Abstract
Differentiation of B cells into plasma cells requires X-box binding protein–1 (XBP-1). In the absence of XBP-1, B cells develop normally, but very little immunoglobulin is secreted. XBP-1 controls the expression of a large set of genes whose products participate in expansion of the endoplasmic reticulum (ER) and in protein trafficking. We define a new role for XBP-1 in exerting selective translational control over high and sustained levels of immunoglobulin M (IgM) synthesis. XBP-1−/− and XBP-1+/+ primary B cells synthesize IgM at comparable levels at the onset of stimulation with lipopolysaccharide or CpG. However, later there is a profound depression in synthesis of IgM in XBP-1−/− B cells, notwithstanding similar levels of μmRNA. In marked contrast, lack of XBP-1 does not affect synthesis and trafficking of other glycoproteins, or of immunoglobulin light chains. Contrary to expectation, degradation of proteins from the ER, using TCRα or US11-mediated degradation of class I major histocompatibility complex molecules as substrates, is normal in XBP-1−/− B cells. Furthermore, degradation of membrane μ was unaffected by enforced expression of XBP-1. We conclude that in primary B cells, the XBP-1 pathway promotes synthesis and secretion of IgM, but does not seem to be involved in the degradation of ER proteins, including that of μ chains themselves.
X-box binding protein–1 (XBP-1) is required for differentiation of B cells into plasma cells. XBP-1−/− B cells develop normally to maturity, but fail to differentiate into plasma cells (1). XBP-1 is a key component of the unfolded protein response (UPR)—a signaling pathway that emanates from the ER; its activation induces the transcription of a large number of target genes that is believed to be essential for quality control of newly synthesized proteins (2–4).
When proteins emerge into the ER, they undergo posttranslational modifications, such as N-linked glycosylation and disulfide bond formation. These modifications, assisted by several chaperones, are required for proper folding and assembly of newly synthesized ER proteins. When the amount of client proteins exceeds the folding capacity of the ER, a state of ER stress ensues that triggers the UPR (2). The UPR activates corrective measures to alleviate the stress conditions in the ER. In mammalian cells, the UPR is driven by at least three transducers: PKR-like endoplasmic reticulum eIF2α kinase (PERK), activating transcription factor 6 (ATF6), and IRE1 (2). When activated, PERK, a serine/threonine kinase, phosphorylates eIF2α, and so reduces the rate of translation to attenuate the protein load in the secretory system. ATF6, when properly engaged, is cleaved and its cytosolic portion translocates to the nucleus, where it activates transcription of numerous target genes, including XBP-1. IRE1, similarly to PERK, is activated by autotransphosphorylation. Activated IRE1 splices XBP-1 mRNA, using an unconventional mechanism similar to that described for Ire1p/Hac1 in yeast (5). The spliced XBP-1 mRNA gives rise to a polypeptide comprised of the original NH2-terminal DNA binding domain and an additional transactivation domain in the COOH terminus. Spliced XBP-1 (XBP-1s) is a potent transcription factor that activates the expression of ER chaperones and promotes the biogenesis of ER membranes (4, 6–8).
Differentiation into plasma cells involves a remarkable remodeling of the secretory pathway. The ER undergoes massive expansion to accommodate the large quantities of newly synthesized Ig, and to ensure successful assembly of the monomeric Ig subunits into multimeric complexes in preparation for secretion (9, 10). The transition of B cells into plasma cells provokes the UPR, as indicated by XBP-1 mRNA splicing and the up-regulation of ER chaperones (10). Furthermore, ectopic expression in mature B cells of XBP-1s, but not the unspliced form of XBP-1, promotes expansion of the ER, an increase in mitochondrial mass and total organelle content, and an overall increase in cell size (11).
