Abstract
Ku70 is a protein that finds itself at the heart of several important cellular processes. It is essential to the Non-Homologous End Joining pathway as a part of the DNA-end binding complex, required for proper maintenance of telomeres and contributes to the DNA damage recognition and regulation of apoptosis. Forces that regulate Ku70 are therefore likely to have large consequences on the physiologic state of the cell. We report here that transient expression of the small protein SUMO resulted in a surprising increase in the abundance of Ku70. Surprisingly, the direct SUMOylation of Ku70 does not appear to be required for this effect. Rather, Ku70 appears to be stabilized through indirect effects on the rate of degradation. The same outcome was obtained by raising the expression of enzymes that promote SUMOylation. It is likely that many other proteins will be similarly regulated, providing a general control of cellular state.
Introduction
The Ku70 protein contributes to several important cellular processes. The heterodimer of Ku70 and Ku80 is a component of the general Non-Homologous End Joining (NHEJ) pathway utilized in all eukaryotic cells as one major method of repairing double-strand DNA breaks [1–2]. Our own interest in this protein extends from its essential role in V(D)J recombination, the rearrangement of the antigen receptor genes in vertebrates [3]. Aside from NHEJ, the Ku proteins play structural and regulatory roles in telomere maintenance with effects that include transcriptional silencing of telomere-proximal genes. The Ku70 protein appears to have a direct role in transcriptional activation and repression of a number of other genes, independent of its interaction with Ku80 [4]. Furthermore, Ku70 has a signaling role by recruiting the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) to the broken ends of DNA molecules and is itself phosphorylated. A role in modulating the DNA damage sensors ATM and ATR has also recently been reported [5]. While the previous functions are largely nuclear, Ku70 has a specific activity in the cytoplasm favoring cell survival by binding and sequestering the pro-apoptosis protein Bax [6]. Finally, for reasons that are still not clear, Ku70 and Ku80 are antigenic targets of the immune system in people suffering from certain autoimmune diseases.
Given the large number of functions for the Ku70 proteins, one might expect that the protein itself is likely to be regulated. Several mechanisms are already recognized to impinge upon Ku70. The steady state level of the protein appears to be regulated by heterodimerization with Ku80, intracellular localization [7], and post-translational modification. Phosphorylation and acetylation are already known modifiers. In addition, the proteasome-mediated degradation of Ku70 appears to be induced during drug-induced apoptosis and acts through ubiquitylation of Ku70 [8]. Curiously, the level of Ku proteins within each cell varies substantially between species. Human cell lines contain greater than ten-fold more Ku proteins than rodent-derived lines do, for reasons that remain obscure. One manifestation of the general importance of the Ku proteins is the observation that deletion of the Ku70 gene in mice, aside from interfering with immune function and DNA repair, is accompanied by growth retardation.
Modifying a protein by the covalent addition of small peptides such as ubiquitin and other ubiquitin-like peptides (including the SUMO family) can have profound effects on the protein and the cell. Certain types of poly-ubiquitylation can signal proteasome-mediated degradation. Other forms of ubiquitylation are commonly employed in the regulation of DNA repair. Recent work in S. cerevisiae indicates that proteasome particles may be associated with sites of DNA repair [9]. The SUMO family (with four paralogues in humans) preserves the general structural fold of ubiquitin but had already diverged in sequence and function in the simplest eukaryotes. Mono-ubiquitylation and SUMOylation can each lead to separate consequences regarding protein localization, complex assembly or conformation [10]. We recently observed that one important protein in the NHEJ pathway, XRCC4, depends upon transient SUMOylation for localization to the nucleus [11]. Several other proteins involved in DNA repair or maintenance are also regulated by SUMO. Furthermore, the RING motif in RAG1 raises the possibility that ubiquitin or SUMO are involved in V(D)J recombination [12]. Here we report that raising the cellular level of SUMO or increasing the degree of protein SUMOylation stabilizes Ku70.
