Abstract
Hepatocellular carcinomas (HCCs) mainly develop from liver cirrhosis and severe liver fibrosis that are established with long-lasting inflammation of the liver. Silencing of the suppressor of the cytokine signaling-1 (SOCS1) gene, a negative regulator of cytokine signaling, by DNA methylation has been implicated in development or progress of HCC. However, how SOCS1 contributes to HCC is unknown. We examined SOCS1 gene methylation in >200 patients with chronic liver disease and found that the severity of liver fibrosis is strongly correlated with SOCS1 gene methylation. In murine liver fibrosis models using dimethylnitrosamine, mice with haploinsufficiency of the SOCS1 gene (SOCS1−/+ mice) developed more severe liver fibrosis than did wild-type littermates (SOCS1+/+ mice). Moreover, carcinogen-induced HCC development was also enhanced by heterozygous deletion of the SOCS1 gene. These findings suggest that SOCS1 contributes to protection against hepatic injury and fibrosis, and may also protect against hepatocarcinogenesis.
Keywords: cytokine, STAT, TGF-β, DNA methylation, hepatitis C virus
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers with poor prognosis. Chronic infection with hepatitis C virus (HCV) is a major risk factor for development of HCC. In general, HCC develops only after one or more decades of HCV infection, and the increased risk is restricted largely to patients with cirrhosis or advanced fibrosis. Although HCV-infected persons with mild or nonhepatic fibrosis are not likely to get HCC, once cirrhosis is established, HCC develops at an annual rate of 1–4% per year (1), and there is a strong correlation between liver fibrosis and HCC development (2).
Suppressor of cytokine signaling-1 (SOCS1), also known as JAK binding protein, is an intracellular protein that inhibits JAK-analyzed mediated cytokine signaling by binding to JAKs. (3–5). SOCS1-deficient mice (SOCS1−/−) die within 3 wk after birth with fulminant hepatitis accompanied with serious fatty degeneration (6, 7). The SOCS1 gene has been implicated as an antioncogene in hepatocarcinoma. Yoshikawa et al. reported aberrant methylation in the CpG island of SOCS1 that correlated with its transcription silencing in HCC cell lines (8). The incidence of aberrant methylation was 65% in the 26 human primary HCC tumor samples analyzed. However, the molecular basis for the development of HCC by SOCS1 gene silencing has not been clarified.
Because HCC development follows severe liver fibrosis, we examined SOCS1 methylation status in chronic liver injury to determine if SOCS1 genetic alteration is involved in liver fibrosis progression as well as in HCC. We found that SOCS1 gene methylation occurred at the hepatitis stage before the onset of HCC and highly correlated with liver fibrosis. Furthermore, using mouse models, we demonstrated that SOCS1 gene deletion augments fibrosis associated with liver damage, probably due to signal transducer and activator of transcription (STAT1) 1 hyperactivation. We propose that the SOCS1 gene is necessary for preventing hepatocarcinogenesis developed from chronic liver injury and liver cirrhosis.
Materials and Methods
Patients and Histology.
A needle biopsy technique was used to obtain liver biopsy specimens from HCC patients in the Department of Internal Medicine of Kurume University Hospital and the Nagata Hospital. Tissue biopsies were stored in −70°C until analysis. Informed consent was obtained from all the patients before biopsies were conducted. Histological evaluations of the liver were based on 5–30-mm-long liver specimens obtained by percutaneous biopsy using 0.8–1.4-gauge needles. The specimens were fixed in a 10% formalin buffer and stained with hematoxylin-eosin. Biopsy specimens with signs of rejection were excluded, and histological results were classified into four categories as follows: normal, lobular hepatitis, chronic hepatitis (defined by piecemeal necrosis or fibrosis), and cirrhosis. The METAVIR score was used to classify biopsy specimens, using simplified scores for fibrosis (from F0, no fibrosis, to F4, cirrhosis; reference 9).
Methylation-specific PCR.
Genomic DNA was obtained from liver biopsy samples using a standard method and bisulfite modification of genomic DNA was performed as described previously (10, 11). The bisulfite-treated DNA was amplified with either a methylation-specific or unmethylation-specific primer as described previously (8). To investigate the methylation state of SOCS1 CpG DNA, we also performed methylation assays at CpG-rich regions around the NotI landmark sites after the genomic region in question had been digested with HpaII, which is sensitive to cytosine methylation in the CCGG recognition sequence as described previously (12).