When proteins fail to assemble or fold correctly in the ER, a quality control mechanism identifies these misfits and targets them for degradation. The process of ER degradation often entails dislocation of the substrates from the ER to the cytoplasm, where the misfolded protein is ubiquitinated and destroyed by the proteasome. The UPR in yeast is linked tightly to degradation of ER proteins. Strains deleted for genes encoding Ire1p or Hac1p cannot properly dispose of misfolded ER proteins (12, 13). By analogy with yeast, circumstantial evidence implicates XBP-1 in control of degradation of misfolded ER proteins in mammalian cells (14). Transcriptional profiling analysis that compared WT and XBP-1−/− B cells indicated that XBP-1 exerts control over many genes implicated in the degradation of ER proteins, including ER degradation enhancing α-mannosidase-like protein (EDEM; an ER lectin required for degradation of certain misfolded substrates), E3 ubiquitin ligases, proteasome subunits, and many others (11).
Despite the seemingly crucial role of XBP-1 in setting the proper conditions for the secretory pathway in plasma cells, evidence that directly positions XBP-1 in the ER degradation pathway is missing. Therefore, it is unclear whether XBP-1 is essential for quality control in the ER. In addition, the contribution of XBP-1 to protein trafficking in the secretory pathway of mature and activated B cells is not known.
Here, we investigated the role of XBP-1 in the biosynthesis and secretion of IgM, and in supporting the degradation of misfolded or misassembled ER proteins in primary plasmablasts. We show that XBP-1−/− and XBP-1+/+ B cells synthesize IgM at comparable levels at the onset of mitogenic stimulation with LPS or CpG. However, over a 3-d period of stimulation there is a profound depression of IgM synthesis and secretion in XBP-1−/− B cells, notwithstanding similar μmRNA levels in WT and XBP-1−/− B cells. The lack of XBP-1 does not affect synthesis or trafficking of other glycoproteins or of Ig light chains, nor does it hamper their degradation. We conclude that in primary B cells, the XBP-1 pathway specifically promotes synthesis and secretion of IgM, a new example of translational control, but does not seem to be involved in the degradation of ER proteins.
RESULTS
XBP-1 controls plasmablasts' cell size
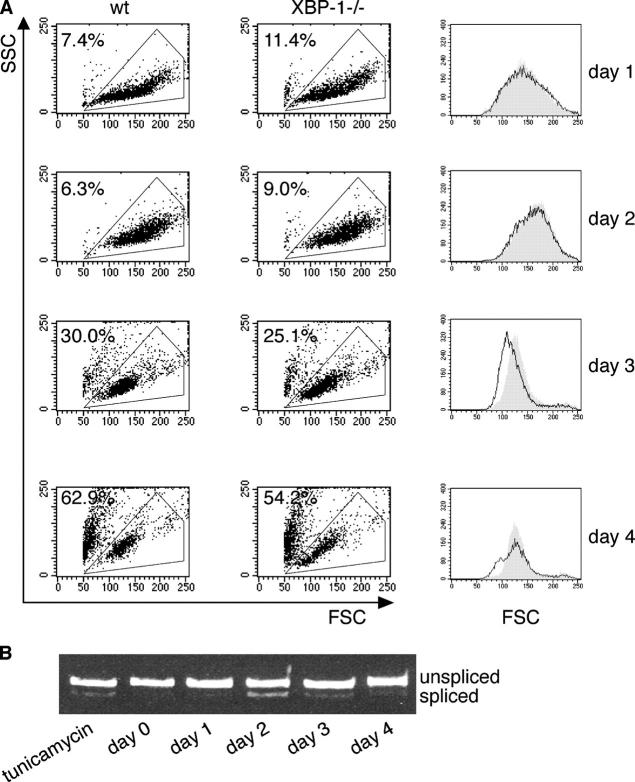
Imposition of expression of XBP-1s in B cell lines, such as RAJI and WEHI231, expands their ER and increases their overall size (11). We investigated whether the absence of XBP-1 would affect the cell size in the course of B cell stimulation by the B cell mitogens, CpG (15), or LPS (16). XBP-1−/− and WT (XBP-1+/+) primary B cells were purified and stimulated with CpG. We saw no significant cell death and no difference in size for the first 48 h of culture (Fig. 1, two top). At day 3, the size of WT cells was larger than that of XBP-1−/−. At day 4, viability of the cells was reduced markedly and decreased to <50% (Fig. 1 A, bottom), a parameter independent of the absence of XBP-1. We analyzed the splicing of XBP-1 mRNA in response to the stimulation of B cells with CpG. Splicing of XBP-1 mRNA was induced at day 2 of CpG stimulation, and continued until termination of the experiment at day 4 (Fig. 1 B). This level of splicing was comparable to the level induced by treatment of naive B cells with 1 μg/ml tunicamycin for 4 h. Similar results were obtained with LPS stimulation, which yielded a larger increase in cell size than did CpG (unpublished data). We conclude that in the course of primary B cell stimulation, XBP-1 splicing promotes the increase in the overall cell size, but does not promote cell survival. Therefore, day 3 of stimulation was selected as the optimal time point for biochemical analyses of the primary plasmablasts, because at this time point, the cells show optimal survival and the effect of XBP-1 deficiency on B cell size is overt.