Experimental Procedures
Antibodies and reagents
Monoclonal mouse antibodies to the FLAG epitope conjugated with horseradish peroxidase [HRP], human Ku70, α-Tubulin, and the rabbit polyclonal antibody to HA were from Sigma-Aldrich (St. Louis, MO). HRP-conjugated monoclonal anti-HA, and anti-T7 antibodies were from Roche Applied Science (Indianapolis, IN) and EMD Bioscience (San Diego, CA), respectively.
DNA manipulations
For transient eukaryotic expression experiments, FLAG-Ku70 or point mutants all based on human Ku70 were cloned into the pCMV-Tag2B vector (Stratagene, La Jolla, CA). All point mutants were made by using a QuikChange II Site-Directed Mutagenesis Kit (Stratagene) and verified by sequencing. Human Ku80 was cloned into the CMV-driven pKH3 vector [37] to encode HA-Ku80.
Plasmids expressing SUMO and SUMO-related enzymes were described previously [32]. EGFP-C1 (Clontech, Mountain View, CA) was used to express EGFP.
Cell culture, transfection and cell sorting
CHO.xrs6 [38, 39] and CHO-K1 (ATCC, Manassas, VA) cells were grown in F-12/DMEM medium (CellGro, Herndon, VA) supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Human HEK293T cells were grown similarly in DMEM media (CellGro). Cells were transfected with plasmids by using Fugene-6 (Roche Applied Bioscience) according to the manufacturer’s protocol.
Cell sorting of HA-SUMO1 and EGFP expressing 293T cells was performed on DakoCytomation (Glostrup, Denmark) MoFLo sorter.
Immunoprecipitation and immunoblotting
Anti-FLAG immunoprecipitation was performed using EZview Red Anti-FLAG M2 affinity gel (Sigma-Aldrich) as described in [32]. HRP-labeled antibodies against FLAG and HA were used to avoid cross-reaction with immunoglobulin. For endogenous human Ku70, 0.5–1 μg of total protein were separated by SDS-PAGE and visualized with anti-Ku70 antibody. ECL and ECL Plus Western blotting reagents (GE Healthcare Piscataway, NJ) were used for detection. Direct immunoblotting of other proteins typically used 20 μg of total protein or as noted.
Immunofluorescence
For immunofluorescence microscopy, cells were grown on cover slips at 2 × 104 cells per slip, fixed with paraformaldehyde, and visualized by using anti-Ku70 monoclonal and anti-HA polyclonal antibodies (Sigma-Aldrich) followed by TRITC-conjugated anti-mouse and FITC-conjugated anti-rabbit antibody (both Invitrogen) staining and counterstaining with DAPI (Sigma-Aldrich). Images were acquired with an Olympus IX81 microscope equipped with a digital camera and standard color filters.
Degradation Time Course
Sub-confluent cell cultures of CHO.xrs6 cells were grown in 60 mm dishes and transfected with FLAG-Ku70 wt or the K556R mutant with or without a SUMO-1 encoding plasmid. Cells were incubated for 36 hours in DMEM/F-12 media supplemented with 10% calf serum and then treated with 20 μg/ml of cycloheximide (Sigma-Aldrich) for 0, 1, 2, 4, and 8 hours before harvest.
Results
Ku70 levels rise, as SUMO is over-expressed
We examined the behavior of Ku proteins under a variety of conditions. The most striking effect is seen in Fig. 1A. FLAG-tagged Ku70 was expressed by transfection of the Chinese Hamster Ovary cell line CHO.xrs6 [13. This cell line does not express Ku80, so the behavior of Ku70 is visualized in the absence of heterodimerization. It should be noted that in the original characterization of this cell line the endogenous Ku70 protein is not detectable by direct immunoblot. Cells were transfected with a plasmid encoding FLAG-Ku70 alone or accompanied by a second plasmid expressing HA-tagged SUMO1. Proteins carrying the FLAG tag were concentrated by immunoprecipitation from lysates containing 500 μg of protein for each lane. No tagged protein is seen in the absence of transfection (lane 1), and a distinct band is visible when FLAG-Ku70 is expressed alone (lane 2). A dramatic increase in the abundance of the Ku70 band was observed when the plasmid encoding SUMO1 was cotransfected (lane 3). The difference in abundance was over five-fold, as determined by densitometry of this and other images. In addition, a minor band above that of Ku70 is also visible in lane 3. This corresponds in size to the SUMOylated form of Ku70, and this interpretation is supported by the immunoblot analysis on the right of Fig. 1A. Equivalent samples were analyzed and visualized using antibody directed towards the HA tag on SUMO1. The most intense band in lane 6 corresponds in size to the upper band of lane 3. Additional bands may represent multiple SUMO moieties on Ku70 or SUMOylated forms of other proteins that copurify.