Mice and Other Animal Models.
The SOCS1-deficient mouse has been described previously (13). For liver fibrosis model, dimethylnitrosamine (DMN) was administrated into 6–8-wk-old mice. Each mouse was given subsequently an intraperitoneal injection of 10 μg DMN per gram of body weight three times a week for 3 wk. The amount of DMN administered was adjusted for body weight each week. 2 d after the third series of injections, venous blood was collected and the mice were killed. Hyaluronic acid and alanine aminotransferase (ALT) were measured using standard methods. The liver was fixed with either 10% formaldehyde for histological examination or frozen immediately in liquid nitrogen for immunoblotting and the extraction of collagen as described previously (14). Liver fibrosis was quantified with Sirius red by digital image analysis as described previously (15). For carcinogenic treatment, 4-wk-old mice were given an intraperitoneal injection of 100 μg diethylnitrosamine (DEN) per gram of body weight once a week for 6 wk (16). 9 mo after the first injection of DEN, these mice were killed, and a 3-mm-thick section from each liver was prepared. An identical tumor from different sections was excluded for counts. The morphology of hepatocellular neoplasms was classified according to cell size, tinctorial properties of the cytoplasm, pattern of growth, and other morphological features as described previously (17). All experiments using these mice were approved by and performed according to the guidelines of the animal ethics committee of Kyushu University. Immunoblot analysis was performed using 20 μg of total liver protein as described previously (18).
Statistical Analysis.
For statistical analysis, we used the Student's t test, and a 95% confidence limit (defined as P < 0.05) was taken to be significant.
Online Supplemental Material.
The results of experiments, which complement the data presented in the paper, are shown in Figs. S1 and S2. Fig. S1 shows sequencing of 58 CpG sites in the 467-bp genomic DNA from patients with bisulfite treatment. Fig. S2 shows a gross image of the liver and sections with hematoxylin-eosin staining of hepatocarcinomas in DEN-treated SOCS1+/− mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20031675/DC1.
Results and Discussion
SOCS1 Methylation Correlates with Liver Fibrosis.
SOCS1 gene deletion in mice results in severe liver inflammation, probably because of hyper IFNγ/STAT1 signaling (7). Severe liver fibrosis due to chronic liver inflammation is shown to be a major cause of HCC development after HCV infection (19). Therefore, we examined the methylation state in the CpG island of the SOCS1 gene in >200 human liver biopsy samples with HCV-associated chronic hepatitis and liver cirrhosis before the onset of HCC. Among these, 209 samples of DNA could be evaluated for methylation status. Representative methylation-specific PCR results are shown in Fig. 1 A. Sequencing analysis of 58 CpG sites in the 467-bp genomic DNA with bisulfite treatment revealed that frequent methylation occurred in the latter half (nucleotides 200–400) of the region, whereas no methylation was detected in the PCR product determined as methylation negative by methylation-specific PCR (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20031675/DC1). We confirmed the reduced expression of the SOCS1 gene in methylation positive livers using real-time PCR (Fig. 1 B). We also confirmed that SOCS1 expression in the HCC region was lower than that in non-HCC region (Fig. 1 B).
Figure 1.
SOCS1 methylation in the liver of HCV positive patients and relationship to liver fibrosis and HCC. (A) Examples of methylation-specific PCR of SOCS1. Primer sets used for amplification are designated as unmethylated (UM) or methylated (M). PCR was performed using bisulfite-treated DNA from the paraffin-embedded liver tissue and PLC/PFR/5 cells (hepatoma cell line) as methylated positive control. (B) Quantification of SOCS1 mRNA levels by real-time RT-PCR in liver samples of non-HCC (−) and HCC (+) regions with SOCS1 gene methylation (n = 15) and unmethylation (n = 17). Methylation status was examined in non-HCC regions. Each group was normalized to the G3PDH values. (C) Summary of methylation status in patients and the HCC development among these patients within 12 yr. (D) The number of patients of each fibrosis score in methylation positive or negative patient groups and the percentage of methylation positive patients at each stage were plotted. (E) Serum platelet counts in patients from methylation positive or negative patient groups were scored. The bar indicates the standard deviation.