Figure 1.
XBP-1 controls plasmablasts' cell size. (A) WT or XBP-1−/− B cells were purified from splenocytes by magnetic depletion with anti-CD43. Cells were plated at 106 cells/ml and stimulated with CpG. Flow cytometry analysis was performed every 24 h, and live cells were gated based on their forward and side scattering. Cells were replated at 106 cells/ml density and were analyzed the next day. Line graphs of the gated cells at the forward scatter channel (FSC) are shown in the right column. Gray, WT cells; white, XBP-1−/− cells. The percentage of dead cells is indicated. (B) Cells were stimulated with CpG for 4 d. RNA was extracted at the indicated times, and splicing of XBP-1 mRNA was analyzed by RT-PCR. Tunicamycin treatment (1μg/ml, 4 h) of naive B cells was used as a positive control.
High levels of biosynthesis and secretion of IgM require XBP-1
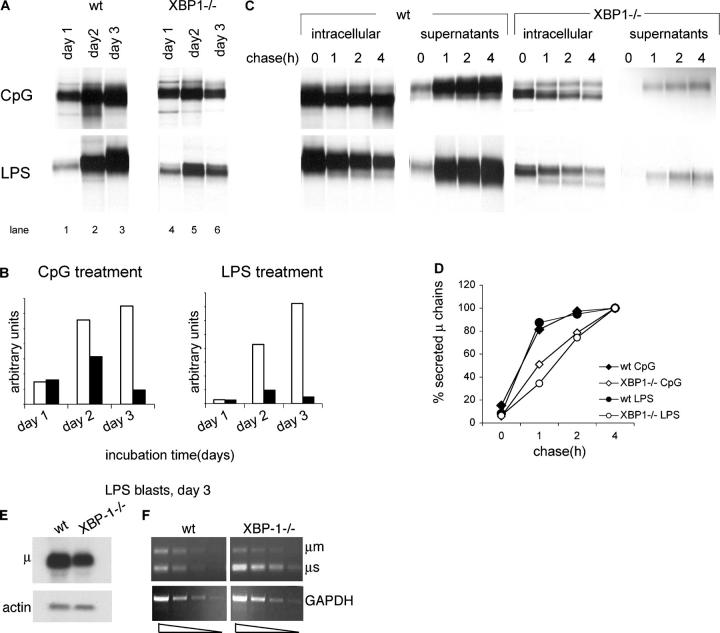
We followed the biosynthesis of IgM in the course of B cell stimulation with LPS or CpG by pulse-chase analysis. The initial level of synthesis of IgM was similar for LPS- or CpG-stimulated B cells. However, at day 3 of stimulation—coincident with the difference in cell size—XBP-1−/− cells showed a steep reduction in the level of IgM synthesis, whereas XBP-1+/+ cells continued to increase their rate of IgM synthesis (Fig. 2 A, compare lane 3 with lane 2 and lane 6 with lane 5). We estimate that in XBP-1−/− B cells the level of synthesis of IgM per cell was reduced by ∼15-fold for LPS stimulation and sevenfold for CpG stimulation by day 3 (Fig. 2 B). The kinetics for IgM secretion were reduced correspondingly in XBP-1−/− cells (Fig. 2 C). Even though ratios of μ chain synthesis differed between CpG and LPS stimulation, secretion rates of IgM were similar for CpG- or LPS-treated XBP-1−/− cells (Fig. 2 D). Hence, XBP-1 was not required for the initial induction of μ synthesis in response to LPS or CpG stimulation, but participates later to support the up-regulation of IgM synthesis and to promote its secretion; these are consistent with the proposed role for XBP-1 in driving the expansion of the ER (11).
