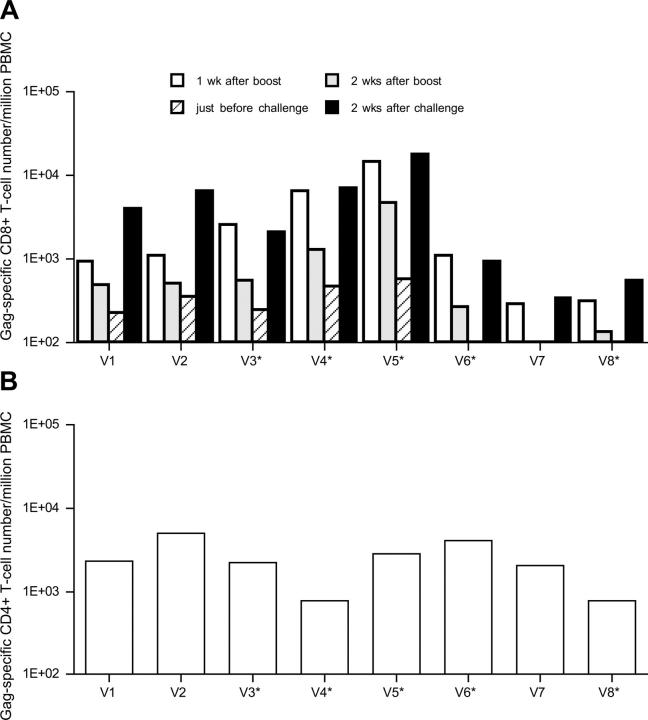
We measured virus-specific T cell levels in the vaccinated macaques by flow cytometric detection of antigen-specific IFN-γ induction. SIV- and Gag-specific T cell responses were examined in PBMCs at weeks 2 and 6 after the DNA vaccination, respectively, but no responses to either SIV or Gag were detectable in any of the vaccinated macaques (not depicted). After the SeV boost, however, we found induction of Gag-specific CD8+ T cells in all of the vaccinees (Fig. 1 A). The levels differed among the macaques, with five (V1, V2, V3, V4, and V5) maintaining detectable levels of Gag-specific CD8+ T cells until challenge. The SeV boost also induced Gag-specific CD4+ T cells in all eight vaccinees (Fig. 1 B).
Figure 1.
Gag-specific T cell frequencies in vaccinated macaques. Macaques V1, V2, V3, and V4 were boosted with a replication-competent SeV-Gag, whereas macaques V5, V6, V7, and V8 were boosted with a replication-defective F(−)SeV-Gag. *, macaques that controlled SIV replication after challenge. (A) Gag-specific CD8+ T cell frequencies per million PBMCs. The frequencies at week 7 after vaccination (1 wk after boost), at week 8 after vaccination (2 wk after boost), at week 19 after vaccination (just before challenge), and at week 2 after challenge (2 wk after challenge) are shown. (B) Gag-specific CD4+ T cell frequencies per million PBMCs at week 7 after vaccination (1 wk after boost). The frequencies were calculated by subtracting the IFN-γ+ T cell frequencies after nonspecific Vv control stimulation from those after Gag-specific Vv Gag stimulation. The background IFN-γ+ T cell frequencies after nonspecific stimulation were <2.0 × 102.
Control of SIVmac239 Replication in Five of Eight Vaccinees.