As shown in Fig. 1 C, aberrant methylation of the SOCS1 gene was present in 45% (94/209) of hepatitis patients detected by methylation-specific PCR (8). These results were confirmed by digesting genome DNA with methylation-sensitive enzymes (unpublished data and reference 12). These data indicate that DNA methylation in the liver occurred before HCC onset in a substantial percentage of hepatitis patients.
Next, we compared the grade of fibrosis and SOCS1 methylation. The progression of liver fibrosis was classified into four stages, from F1 to F4. As shown in Fig. 1 D, the frequency of SOCS1 methylation was 32% in the F1 stage, and it increased to nearly 60% in the F4 stage. The proportion of methylation significantly increased with the advancement of liver fibrosis, suggesting that the frequency of SOCS1 methylation correlated with the severity of liver fibrosis (Fig. 1 D). It has been reported that the platelet count accurately predicts significant HCV-related fibrosis (20). As shown in Fig. 1 E, the decrease in the platelet number was greater in SOCS1 methylation-positive patients (P < 0.01). These data reveal that SOCS1 methylation in the liver correlates with the liver fibrosis induced by hepatitis. Thus, we propose that at the earlier stages of chronic liver injury, SOCS1 methylation occurs, and the SOCS1 gene methylation ratio increases as the stage of fibrosis progresses.
SOCS1 Haploinsufficiency Enhanced Liver Fibrosis in Mice.
To verify the relationship between reduced expression of SOCS1 and liver fibrosis, we used a DMN-induced liver fibrosis model and mice carrying a heterozygous deletion of the SOCS1 gene (SOCS1−/+) as a model of SOCS1 gene silencing. An intraperitoneal injection of DMN has been shown to induce fatty degeneration of hepatocytes, activation and proliferation of hepatic stellate cells (also called Ito cells and lipocytes), infiltration by macrophages (possibly Kupffer cells), and secretion of TGF-β, which promotes fibrosis. First, we examined the mortality rate after injection of DMN (Fig. 2 A). Approximately 50% of mice with haploinsufficiency of the SOCS1 gene (SOCS1−/+ mice) died within 3 wk, whereas >90% of SOCS1+/+ mice survived. Serum ALT levels in SOCS1−/+ mice were higher than those in SOCS1+/+ mice, suggesting that more severe liver damage occurred in SOCS1−/+ mice. As shown in Fig. 2 C, DMN treatment resulted in tissue remodeling with a matrix deposition mainly in the centrilobular area and portal tract, as assessed histologically (Azan staining and Sirius red staining) or by immunohistostaining using specific antibodies against α-smooth muscle actin (SMA), TGF-β1, and fibronectin (Fig. 2 C). These histological analyses indicate that SOCS1 heterozygous deletion cooperatively enhanced fibrosis. We also estimated fibrosis by quantitative measurement of Sirius red staining (Fig. 2 D) and by measurement of serum hyaluronate levels (Fig. 2 E), a serum marker of the progression of liver fibrosis in humans (21). These data indicate that SOCS1 gene haploinsufficiency augmented DMN-induced liver fibrosis. These findings also support the current notion that hepatic stellate cells are transformed into myofibroblasts (expressing SMA) and secrete TGF-β, thereby stimulating the production of an extracellular matrix and probably the proliferation of these cells.
Figure 2.
Liver fibrosis is enhanced in SOCS1−/+ mice. (A) Survival rate in DMN-treated mice. DMN (10 μg/g body weight) was intraperitoneally injected on the indicated day (arrows), and mortality was examined. The numbers of mice examined and survived on day 21 are shown. (B) Serum ALT values were significantly greater in SOCS1−/+ than in SOCS1+/+ mice after 3 wk of DMN treatment. (C) Histological and immunohistological analysis of liver from mice treated with DMN. Sections of livers were examined using Azan and Sirius red staining and immunohistostaining with antibodies against α-smooth muscle actin (SMA), TGF-β1, and fibronectin. (D) The surface area stained with Sirius red was quantified using digital image analysis. (E) Serum hyaluronate levels in DMN-treated mice. Blood was collected 3 wk after DMN treatment. (B, D, and E) SOCS1−/+ (n = 6) and SOCS1+/+ (n = 8).