Figure 2.
High levels of biosynthesis and secretion of IgM require XBP-1. (A) 106 live cells, as determined by trypan blue exclusion, were pulse-labeled with [35S]methionine for 30 min. Cells were lysed in 1% SDS, and lysate was diluted to 0.07% SDS with NP-40 lysis mix followed by immunoprecipitation with anti-μ antibodies and analysis by SDS-PAGE (10%). (B) Autoradiograms were quantified by phosphoimager. Empty bars, WT B cells; black bars, XBP1−/− B cells. (C) Cells stimulated for 3 d with LPS or CpG were pulse-labeled with [35S]methionine for 30 min and chased for up to 4 h. Cells were lysed in 1% SDS; lysate was diluted to 0.07% SDS with NP-40 lysis mix followed by immunoprecipitation with anti-μ antibodies. At each time point, IgM also was recovered from the media by immunoprecipitated with anti-μ antibodies. Immunoprecipitates were analyzed by SDS-PAGE (10%). Each lane represents material from 106 live cells. (D) Autoradiograms were quantified by phosphoimager. Secreted μ chains were expressed as percentage from μ chains recovered after 4-h chase. (E) RNA was extracted from day 3 LPS-treated WT or XBP-1−/− B cells and analyzed by Northern blotting. Quantification indicates a 2.6-fold reduction in μ mRNA levels in XBP-1−/− cells. (F) Semi-quantitative RT-PCR analysis of cDNA prepared from RNA of day 3 LPS-treated WT or XBP-1−/− B cells. Threefold dilution series of the cDNA were used as input material for the PCR with primers specific for μ chains or GAPDH as a reference. The analysis indicates a less than threefold reduction in μ mRNA levels in XBP-1−/− cells.
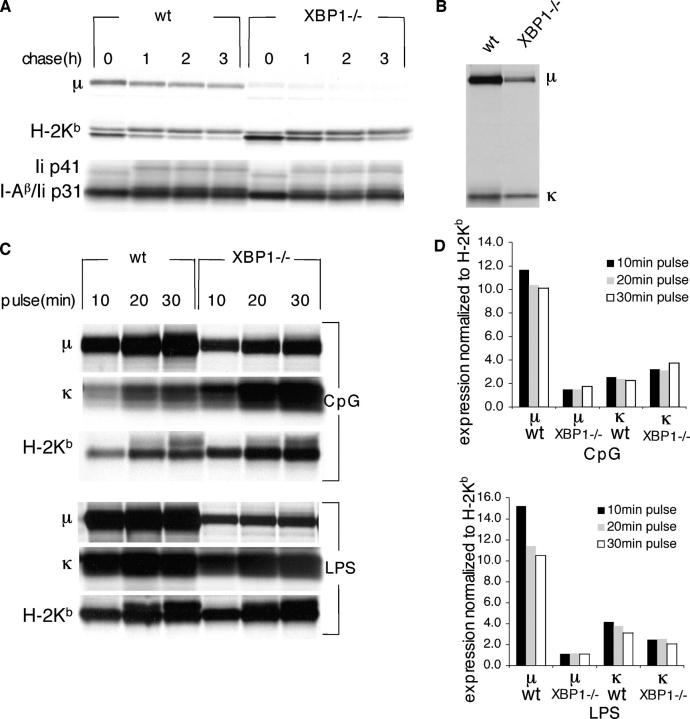
In the course of differentiation into plasma cells, the synthesis of light chains is induced strongly to allow proper assembly with the heavy chains. Therefore, we also measured the effect of XBP-1 deficiency on the induction of synthesis of the κ light chain. B cells were cultured with CpG or LPS for 3 d. Cells (1 × 106) were pulse-labeled for 10, 20, or 30 min. The level of synthesis of μ chains and κ chains were normalized to the synthesis of class I MHC product. Although μ chains were induced in the WT cells by ∼10-15 fold for LPS and by approximately ninefold for CpG stimulation, neither of these treatments resulted in an equivalent induction of the κ light chains (Fig. 3, C and D).