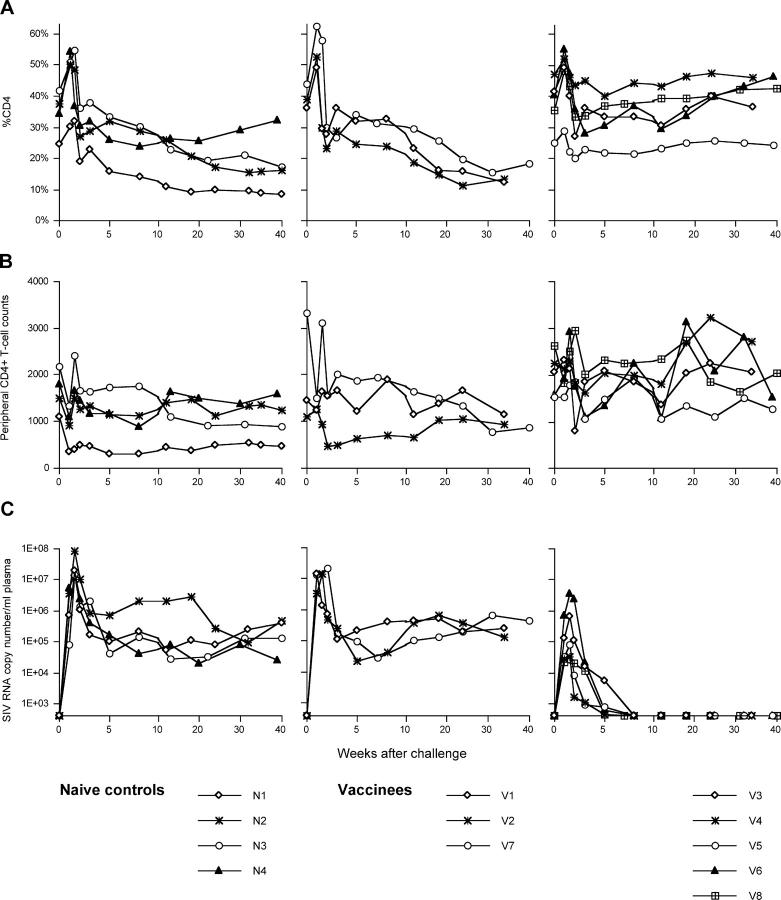
These vaccinated macaques were challenged intravenously with 1,000 TCID50 of SIVmac239 at week 19 after the DNA prime (13 wk after the SeV boost). The unvaccinated control macaques had high peak viremia (>107 SIV RNA copies/ml plasma) on day 10 after challenge and maintained relatively high plasma viral concentrations (104–106 SIV RNA copies/ml plasma; Fig. 2). Three of them showed gradual loss of percent CD4 in peripheral T lymphocytes. In contrast, five vaccinated macaques (V3, V4, V5, V6, and V8) controlled replication of this highly pathogenic challenge virus. In these macaques, plasma viremia became undetectable after week 5 and peripheral CD4+ T cells were maintained. The other three vaccinees (V1, V2, and V7) failed to control virus replication and showed gradual loss of percent CD4 in peripheral T lymphocytes similar to the naive control animals. One of them (macaque V2) was killed at week 42 because of dyspnea, loss of body weight, and loss of peripheral CD4+ T cells (4.4%, 97 cells/μl at week 42). Autopsy revealed that this animal developed AIDS with Pneumocystis carinii pneumonia.
Figure 2.
Changes in peripheral CD4+ T cell levels and plasma viral loads after SIVmac239 challenge. (A) Percents of CD4+ T cells in peripheral blood. (B) CD4+ T cell counts in peripheral blood. (C) Plasma viral loads (SIV RNA copy number/ml). The left panels show the naive controls (N1, N2, N3, and N4), the middle panels show the vaccinees that failed to control SIV replication (V1, V2, and V7), and the right panels show the vaccinees that controlled SIV replication (V3, V4, V5, V6, and V8). The portion until week 10 after challenge is enlarged.
At week 2 after challenge, we detected anamnestic Gag-specific CD8+ T cell responses in all of the vaccinated macaques, indicating efficient secondary responses during the acute phase of infection (Fig. 1 A). These levels varied from macaque to macaque. Macaque V5 showed the highest level of Gag-specific CD8+ T cells and macaque V7 showed the lowest. No significant difference in the levels was observed between the macaques that controlled viral replication and those that did not. The magnitude of the total prechallenge Gag-specific CD8+ T cell or CD4+ T cell responses did not appear to correlate with the level of control. We examined plasma-neutralizing activities against SIVmac239 as described previously (31), but found no neutralizing activities in any of the controls or the vaccinees at weeks 5 or 12 after challenge (not depicted), indicating that neutralizing antibodies were not essential for the control of SIV replication observed in this experiment.
Rapid Selection of CTL Escape Variants in the Vaccinees That Controlled SIVmac239 Replication.