Fig. 1. Ku protein levels respond to SUMO1 expression.
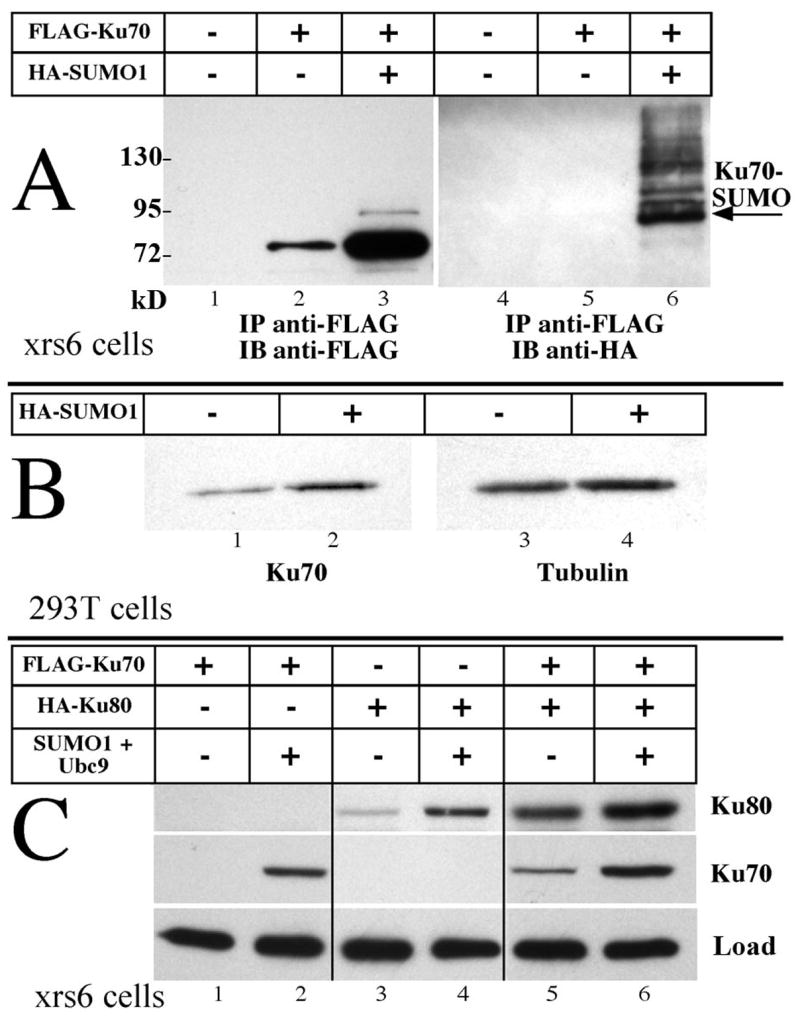
A) Plasmids encoding Ku70 and SUMO1 were variously transfected into CHO.xrs6 cells that lack endogenous Ku80. 500 μg of protein in lysates collected at 48 hours were immunoprecipitated with anti-FLAG agarose, analyzed by SDS-PAGE and visualized with anti-FLAG (lanes 1–3) or anti-HA antibodies (lanes 4–6). The arrow designates the band corresponding to singly SUMOylated Ku70. Unmodified Ku70 increased over five-fold in the presence of additional SUMO1. Protein masses of size markers are indicated in kD. B) Human cell line HEK293T was transfected with plasmids encoding EGFP alone or together with SUMO1. At 48 hours, EGFP positive cells were sorted and harvested. Endogenous Ku70 (lanes 1, 2) and tubulin as loading control (lanes 3, 4) were visualized directly by immunoblotting 1 μg of protein from each lysate. Endogenous Ku70 increased 2–3 folds while tubulin was unaffected. C) Ku70 and Ku80, variously expressed with Ubc9 and SUMO1, show additive effects. Equal loads of 20 μg of protein in each lane were visualized for the two tagged proteins. Lanes from a single gel were assembled for this image. Both Ku70 and Ku80 increase with SUMO1.