Next, we used another chronic liver injury model induced by a choline-deficient, L-amino acid–defined diet. As shown in Fig. 3 (A and B), liver fibrosis, as assessed by Azan and Sirius red staining, occurred more markedly in SOCS1−/+ mice than in the control mice (SOCS1+/+). In addition, the amount of total collagen, a marker of fibrosis, in SOCS1−/+ mice was higher than that in the control mice (Fig. 3 C, SOCS1+/+). Again, SOCS1 gene haploinsufficiency enhanced diet-induced liver fibrosis. These data indicate that loss of the SOCS1 gene in the liver enhances the liver fibrosis induced by liver damages.
Figure 3.

Fibrosis of the liver induced by a choline-deficient, L-amino acid–defined diet. (A) Histology of livers from indicated genotypes of mice fed a choline-deficient, L-amino acid-defined diet for 12 wk. Liver sections were examined histologically using Azan and Sirius red staining. (B) The surface area stained with Sirius red was quantitated using digital image analysis (n = 4). (C) Total collagen content of the liver of mice fed a choline-deficient diet for 4 or 12 wk. The total collagen content is shown with SD (n = 4).
Correlation between STAT Activation and DMN-induced Liver Fibrosis.
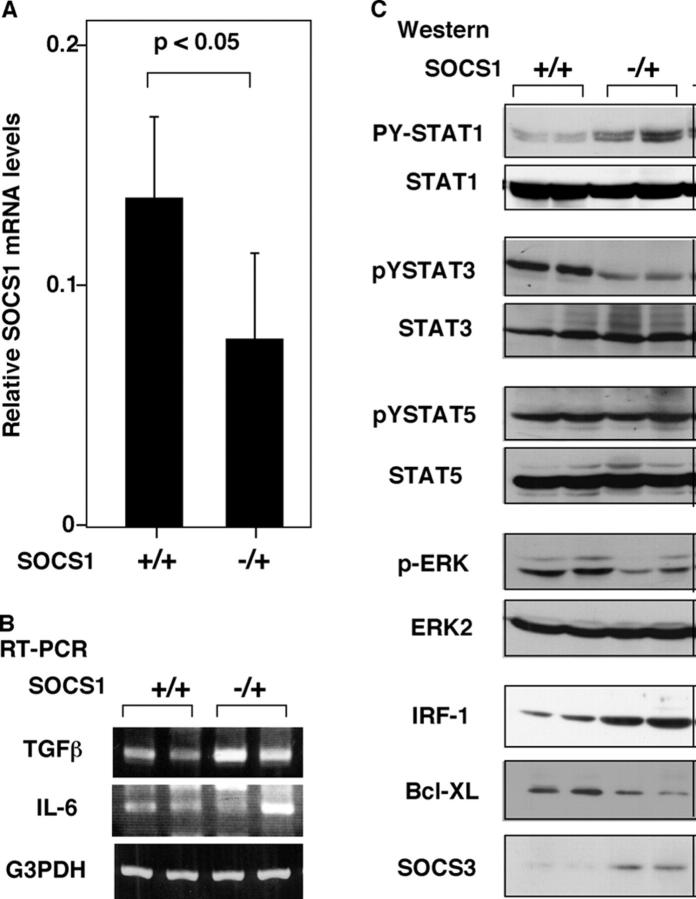
After injection of DMN, plasma levels of a wide variety of cytokines, including IL-6 and IFNγ, were dramatically elevated (22). To elucidate the molecular basis of induction of liver fibrosis by SOCS1 gene silencing, we examined the activation status of intercellular cytokine-signaling molecules using Western blotting. As shown in Fig. 4 (A and B), RT-PCR confirmed the reduction of SOCS1 mRNA levels and elevated TGF-β production in DMN-treated SOCS1−/+ mice. Western blotting analysis indicated that DMN injection induced phosphorylation of STAT1, STAT3, STAT5, and ERK (Fig. 4 C). The most striking correlation to fibrosis was elevated STAT1 activation and reduced STAT3 activation in SOCS1−/+ mice, whereas STAT5 phosphorylation was almost equal between SOCS1−/+ and SOCS1+/+ mice. STAT1 may promote liver injury by inducing IRF1, whereas STAT3 protects cells from apoptosis by inducing Bcl-XL. Consistent with strong STAT1 activation and decreased STAT3 activation in SOCS1−/+ mice, induction of the Bcl-XL protein was diminished, whereas the induction of the IRF-1 protein was significantly enhanced in these mice. Interestingly, SOCS3 expression was enhanced in SOCS1−/+ mice. Because SOCS3 is a negative regulator of gp130-mediated signals (23), elevated expression of SOCS3 may explain the reduced STAT3 activation in these mice.