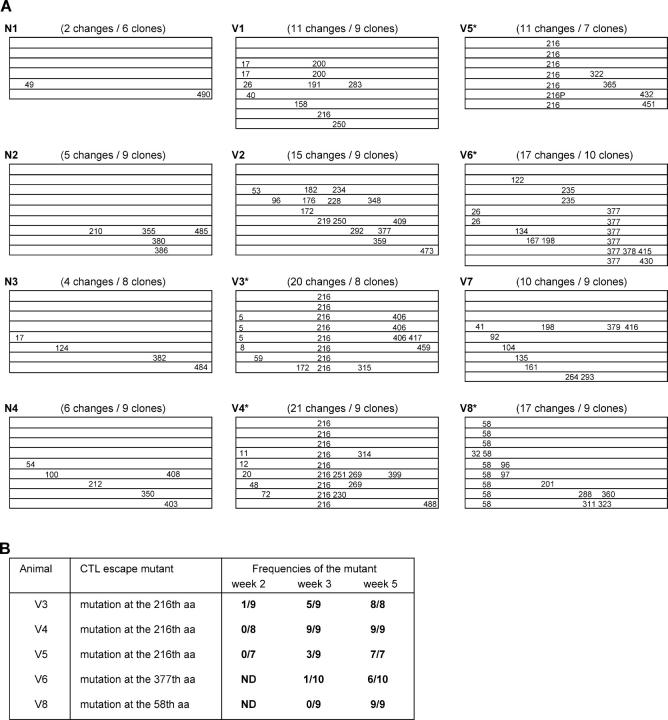
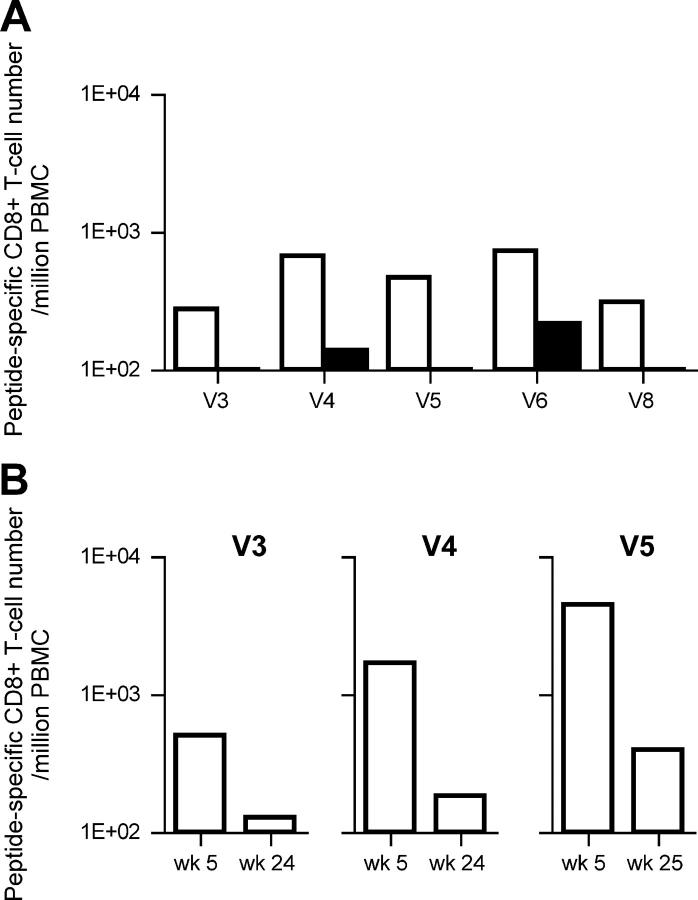
To determine whether vaccine-induced Gag-specific T cell responses exerted a selective pressure on the virus, we sequenced the SIV gag region in the viral genomes obtained from plasma RNA at week 5 after challenge (Fig. 3 A). The numbers of aa changes per clone in the vaccinated macaques were significantly higher than those in the unvaccinated (mean: unvaccinated, 0.51; vaccinated, 1.75; P = 0.0006 by t test). This may reflect the immune pressure by vaccine-induced Gag-specific T cell responses. Interestingly, all of the macaques that controlled SIVmac239 replication (V3, V4, V5, V6, and V8), but not those unable to control the virus, showed consistent aa changes in Gag (Fig. 3 A). Among them, three macaques (V3, V4, and V5) had a common aa change, leucine (L) to serine (S) at the 216th aa in Gag. We then examined peptide-specific T cell responses after the SeV boost and found, in these three macaques but not in the other vaccinees, efficient expansion of CD8+ T cells specific for an epitope (Gag206–216; IINEEAADWDL) spanning from the 206th to the 216th aa in SIVmac239 Gag. Interestingly, these three macaques showed no or diminished recognition of the mutant peptide, IINEEAADWDS (Gag206–216L216S; Fig. 4 A), indicating that this mutant likely represents an escape variant. Sequence analysis of viral genomes from the three macaques that made responses to this epitope at weeks 2 and 3 revealed that this CTL escape mutant became dominant around week 3 after challenge (Fig. 3 B). Thus, in these three macaques with high levels of Gag206–216-specific CD8+ T cells, the wild-type challenge virus disappeared quickly and only the CTL escape mutant was detectable in plasma at week 5. These three macaques had high levels of Gag206–216-specific CD8+ T cells 3 wk after challenge. However, these levels were considerably reduced in the chronic phase (Fig. 4 B).
Figure 3.