While a SUMOylated form of Ku70 appears to be present, the vast majority of the increase represents unmodified protein. In fact, the SUMOylated form is only visible when substantial loads of Ku70 are applied to the gel, as in panel A. These points are reiterated in Fig. 1B. Here we repeat the basic observation using a human cell line, HEK293T, visualizing the endogenous Ku70 protein in cells where Ku80 is normal. Cells were transfected with a plasmid encoding EGFP to allow sorting. One sample was additionally cotransfected with the SUMO1 expression plasmid. A distinct increase in Ku70 abundance was seen in the presence of additional SUMO1 (lane 2) compared to the equivalent load from cells transfected with the EGFP plasmid alone (lane 1). Tubulin did not differ between these two samples (lanes 3, 4), and a second DNA repair protein, XRCC4, also did not change in abundance in similar experiments [11].
Ku80 abundance parallels that of Ku70, as seen in Fig. 1 panel C. Here, the CHO.xrs6 cell line was transfected with plasmids expressing FLAG-Ku70, HA-Ku80 and the combination of SUMO1 (not tagged) and Ubc9 (the SUMO E2 conjugating protein). 20 μg of total protein was loaded directly in each lane and visualized with antibody against each tag. In the absence of Ku80, Ku70 levels changed from undetectable to distinctly positive upon the addition of the SUMOylating plasmids (lanes 1, 2). Similarly, Ku80 levels were substantially greater with additional SUMO (lanes 3, 4). Finally, both effects were additive as each of Ku70 and Ku80 were increased in abundance when coexpressed with the other protein, and these levels further increased with additional SUMO (lanes 5, 6). While the behavior of Ku70 is clearly dependent on the addition of SUMO, the rise in Ku80 levels may owe to two effects. A direct stabilization of Ku80 akin to that of Ku70 is one component, but we cannot ignore that the stabilization of endogenous Ku70 (invisible in these blots) may also contribute to the apparent stabilization of Ku80.
Ku70 levels are determined by protein stabilization
Increased levels of cellular SUMO1 were associated with an increase in the abundance of the unmodified form of Ku70 as well as a small fraction of a modified form (Fig. 1A lane 3 and 6). Is the effect on protein levels dependent on the direct SUMOylation of Ku70? We mutated 10 lysines of Ku70 (data not shown) to identify and eliminate the target of SUMOylation. SUMOylation frequently occurs at lysines within a consensus sequence motif ΨKXE where Ψ represents a large hydrophobic residue [14]. One mutant, K556R, reduced the level of the direct SUMOylation product by at least 70%, as seen in Fig. 2A. Despite reduction of direct SUMOylation, the mutant retained the same increase in abundance seen previously with the wild type proteins (compare Fig. 2A lanes 1 to 2 and 3 to 4). A change in steady state levels is explained by a change either in the rate of synthesis or degradation. Fig. 2B shows the analysis of degradation achieved by the method of cycloheximide washout. CHO.xrs6 cells, transfected with a Ku70 construct either alone, or accompanied by a SUMO1 plasmid, were allowed to grow for 36 hours before the addition of cycloheximide. This treatment halts further protein synthesis, allowing degradation to continue at its normal rate. The disappearance of the Ku70 band is presented on a semi-log plot and the single exponential that best fits the data is shown as dotted lines. Both wild type Ku70 and the K556R mutant display similar kinetics. In the absence of SUMO, wild type Ku70 degraded with T1/2 of 3.7 hours. In the presence of the SUMO plasmid, the T1/2 lengthened to 8.4 hours. Similarly, the K556 mutant T1/2 values were 2.75 and 8.2 hours.