Figure 4.
Activation of STATs and proapoptotic and antiapoptotic proteins in DMN-induced liver fibrosis. (A) Expression levels of SOCS1 mRNA were quantitated using real-time RT-PCR. The expression was normalized as a ratio using G3PDH mRNA as a housekeeping gene (n = 6). Each group of mice was sacrificed 3 wk after treatment with DMN. (B) RT-PCR analysis for TGF-β and IL-6 levels. Total RNA were prepared and analyzed using indicated primer sets. (C) Activation of STATs, SOCS1, and proapoptotic or antiapoptotic proteins in the liver. Total liver protein extracts were prepared and analyzed by Western blotting using indicated antibodies. Data are representative of three independent experiments with similar results. p, phosphorylated form.
In the ConA-induced hepatitis model, as in the DMN-induced fibrosis model, both STAT1 and STAT3 are activated (24). Using STAT1-, IFNγ-, and IL-6–deficient mice, Hong et al. proposed that STAT1 promotes liver injury by inducing the expression of proapoptotic IRF-1 protein, whereas STAT3 activation prevents liver damage by inducing antiapoptotic signals, such as Bcl-XL (24). Our data on STAT1 and STAT3 activation induced by DMN treatment are consistent with that proposal. Increased activation of STAT1 induces IRF-1 and reduced activation of STAT3 leads decrease in Bcl-XL expression, and these, at least in part, may account for a severe liver injury in SOCS1−/+ mice. Therefore, SOCS1 deletion may cooperatively promote liver damage and fibrosis by activation of STAT1 and repression of STAT3.
Hepatocarcinogenesis in SOCS1−/+ Mice.
Next, we examined if the SOCS1 loss of heterozygosity promotes hepatocarcinogenesis. We induced liver tumors by a well-established chemical carcinogenesis protocol using DEN. Because nitrate and nitrosamine synthesis are increased in viral hepatitis (25), DEN-induced HCC is thought to be an adequate model of hepatocarcinogenesis due to viral hepatitis. After 9 mo, liver tumors were developed. The number of tumors, including HCC in the SOCS−/+ mice, was higher than the number of tumors in the control mice (Fig. 5). As has been found in human HCC, male mice developed more tumors than did female. Large tumor nodules were macroscopically visible in SOCS1−/+ mice (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20031675/DC1). These tumors, whitish in color and discriminated from surrounding tissues, were consisted of hyperplastic nodules with no or mild dysplasia and typical HCCs that had a trabecular structure with or without fatty degeneration (Fig. S2). These data indicate that SOCS1 gene haploinsufficiency correlates with increased cancer risk in the liver.
Figure 5.
Hepatocarcinogenesis in SOCS1−/+ mice. Liver tumors of SOCS1−/+ mice 9 mo after DEN-induced tumor initiation. The total number of tumors (unshaded bars) including HCC (shaded bars) were scored from 100 series of 3-mm sections from livers of four female (f) and male (m) mice, with the indicated genotypes.
Our work demonstrated that SOCS1 gene haploinsufficiency in mice and SOCS1 gene silencing by DNA methylation in humans highly correlate with increased liver damage, liver fibrosis, and hepatocarcinogenesis, although it is unclear how liver injury contributes to fibrosis and hepatocarcinogenesis at present. Although the mechanism of aberrant gene methylation is unknown, it may be caused by increases in de novo methylation activity or by a defect of the protection mechanism against de novo methylation. However, there is no evidence of imprinting of the SOCS1 gene. Therefore, from data of SOCS1 gene methylation status of chronic hepatitis and liver cirrhosis sample, we propose that the SOCS1 gene is likely to be epigenetically methylated as liver fibrosis progresses, just as the methylation of the p16 (INK4A) tumor suppressor gene occurs during liver fibrosis (26).
It has been shown that HCC development of a person with HCV chronic infection is correlated with the severity of liver fibrosis. However, we found that SOCS1 methylation (P = 0.0015) is highly correlated with hepatocarcinogenesis, which is comparable to the correlation between fibrosis and HCC (P = 0.0011). These observations support our proposal that SOCS1 methylation can be a useful diagnostic indicator for HCC risk. SOCS1 methylation status may also partly explain the difference in individuals in the clinical course after hepatitis virus infection.