Mutations in SIV gag. (A) Schematic representation of the positions of aa changes in SIV Gag in each macaque after challenge. 6–10 clones of plasmids carrying the whole gag region amplified from plasma RNA at week 5 after challenge were obtained from each macaque and sequenced. Each lane represents the whole gag sequence derived from each clone and the positions of aa changes detected are indicated. Total number of aa changes and number of clones sequenced are shown in the parentheses. All the changes at aa 58 were glutamine to lysine, and all at aa 377 were isoleucine to threonine. All the changes at aa 216 other than the one indicated as 216P were L to S. The 216P represents L to P change at aa 216. (B) Frequencies of the CTL escape mutants at weeks 2, 3, and 5 in the vaccinees that controlled SIV replication. The ratio of the number of the clones with the escape mutation to the number of the sequenced clones is shown.
Figure 4.
Peptide-specific T cell frequencies in the vaccinees that controlled SIV replications. (A) Comparison between the epitope peptide–specific and the variant peptide–specific CD8+ T cell responses. PBMCs at week 10 after vaccination in macaque V3, at week 10 after vaccination in V4, at week 15 after vaccination in V5, at week 3 after challenge in V6, and at week 3 after challenge in V8 were used. The open bars indicate the levels of CD8+ T cells specific for Gag206–216 peptide in V3, V4, and V5, Gag367–381 peptide in V6, and Gag50–65 peptide in V8, respectively. The solid bars indicate the levels of CD8+ T cells specific for Gag206–216L216S peptide in V3, V4, and V5, Gag367–381I377T peptide in V6, and Gag50–65Q58K peptide in V8, respectively. (B) Gag206–216-specific CD8+ T cell levels in macaques V3, V4, and V5 after challenge. The background IFN-γ+ CD8+ T cell frequencies after nonspecific stimulation were <1.0 × 102.
We further examined epitope-specific CD8+ T cell responses in the other two macaques that controlled viral replication. In macaque V6, a mutation leading to a change at the 377th aa (isoleucine to threonine) was observed at week 5 (Fig. 3 A). Analysis of peptide-specific responses revealed that this macaque had a high level of CD8+ T cells specific for a 15-mer peptide corresponding to aa 367–381 in SIVmac239 Gag (Gag367–381) at week 3 after challenge. Stimulation by the mutant Gag367–381 peptide with the substitution (Gag367–381I377T) induced IFN-γ+ CD8+ T cells, but their frequency was lower than that after stimulation by the wild-type Gag367–381 peptide (Fig. 4 A). Additionally, viruses from macaque V8 had a mutation leading to a change at the 58th aa (glutamine to lysine; Fig. 3 A). In this animal, CD8+ T cell responses specific for a 16-mer peptide corresponding to aa 50–65 in SIVmac239 Gag (Gag50–65) were observed at week 3 after challenge. Stimulation by the mutant Gag50–65 peptide with the substitution (Gag50–65Q58K) failed to induce IFN-γ+ CD8+ T cells (Fig. 4 A). Each of these mutants became dominant at approximately week 5 after challenge in the corresponding macaque (Fig. 3 B).
Among the 12 macaques used in the challenge experiment, 8 macaques (2 naive controls and 6 vaccinees) descended from a single male, macaque R90-120 (its sons: N2, V2, and V3; its grandsons: N3, V4, V5, V6, and V7; Table I). Analysis of MHC-I Mamu-A and Mamu-B alleles indicated that four macaques of the eight R90-120 descendants, N2, V3, V4, and V5, share an MHC-I haplotype (90–120-Ia) derived from macaque R90-120. Analysis of MHC-II also suggested that these macaques possibly share an MHC-II haplotype derived from macaque R90-120. Among these macaques possessing the 90–120-Ia haplotype, three (V3, V4, and V5) were vaccinees that controlled SIV replication with high levels of Gag206–216-specific CD8+ T cell responses. The remaining one (naive control macaque N2) showed a detectable level of Gag206–216-specific CD8+ T cell responses at week 3 after challenge, although the level was low (2.5 × 102 cells/million PBMCs). These results strongly suggest that the Gag206–216 epitope is restricted by an MHC-I molecule derived from the 90–120-Ia haplotype.
Diminished Replicative Ability of the CTL Escape Variant SIV.