Fig. 2. SUMO stabilizes Ku70 indirectly.
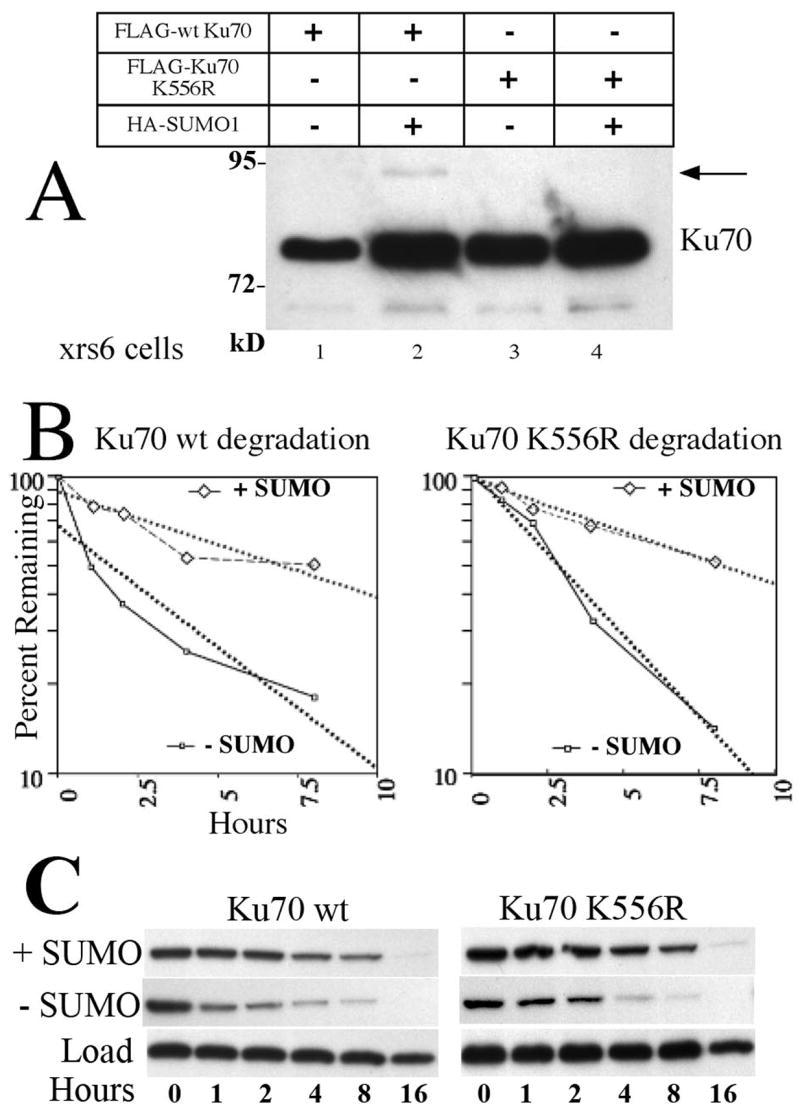
A) FLAG-tagged wild type (wt) Ku70 (lanes 1, 2) increases in abundance when coexpressed with additional SUMO1 in CHO.xrs6 cells. A SUMOylated band is visible (arrow). Mutant K556R (lanes 3, 4) is similarly increased in abundance however no SUMOylated band is visible. Protein was analyzed as in Fig. 1A. B) Degradation time course. CHO.xrs6 cells transfected as in A were treated with cycloheximide to halt further protein synthesis. Subsequent samples taken at intervals were immunoblotted. The disappearance of the Ku70 band is graphed on this semi-log plot and the best fit exponential is fitted (dotted lines). Solid and dashed lines respectively represent Ku70 without or with additional SUMO. Both wild type Ku70 and the K556R mutant display similar kinetics. C) Immunoblot images of the FLAG-tagged proteins depicted in panel B. Samples contain equal total protein.