Acknowledgments
We thank Y. Kawabata for technical assistance, Dr. T. Naka and M. Ohara for comments and suggestions, and the Nagata Hospital for the donation of liver samples.
This work was supported by special Grants-in-Aid from the Ministry of Education, Science, Technology, Sports and Culture of Japan, the Japan Health Science Foundation, Mochida Memorial Foundation, and the Uehara Memorial Foundation.
The online version of this article contains supplemental material.
Abbreviations used in this paper: ALT, alanine aminotransferase; DEN, diethylnitrosamine; DMN, dimethylnitrosamine; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; SMA, smooth muscle actin; SOCS1, suppressor of the cytokine signaling-1; STAT, signal transducer and activator of transcription.
References
- 1.Hoshida, Y., K. Ikeda, M. Kobayashi, Y. Suzuki, A. Tsubota, S. Saitoh, Y. Arase, N. Murashima, K. Chayama, and H. Kumada. 1999. Chronic liver disease in the extremely elderly of 80 years or more: clinical characteristics, prognosis and patient survival analysis. J. Hepatol. 31:860–866. [DOI] [PubMed] [Google Scholar]
- 2.Ikeda, K., S. Saitoh, Y. Suzuki, M. Kobayashi, A. Tsubota, I. Koida, Y. Arase, M. Fukuda, K. Chayama, N. Murashima, and H. Kumada. 1998. Disease progression and hepatocellular carcinogenesis in patients with chronic viral hepatitis: a prospective observation of 2215 patients. J. Hepatol. 28:930–938. [DOI] [PubMed] [Google Scholar]
- 3.Naka, T., M. Narazaki, M. Hirata, T. Matsumoto, S. Minamoto, A. Aono, N. Nishimoto, T. Kajita, T. Taga, K. Yoshizaki, et al. 1997. Structure and function of a new STAT-induced STAT inhibitor. Nature. 387:924–929. [DOI] [PubMed] [Google Scholar]
- 4.Starr, R., T.A. Willson, E.M. Viney, L.J. Murray, J.R. Rayner, B.J. Jenkins, T.J. Gonda, W.S. Alexander, D. Metcalf, N.A. Nicola, and D.J. Hilton. 1997. A family of cytokine-inducible inhibitors of signalling. Nature. 387:917–921. [DOI] [PubMed] [Google Scholar]
- 5.Endo, T.A., M. Masuhara, M. Yokouchi, R. Suzuki, H. Sakamoto, K. Mitsui, A. Matsumoto, S. Tanimura, M. Ohtsubo, H. Misawa, et al. 1997. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 387:921–924. [DOI] [PubMed] [Google Scholar]
- 6.Metcalf, D. 1999. The SOCS-1 story. Exp. Hematol. 27:1715–1723. [DOI] [PubMed] [Google Scholar]
- 7.Naka, T., H. Tsutsui, M. Fujimoto, Y. Kawazoe, H. Kohzaki, Y. Morita, R. Nakagawa, M. Narazaki, K. Adachi, T. Yoshimoto, et al. 2001. SOCS-1/SSI-1-deficient NKT cells participate in severe hepatitis through dysregulated cross-talk inhibition of IFN-gamma and IL-4 signaling in vivo. Immunity. 14:535–545. [DOI] [PubMed] [Google Scholar]
- 8.Yoshikawa, H., K. Matsubara, G.S. Qian, P. Jackson, J.D. Groopman, J.E. Manning, C.C. Harris, and J.G. Herman. 2001. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat. Genet. 28:29–35. [DOI] [PubMed] [Google Scholar]
- 9.Bedossa, P., and T. Poynard. 1996. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 24:289–293. [DOI] [PubMed] [Google Scholar]
- 10.Herman, J.G., J.R. Graff, S. Myohanen, B.D. Nelkin, and S.B. Baylin. 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA. 93:9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merlo, A., J.G. Herman, L. Mao, D.J. Lee, E. Gabrielson, P.C. Burger, S.B. Baylin, and D. Sidransky. 1995. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med. 1:686–692. [DOI] [PubMed] [Google Scholar]