Figure 3.
XBP-1 does not affect the synthesis of κ light chain or synthesis and trafficking of class I and class II MHC. Cells stimulated for 3 d with CpG were pulse-labeled with [35S]methionine for 30 min and chased for up to 3 h. Cells were lysed in lysis buffer containing 0.5% NP-40. Under these conditions, MHC class I and class II complexes are preserved. (A) 90% of input were immunoprecipitated with P8 to recover H-2Kb. Class II I-A were immunoprecipitated with JV2 antibody. Immunoprecipitates were analyzed by SDS-PAGE (10%). Note the nonspecific recovery of μ chains only from WT cells. (B) 10% of input was immunoprecipitated with anti-μ antibodies. (C) Cells were stimulated for 3 d with CpG or LPS. 106 cells were pulse-labeled with [35S]methionine for 10, 20 or 30 min. Cells were lysed in 1% SDS, and the lysate was diluted to 0.07% SDS with NP-40 lysis mix. 70% of lysis mix was immunoprecipitated with P8, 20% was immunoprecipitated with anti-μ, and 10% was immunoprecipitated with anti-κ. Immunoprecipitates were analyzed by SDS-PAGE (12%). (D) Autoradiograms were quantified by phosphoimager. Expression levels of μ and κ were normalized to the corresponding levels of H-2Kb.
XBP-1 may exert control over μ chain synthesis at the level of transcription or by affecting posttranscriptional processes, such as translation or targeting of nascent chains to the ER (17). We compared, by semiquantitative PCR and Northern blot analysis, the levels of μ mRNA extracted from equal numbers of live cells treated for 3 d with LPS. We observed only a slight reduction in μ transcript levels in XBP-1−/− cells, as measured by Northern blotting or by semiquantitative RT-PCR (Fig. 2, E and F). We conclude that XBP-1 controls μ chain synthesis by a mechanism that is largely posttranscriptional.
XBP-1 does not affect the synthesis and trafficking of class I and class II MHC
Transcription profiling conducted for XBP-1 in B cells indicates regulation over a large number of genes whose products are implicated in different aspects of the secretory pathway (4, 11). Therefore, the absence of XBP-1 was predicted to affect the secretory pathway generally, rather than affecting immunoglobulin synthesis and secretion specifically. We examined the biosynthesis and trafficking of class I MHC and class II MHC products to test this prediction. CpG-stimulated B cells were pulse-labeled and MHC products were recovered from them. In contrast to the μ chains (Fig. 3 B), class I MHC heavy chains (HCs) were synthesized at comparable levels in XBP-1−/− and XBP-1+/+ cells (Fig. 3 A, top). Trafficking of HCs, as inferred from their N-linked glycan modifications, was not affected by the absence of XBP-1. We also followed the synthesis, assembly, and trafficking of class II MHC products. Levels of class II MHC synthesis also were similar in XBP-1−/− and XBP-1+/+ cells. Assembly of class II MHC with the invariant chain and maturation of the complex, as assessed by the shift in electrophoretic mobility, were indistinguishable for both cell types (Fig. 3 A, bottom).