SUMO1 and SUMO2 produce similar effects
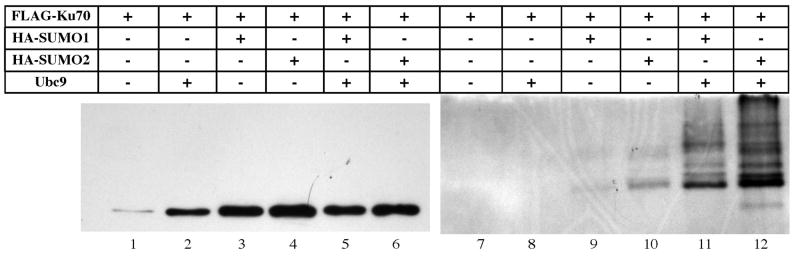
The various SUMO family members can differ in target specificity, so we tested whether SUMO2 would elicit the same response as SUMO1. Fig. 3 indicates that the two differed in the degree of direct SUMOylation of Ku70, but both yielded the same phenomenon with regard to protein stabilization. Lane 1 reveals the low level of Ku70 in CHO.xrs6 cells seen in previous figures, when transiently expressed without additional SUMO. The level of Ku70 increased (lane 2) when Ubc9 was cotransfected, and even more so when SUMO1 was transfected (lane 3). A similar effect, perhaps even to a slightly higher degree, was obtained with a SUMO2 expressing plasmid (lane 4). Simultaneous cotransfection of a SUMO plasmid along with Ubc9 did not have any further effect on the abundance of Ku70 (lanes 5, 6) although the presence of Ubc9 did increase the level of the SUMOylated products (lanes 9–12).
Fig. 3. Ku70 stabilized by SUMO2.
FLAG-Ku70 was transfected alone (lane 1 and 7) or with combinations of Ubc9, SUMO1 or SUMO2 into CHO.xrs6 cells. Each sample was immunoprecipitated with anti-FLAG conjugated agarose beads and immunoblotted. Samples 1–6 were visualized with antibody directed against the FLAG epitope while samples 7–12 were visualized with antibody directed against the HA tag on the SUMO proteins. While SUMO1 and SUMO2 alone (lanes 3 and 4) stabilized Ku70 to a similar degree, the addition of Ubc9 increased the abundance of the SUMOylated form (lanes 11 and 12) without any additional stabilization of Ku70 (lanes 5 and 6).
In an analogous manner, we tested the effect of various SUMO E3 enzymes by cotransfecting FLAG-Ku70 with plasmids expressing each of PIAS1, PIASXβ, and RANBP2. Each of these raised Ku70 levels comparably to the SUMO1 plasmid (Supplemental data).
Immunofluorescence of Human cells responding to SUMO
Ku70 is sufficiently abundant to be visualized directly in human cells. Figure 4 shows a series of epifluorescence images of HEK293T cells variously treated and stained. The otherwise untouched cell line was stained for Ku70 (panel A) producing a uniform, predominantly nuclear, distribution. Cells transfected to express additional SUMO1, and stained only for Ku70 (panel B) now show a mixture of bright staining cells against the background similar to that above. Such a distribution would be expected since transfection does not deliver plasmid to every cell. We chose to label these cells with only one fluorescent antibody to reassure ourselves that the fluorescence observed in the Ku channel could not be explained by leakage of the HA-SUMO signal through the optical color filters. The remaining panels show cells transfected with the HA-SUMO1 expressing plasmid, and doubly stained for Ku70 and the HA epitope. Panels C and E illustrate the distribution of Ku70, while panels D and F show the HA-SUMO1. The correlation within each pair provides compelling evidence that increased expression of SUMO1 leads to a stabilization of endogenous Ku70 in human cells, much as we showed in Fig. 2B. We also note that some cytoplasmic staining of Ku70 can be detected, although there is no clear redistribution in the presence of additional SUMO.
Fig. 4. Immunofluorescence stains of HEK293T cells.