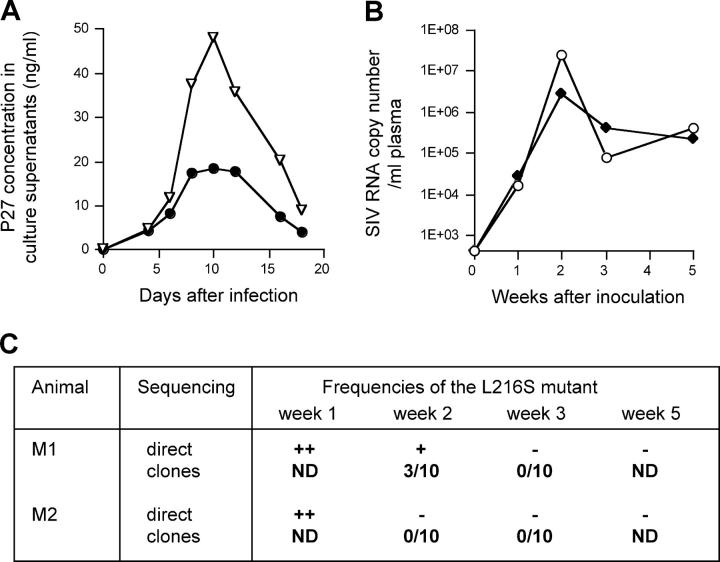
We then explored the hypothesis that the escape mutation selected by the vaccine-induced Gag206–216-specific CTL resulted in a loss of viral fitness. We constructed a molecular clone of the escape mutant SIV, referred to as SIVmac239G216S, with a mutation resulting in the L to S substitution at the 216th aa in Gag. The mutant SIV was replication competent in vitro but showed lower levels of proliferation kinetics in PBMC culture compared with the wild-type SIVmac239 (Fig. 5 A). To compare the SIVmac239G216S replication kinetics with the wild-type in macaques, two macaques (M1 and M2, neither of them descended from macaque R90-120) were coinoculated intramuscularly with 5 mg of the SIVmac239 molecular clone DNA (pBRmac239) and 5 mg of the SIVmac239G216S molecular clone DNA (pBRmac239G216S; Fig. 5, B and C). Both viral genomes were detected at comparable levels in plasma from both of the macaques at week 1 after the inoculation. After that, however, the mutant SIVmac239G216S disappeared and the wild-type SIVmac239 became dominant. Neither of the macaques showed Gag206–216-specific CD8+ T cell responses at week 3 (not depicted). These results indicate that the L to S change at the 216th aa in Gag is disadvantageous for SIV replication in the absence of Gag206–216-specific CD8+ T cell responses in macaques.
Figure 5.
Comparison of replication efficiencies between the wild-type SIVmac239 and the escape variant SIVmac239G216S. (A) Replication kinetics of SIVmac239 (▿) and SIVmac239G216S (•) in macaque PBMCs. MT4 cells were transfected with pBRmac239 and pBRmac239G216S to obtain SIVmac239 and SIVmac239G216S, respectively. PBMCs were infected with the viruses at a multiplicity of infection of 0.0002 and concentrations of SIV Gag p27 in their culture supernatants were measured by ELISA (Beckman Coulter). A representative result from three independent experiments is shown. (B) Plasma viral loads (SIV RNA copy number/ml) in macaques M1 (○) and M2 (♦) after inoculation with both of the wild-type SIVmac239 molecular clone DNA and the mutant SIVmac239G216S molecular clone DNA. (C) Frequencies of the mutant viral genome in plasma in the macaques inoculated with both of the wild-type SIVmac239 molecular clone DNA and the mutant SIVmac239G216S molecular clone DNA. In case of direct sequencing of the PCR products (indicated by direct), ++ indicates detection of both the wild-type and the mutant at comparable levels, + indicates detection of the wild-type predominantly and the mutant slightly, and − indicates detection of the wild-type only. In case of sequencing clones (indicated by clones), the ratio of the number of the mutant clones to the number of the sequenced clones is shown.
Discussion
In this study, we present evidence indicating that vaccine-induced CTLs control SIVmac239 replication in rhesus macaques. Each of the macaques that controlled viral replication had a mutation in Gag leading to an aa change in a CTL epitope by week 5 after challenge, reflecting strong CTL-induced selective pressure. This finding lends support to the notion that epitope-specific CTL responses played a central role in the control of replication of the SIVmac239 challenge virus because it was difficult to detect the challenge virus at week 5 after challenge.
Among the 12 macaques used in the challenge experiment, 8 macaques descended from macaque R90-120 and 4 of them shared an MHC-I haplotype, 90–120-Ia. Among the four, not the naive (N2) but the three vaccinees (V3, V4, and V5) controlled SIV replication and selected for the same Gag206–216-specific CTL escape variant with L to S change at the 216th aa in Gag. Therefore, we examined the reproducibly selected escape variant SIVmac239G216S intensively and found that in the absence of Gag206–216-specific CD8+ T cell responses, its replication efficiency is diminished compared with the wild-type SIVmac239 in vivo as well as in vitro. The rapid selection of the escape variant with lower viral fitness in the vaccinees with Gag206–216-specific CTLs indicates that the vaccine-induced CTLs exerted strong immune pressure leading to clearance of the wild-type SIVmac239.