- 12.Nagai, H., Y.S. Kim, N. Konishi, M. Baba, T. Kubota, A. Yoshimura, and M. Emi. 2002. Combined hypermethylation and chromosome loss associated with inactivation of SSI-1/SOCS-1/JAB gene in human hepatocellular carcinomas. Cancer Lett. 186:59–65. [DOI] [PubMed] [Google Scholar]
- 13.Marine, J.C., D.J. Topham, C. McKay, D. Wang, E. Parganas, D. Stravopodis, A. Yoshimura, and J.N. Ihle. 1999. SOCS1 deficiency causes a lymphocyte-dependent perinatal lethality. Cell. 98:609–616. [DOI] [PubMed] [Google Scholar]
- 14.Tokuda, A., M. Itakura, N. Onai, H. Kimura, T. Kuriyama, and K. Matsushima. 2000. Pivotal role of CCR1-positive leukocytes in bleomycin-induced lung fibrosis in mice. J. Immunol. 164:2745–2751. [DOI] [PubMed] [Google Scholar]
- 15.Canbay, A., M.E. Guicciardi, H. Higuchi, A. Feldstein, S.F. Bronk, R. Rydzewski, M. Taniai, and G.J. Gores. 2003. Cathepsin B inactivation attenuates hepatic injury and fibrosis during cholestasis. J. Clin. Invest. 112:152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole, T.M., and N.R. Drinkwater. 1996. Two genes abrogate the inhibition of murine hepatocarcinogenesis by ovarian hormones. Proc. Natl. Acad. Sci. USA. 93:5848–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward, J.M., N. Konishi, and B.A. Diwan. 1990. Renal tubular cell or hepatocyte hyperplasia is not associated with tumor promotion by di(2-ethylhexyl)phthalate in B6C3F1 mice after transplacental initiation with N-nitrosoethylurea. Exp. Pathol. 40:125–138. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida, H., N. Kato, Y. Shiratori, M. Otsuka, S. Maeda, J. Kato, and M. Omata. 2001. Hepatitis C virus core protein activates nuclear factor kappa B-dependent signaling through tumor necrosis factor receptor-associated factor. J. Biol. Chem. 276:16399–16405. [DOI] [PubMed] [Google Scholar]
- 19.El-Serag, H.B. 2002. Hepatocellular carcinoma and hepatitis C in the United States. Hepatology. 36:S74–S83. [DOI] [PubMed] [Google Scholar]
- 20.Chitturi, S., and J. George. 2000. Predictors of liver-related complications in patients with chronic hepatitis C. Ann. Med. 32:588–591. [DOI] [PubMed] [Google Scholar]
- 21.Tanikawa, K. 1994. Serum marker for hepatic fibrosis and related liver pathology. Pathol. Res. Pract. 190:960–968. [DOI] [PubMed] [Google Scholar]
- 22.Schook, L.B., J.F. Lockwood, S.D. Yang, and M.J. Myers. 1992. Dimethylnitrosamine (DMN)-induced IL-1 beta, TNF-alpha, and IL-6 inflammatory cytokine expression. Toxicol. Appl. Pharmacol. 116:110–116. [DOI] [PubMed] [Google Scholar]
- 23.Yasukawa, H., A. Sasaki, and A. Yoshimura. 2000. Negative regulation of cytokine signaling pathways. Annu. Rev. Immunol. 18:143–164. [DOI] [PubMed] [Google Scholar]
- 24.Hong, F., B. Jaruga, W.H. Kim, S. Radaeva, O.N. El-Assal, Z. Tian, V.A. Nguyen, and B. Gao. 2002. Opposing roles of STAT1 and STAT3 in T cell-mediated hepatitis: regulation by SOCS. J. Clin. Invest. 110:1503–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, R.H., B. Baldwin, B.C. Tennant, and J.H. Hotchkiss. 1991. Elevated formation of nitrate and N-nitrosodimethylamine in woodchucks (Marmota monax) associated with chronic woodchuck hepatitis virus infection. Cancer Res. 51:3925–3929. [PubMed] [Google Scholar]
- 26.Kaneto, H., S. Sasaki, H. Yamamoto, F. Itoh, M. Toyota, H. Suzuki, I. Ozeki, N. Iwata, T. Ohmura, T. Satoh, et al. 2001. Detection of hypermethylation of the p16(INK4A) gene promoter in chronic hepatitis and cirrhosis associated with hepatitis B or C virus. Gut. 48:372–377. [DOI] [PMC free article] [PubMed] [Google Scholar]