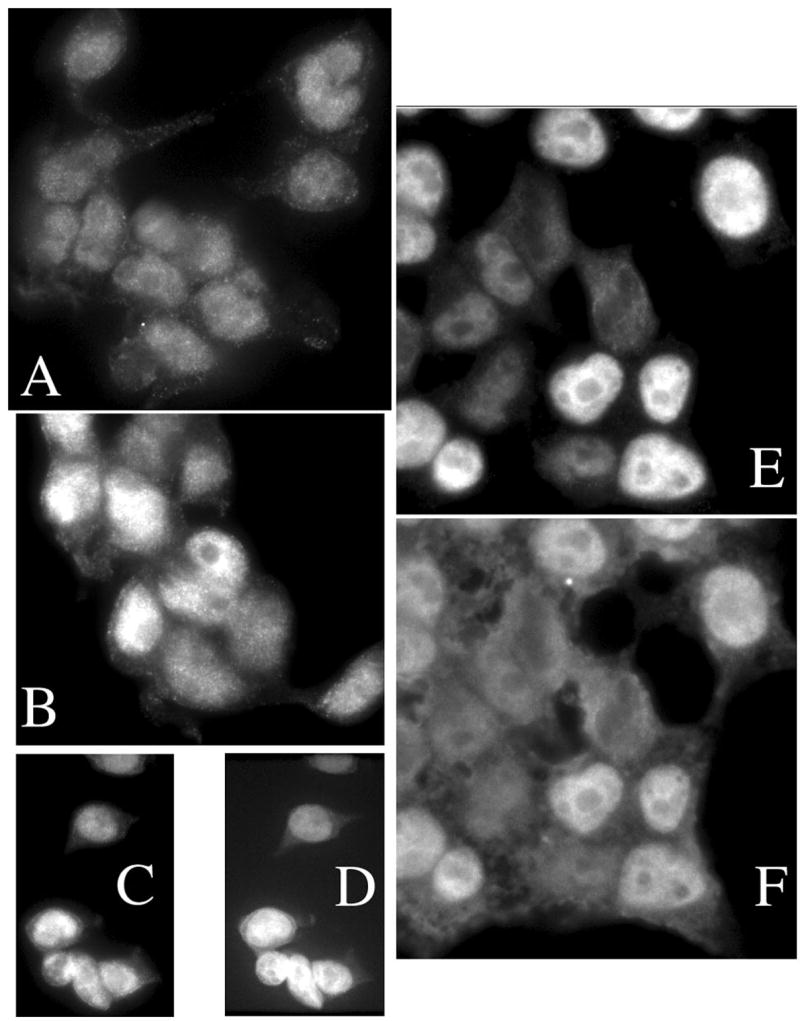
A) Human HEK293T cells were stained for endogenous Ku70 using TRITC conjugated secondary antibody in the absence of transfected SUMO plasmids. A uniform, predominantly nuclear, distribution is seen. B) Cells were transfected to express SUMO1, and stained for Ku70 alone, as above. A mixture of bright staining cells appears against the background similar to A. C–F) Cells were transfected to express SUMO1, stained for Ku70 as above and double stained for HA-SUMO1 using FITC conjugated secondary antibody. Cells bright for Ku70 (C and E) were also bright for HA-SUMO1 (D and F).