The emergence of escape variants depends on the balance between CTL-induced immune pressure and viral fitness costs (32). Viral escape from CTLs during the acute phase of natural immunodeficiency virus infections has been observed in Tat, Nef, Vpr, and Env (33–36). Escape variants with mutations in the structural protein Gag have been also reported (37), but it has been shown that they mostly diminish viral fitness and require multiple additional compensatory mutations to restore their replicative competence (38–41). Indeed, the Gag206–216-specific CTL escape variant selected in macaques V3, V4, and V5 diminished viral replication. Therefore, our results suggest that the vaccine-induced CTLs were crucial to the rapid containment of replication of the challenge virus and selected for the virus with diminished replicative ability. Without compensatory mutations, the crippled virus might be easily controlled by the immune system.
The macaques used in our challenge experiment were non-Indian rhesus and the setpoint plasma viral loads in the naive control group might be lower than those usually observed in SIVmac239-infected Indian rhesus. However, the viral loads are higher than those typically observed in untreated humans infected with HIV-1 and equivalent to viral loads seen in SIVsmE660-infected Indian rhesus (16, 42). Indeed, all of the naive animals failed to control the virus replication after SIVmac239 challenge, indicating that CTLs are unable to contain and clear the virus in natural SIVmac239 infections of our non-Indian rhesus macaques. Thus, this study provides clear evidence demonstrating that vaccine induction of effective CTLs that can cripple the virus can result in the containment of replication of a neutralization-resistant, highly pathogenic immunodeficiency virus that is unable to be contained in the natural chronic course of infections. In conclusion, our results show that vaccine-induced CTLs can control SIVmac239 replication and indicate that induction of highly effective CTLs might be critical for the vaccine-based containment of immunodeficiency virus replication.
Acknowledgments
We thank M.A. Martin for providing SHIVMD14YE DNA, T. Kodama and R.C. Desrosiers for providing SIVmac239, Y. Ami, F. Ono, K. Komatsuzaki, K. Oto, H. Oto, H. Ogawa, K. Hanari, A. Kono, H. Akari, R. Mukai, A. Yamada, and K. Terao for assistance in the animal experiments, and M. Takiguchi, A. Kato, A. Okano, M. Matsuda, W. Sugiura, N. Yamamoto, A. Kojima, T. Sata, T. Takemori, T. Kurata, and A. Nomoto for their help.
This work was supported by the Ministry of Health, Labor, and Welfare, the Human Sciences Foundation and the Ministry of Education and Science in Japan, and by the National Institutes of Health.
Abbreviations used in this paper: aa, amino acid(s); B-LCL, B lymphoblastoid cell line; DGGE, denaturing gradient gel electrophoresis; L, leucine; nt, nucleotide; RSCA, reference strand–mediated conformation analysis; S, serine; SeV, Sendai virus; SHIV, simian HIV; SIV, simian immunodeficiency virus; VSV-G, vesicular stomatitis virus G; Vv, vaccinia virus.
References
- 1.Koup, R.A., J.T. Safrit, Y. Cao, C.A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D.D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrow, P., H. Lewicki, B.H. Hahn, G,M. Shaw, and M.B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogg, G.S., X. Jin, S. Bonhoeffer, P.R. Dunbar, M.A. Nowak, S. Monard, J.P. Segal, Y. Cao, S.L. Rowland-Jones, V. Cerundolo, et al. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 279:2103–2106. [DOI] [PubMed] [Google Scholar]
- 4.Matano, T., R. Shibata, C. Siemon, M. Connors, H.C. Lane, and M.A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72:164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz, J.E., M.J. Kuroda, S. Santra, V.G. Sasseville, M.A. Simon, M.A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B.J. Scallon, et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 283:857–860. [DOI] [PubMed] [Google Scholar]
- 6.Jin, X., D.E. Bauer, S.E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C.E. Irwin, J.T. Safrit, J. Mittler, L. Weinberger, et al. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus–infected macaques. J. Exp. Med. 189:991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker, B.D., and B.T. Korber. 2001. Immune control of HIV: the obstacles of HLA and viral diversity. Nat. Immunol. 2:473–475. [DOI] [PubMed] [Google Scholar]
- 8.MaMichael, A.J., and T. Hanke. 2003. HIV vaccines 1983–2003. Nat. Med. 9:874–880. [DOI] [PubMed] [Google Scholar]
- 9.Gea-Banacloche, J.C., S.A. Migueles, L. Martino, W.L. Shupert, A.C. McNeil, M.S. Sabbaghian, L. Ehler, C. Prussin, R. Stevens, L. Lambert, et al. 2000. Maintenance of large numbers of virus-specific CD8+ T cells in HIV-infected progressors and long-term nonprogressors. J. Immunol. 165:1082–1092. [DOI] [PubMed] [Google Scholar]
- 10.Barouch, D.H., S. Santra, J.E. Schmitz, M.J. Kuroda, T.-M. Fu, W. Wagner, M. Bilska, A. Craiu, X.X. Zheng, G.R. Krivulka, et al. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 290:486–492. [DOI] [PubMed] [Google Scholar]
- 11.Amara, R.R., F. Villinger, J.D. Altman, S.L. Lydy, S.P. O'Neil, S.I. Staprans, D.C. Montefiori, Y. Xu, J.G. Herndon, L.S. Wyatt, et al. 2001. Control of a mucosal challenge and prevention of AIDS in rhesus macaques by a multiprotein DNA/MVA vaccine. Science. 292:69–74. [DOI] [PubMed] [Google Scholar]
- 12.Rose, N.F., P.A. Marx, A. Luckay, D.F. Nixon, W.J. Moretto, S.M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J.K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell. 106:539–549. [DOI] [PubMed] [Google Scholar]
- 13.Matano, T., M. Kano, H. Nakamura, A. Takeda, and Y. Nagai. 2001. Rapid appearance of secondary immune responses and protection from acute CD4 depletion after a highly pathogenic immunodeficiency virus challenge in macaques vaccinated with a DNA-prime/Sendai viral vector-boost regimen. J. Virol. 75:11891–11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiver, J.W., T.M. Fu, L. Chen, D.R. Casimiro, M.E. Davies, R.K. Evans, Z.Q. Zhang, A.J. Simon, W.L. Trigona, S.A. Dubey, et al. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature. 415:331–335. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg, M.B., and J.P. Moore. 2002. AIDS vaccine models: challenging challenge viruses. Nat. Med. 8:207–210. [DOI] [PubMed] [Google Scholar]
- 16.Egan, M.A., W.A. Charini, M.J. Kuroda, J.E. Schmitz, P. Racz, K. Tenner-Racz, K. Manson, M. Wyand, M.A. Lifton, C.E. Nickerson, et al. 2000. Simian immunodeficiency virus (SIV) gag DNA-vaccinated rhesus monkeys develop secondary cytotoxic T-lymphocyte responses and control viral replication after pathogenic SIV infection. J. Virol. 74:7485–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen, T.M., L. Mortara, B.R. Mothe, M. Liebl, P. Jing, B. Calore, M. Piekarczyk, R. Ruddersdorf, D.H. O'Connor, X. Wang, et al. 2002. Tat-vaccinated macaques do not control simian immunodeficiency virus SIVmac239 replication. J. Virol. 76:4108–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton, H., T.U. Vogel, D.K. Carter, K. Vielhuber, D.H. Fuller, T. Shipley, J.T. Fuller, K.J. Kunstman, G. Sutter, D.C. Montefiori, et al. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen, T.M., P. Jing, B. Calore, H. Horton, D.H. O'Connor, T. Hanke, M. Piekarczyk, R. Ruddersdorf, B.R. Mothe, C. Emerson, et al. 2002. Effects of cytotoxic T lymphocytes (CTL) directed against a single simian immunodeficiency virus (SIV) Gag CTL epitope on the course of SIVmac239 infection. J. Virol. 76:10507–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kano, M., T. Matano, A. Kato, H. Nakamura, A. Takeda, Y. Suzaki, Y. Ami, K. Terao, and Y. Nagai. 2002. Primary replication of a recombinant Sendai viral vector in macaques. J. Gen. Virol. 83:1377–1386. [DOI] [PubMed] [Google Scholar]
- 21.Takeda, A., H. Igarashi, H. Nakamura, M. Kano, A. Iida, T. Hirata, M. Hasegawa, Y. Nagai, and T. Matano. 2003. Protective efficacy of an AIDS vaccine, a single DNA-prime followed by a single booster with a recombinant replication-defective Sendai virus vector, in a macaque AIDS model. J. Virol. 77:9710–9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata, R., F. Maldarelli, C. Siemon, T. Matano, M. Parta, G. Miller, T. Fredrickson, and M.A. Martin. 1997. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J. Infect. Dis. 176:362–373. [DOI] [PubMed] [Google Scholar]
- 23.Kato, A., Y. Sakai, T. Shioda, T. Kondo, M. Nakanishi, and Y. Nagai. 1996. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells. 1:569–579. [DOI] [PubMed] [Google Scholar]
- 24.Li, H.O., Y.F. Zhu, M. Asakawa, H. Kuma, T. Hirata, Y. Ueda, Y.S. Lee, M. Fukumura, A. Iida, A. Kato, et al. 2000. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J. Virol. 74:6564–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kestler, H., D.J. Ringler, K. Mori, D.L. Panicali, P.K. Sehgal, M.D. Daniel, and R.C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 65:651–662. [DOI] [PubMed] [Google Scholar]