Discussion
The data presented here show that Ku70 protein is stabilized, and accumulates to levels severalfold higher than before, in cells manipulated to increase the activity of SUMO. How does this occur and what are the physiologic implications? The simplest pathway would involve the direct SUMOylation of Ku70. Since protein degradation frequently is regulated by ubiquitylation, one can imagine that these two alternative modifications may compete for the same substrate, and that raising the level of SUMO reduces the frequency of ubiquitylation. This would result in a net stabilization. However, such direct competition has not often been detected, and would only stabilize the SUMOylated form of the protein. Our data indicate that the unmodified form accumulates, and that direct modification of Ku70 does not contribute to this stabilization. Only a small fraction of the Ku70 appears to be SUMOylated (e.g. Fig. 1A) and the degree of Ku70 stabilization does not correlate with the degree of modification (Figs. 2 and 3). Rather, indirect mechanisms seem to be responsible. We already appreciate that ubiquitylation is regulated. DNA damage, for example, can activate E3 ligases leading to degradation or stabilization of proteins involved in DNA repair [9]. Recent studies show how networks of modifying proteins can propagate a signal through several steps, modifying the modifiers, in the pathway that leads to stabilization of p53 [15]. SUMO too has already been determined to modify proteins involved in DNA repair [11]. The absolute level of SUMO proteins in a cell is one potential feature that can be regulated. Current evidence suggests that the majority of SUMO1 exists in the conjugated form, while SUMO2 and -3 maintain free pools [16], and this is another aspect that can be regulated. Finally, the cascade of enzymes engaged in SUMOylation may be regulated. Recent evidence indicates that cellular stresses may alter all of these. The proposal that best fits our data with regard to the indirect stabilization of Ku70 is that the degradation rate of the protein is determined by both ubiquitylation and deubiquitylation. SUMOylation may regulate one or more of the enzymes that determine Ku70 degradation. If SUMOylation of an ubiquitylating E3 protein reduces its activity on Ku70, Ku70 will be stabilized. Similarly, activating or stabilizing a deubiquitylating enzyme could also stabilize Ku70. Any protein that is targeted by the same enzymes would be expected to be subject to the same effect, thereby coordinating multiple proteins. Since many proteins may mediate ubiquitylation, deubiquitylation, SUMOylation and deSUMOylation, and these may affect each other, predicting the outcome of any single manipulation may be challenging.
Supplementary Material
Acknowledgments
We are grateful for technical help with microscopy from Dr. Verkhusha lab (AECOM). RANBP2/Nup358 was recloned from a plasmid kindly provided by C. Lima (MSKCC). V.Y. was partially supported by NCI training grant T32 CA09173. M.S. was partially supported by NIH R01 AI 41711. S. M. was partially supported by Pilot Grant P20CA10373 and American Heart Association (AHA) Grant-in-Aid. V. G. is a recipient of AHA pre-doctoral fellowship (0515135B.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gullo C, Au M, Feng G, Teoh G. The biology of Ku and its potential oncogenic role in cancer. Biochim Biophys Acta. 2006;1765(2):223–234. doi: 10.1016/j.bbcan.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85(11):1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–32. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 4.Shi L, et al. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 2007;35(7):2302–2310. doi: 10.1093/nar/gkm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomimatsu N, et al. Ku70/80 modulates ATM and ATR signaling pathways in response to DNA double strand breaks. J Biol Chem. 2007;282(14):10138–10145. doi: 10.1074/jbc.M611880200. [DOI] [PubMed] [Google Scholar]
- 6.Gomez JA, et al. Bax-inhibiting peptides derived from Ku70 and cell-penetrating pentapeptides. Biochem Soc Trans. 2007;35:797–801. doi: 10.1042/BST0350797. [DOI] [PubMed] [Google Scholar]
- 7.Koike M, Koike A. The Ku70-binding site of Ku80 is required for the stabilization of Ku70 in the cytoplasm, for the nuclear translocation of Ku80, and for Ku80-dependent DNA repair. Exp Cell Res. 2005;305(2):266–276. doi: 10.1016/j.yexcr.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 8.Gama V, et al. Involvement of the ubiquitin pathway in decreasing Ku70 levels in response to drug-induced apoptosis. Exp Cell Res. 2006;312(4):488–499. doi: 10.1016/j.yexcr.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Krogan NJ, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16(6):1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 10.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6(8):599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 11.Yurchenko V, Xue Z, Sadofsky MJ. SUMO Modification of human XRCC4 regulates its localization and function in DNA Double-Strand Break repair. Mol Cell Biol. 2006;26(5):1786–1794. doi: 10.1128/MCB.26.5.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yurchenko V, Xue Z, Sadofsky M. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Gen Dev. 2003;17(5):581–585. doi: 10.1101/gad.1058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260(5105):207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276(16):12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 15.Brooks CL, Li M, Gu W. Mechanistic Studies of MDM2-mediated Ubiquitination in p53 Regulation. J Biol Chem. 2007;282(31):22804–22815. doi: 10.1074/jbc.M700961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275(9):6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.