- 26.Voss, G., S. Nick, C. Stahl-Hennig, K. Ritter, and G. Hunsmann. 1992. Generation of macaque B lymphoblastoid cell lines with simian Epstein-Barr-like viruses: transformation procedure, characterization of the cell lines and occurrence of simian foamy virus. J. Virol. Methods. 39:185–195. [DOI] [PubMed] [Google Scholar]
- 27.Mackett, M., G.L. Smith, and B. Moss. 1982. Vaccinia virus: a selectable eukaryotic cloning and expression vector. Proc. Natl. Acad. Sci. USA. 79:7415–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori, K., Y. Yasutomi, S. Ohgimoto, T. Nakasone, S. Takamura, T. Shioda, and Y. Nagai. 2001. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J. Virol. 75:4023–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arguello, J.R., A.M. Little, A.L. Pay, D. Gallardo, I. Rojas, S.G. Marsh, J.M. Goldman, and J.A. Madrigal. 1998. Mutation detection and typing of polymorphic loci through double-strand conformation analysis. Nat. Genet. 18:192–194. [DOI] [PubMed] [Google Scholar]
- 30.Knapp, L.A., L.F. Cadavid, M.E. Eberle, S.J. Knechtle, R.E. Bontrop, and D.I. Watkins. 1997. Identification of new Manu-DRB alleles using DGGE and direct sequencing. Immunogenetics. 45:171–179. [DOI] [PubMed] [Google Scholar]
- 31.Shibata, R., C. Siemon, S.C. Czajak, R.C. Desrosiers, and M.A. Martin. 1997. Live, attenuated simian immunodeficiency virus vaccines elicit potent resistance against a challenge with a human immunodeficiency virus type 1 chimeric virus. J. Virol. 71:8141–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, O.O., P.T.N. Sarkis, A. Ali, J.D. Harlow, C. Brander, S.A. Kalams, and B.D. Walker. 2003. Determinant of HIV-1 mutational escape from cytotoxic T lymphocytes. J. Exp. Med. 197:1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, D.A., P.J. Goulder, P. Klenerman, A.K. Sewell, P.J. Easterbrook, M. Troop, C.R. Bangham, and R.E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA. 94:1890–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borrow, P., H. Lewicki, X. Wei, M.S. Horwitz, N. Peffer, H. Meyers, J.A. Nelson, J.E. Gairin, B.H. Hahn, M.B. Oldstone, et al. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTL) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205–211. [DOI] [PubMed] [Google Scholar]
- 35.Allen, T.M., D.H. O'Connor, P. Jing, J.L. Dzuris, B.R. Mothe, T.U. Vogel, E. Dunphy, M.E. Liebl, C. Emerson, N. Wilson, et al. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 407:386–390. [DOI] [PubMed] [Google Scholar]
- 36.O'Connor, D.H., T.M. Allen, T.U. Vogel, P. Jing, I.P. DeSouza, E. Dodds, E.J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, et al. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493–499. [DOI] [PubMed] [Google Scholar]
- 37.Leslie, A.J., K.J. Pfafferott, P. Chetty, R. Draenert, M.M. Addo, M. Feeney, Y. Tang, E.C. Holmes, T. Allen, J.G. Prado, et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289. [DOI] [PubMed] [Google Scholar]
- 38.Kelleher, A.D., C. Long, E.C. Holmes, R.L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, et al. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27–restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peyerl, F.W., D.H. Barouch, W.W. Yeh, H.S. Bazick, J. Kunstman, K.J. Kunstman, S.M. Wolinsky, and N.L. Letvin. 2003. Simian-human immunodeficiency virus escape from cytotoxic T-lymphocyte recognition at a structurally constrained epitope. J. Virol. 77:12572–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedrich, T.C., C.A. Frye, L.J. Yant, D.H. O'Connor, N.A. Kriewaldt, M. Benson, L. Vojnov, E.J. Dodds, C. Cullen, R. Rudersdorf, et al. 2004. Extra-epitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic T-lymphocyte response. J. Virol. 78:2581–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedrich, T.C., E.J. Dodds, L.J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D.T. Evans, R.C. Desrosiers, B.R. Mothe, J. Sidney, et al. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10:275–281. [DOI] [PubMed] [Google Scholar]
- 42.Ling, B., R.S. Veazey, A. Luckay, C. Penedo, K. Xu, J.D. Lifson, and P.A. Marx. 2002. SIV(mac) pathogenesis in rhesus macaques of Chinese and Indian origin compared with primary HIV infections in humans. AIDS. 16:1489–1496. [DOI] [PubMed] [Google Scholar]