We reach the conclusion that in primary B cells, XBP-1 specifically promotes the synthesis, maturation, and secretion of IgM heavy chains, but not the synthesis and maturation of other (glyco)proteins that enter the ER, such as classes I and class II MHC, and Ig light chains. In addition, the trafficking of class I and class II MHC products from the ER to the Golgi complex is not affected by XBP-1. Had XBP-1 been required to set the stage for glycoprotein synthesis generally, then synthesis of IgM and MHC products should be affected equally. Because nascent glycoproteins, regardless of their identity, are held to use the same mechanism to gain access to the ER, the similar levels of synthesis of MHC products argue against a general defect in cotranslational insertion into the ER in the absence of XBP-1, and indicate a role for XBP-1 in translation or ER function that is Ig-specific.
Degradation of ER proteins in plasmablasts does not require XBP-1
XBP-1 controls the expression of several genes that participate in the degradation of ER proteins. EDEM, an ER lectin that is required for the degradation of several misfolded ER proteins, is a specific target of XBP-1 (4). In mouse embryonal fibroblasts that are deficient for XBP-1 or IRE1α, very little, if any, EDEM is present. In these cells, expression of EDEM is restored only when XBP-1 levels are reconstituted. Thus far, EDEM is the only factor in mammalian cells that directly links the UPR to ER degradation (14).
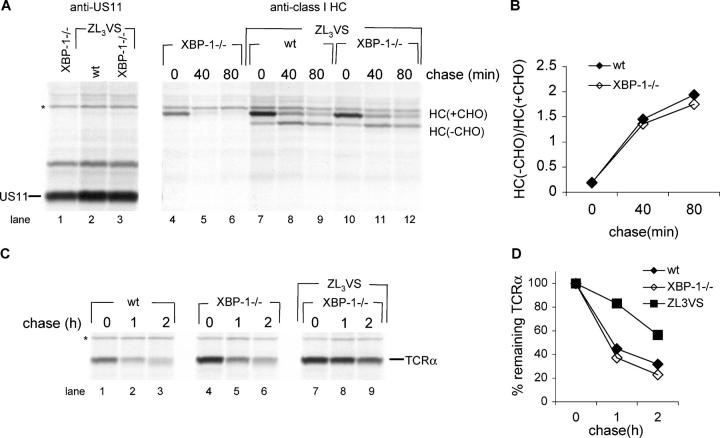
The human cytomegalovirus–encoded glycoprotein US11 targets HCs for dislocation from the ER and degradation by the proteasome. When the proteasome is inhibited, a deglycosylated HC intermediate accumulates in the cytoplasm. This pathway is emblematic of how mammalian cells clear the ER of unwanted proteins. We previously investigated US11-mediated degradation of HC in XBP-1−/− mouse embryonal fibroblasts. Although the presence of XBP-1 was not essential, it did contribute to efficient degradation of class I MHC products (18). Therefore, we tested US11-mediated degradation of HC in XBP-1−/− B cells. To this end, we constructed a bicistronic retroviral vector that expresses US11 and the class I allele HLA-A2. This strategy ensures a fixed ratio between US11 and its cognate class I MHC substrate in the primary B cells that are transduced, regardless of their genotype. Pulse-chase analysis in the presence of the proteasome inhibitor ZL3VS show that the conversion of glycosylated HLA-A2 to the deglycosylated intermediate, as mediated by US11, is similar in XBP-1−/− and in WT B cells (Fig. 4 A, lanes 7–12, quantified in Fig. 4 B). In the absence of proteasome inhibitor, HLA-A2 was degraded rapidly (Fig. 4 A, lanes 4–6). We conclude that in B cells, XBP-1 is not involved in US11-mediated degradation of HCs.
Figure 4.
Degradation of ER proteins in plasmablasts does not require XBP-1. (A) Cells stimulated 24 h days with CpG were transduced with a retrovirus that encodes HLA-A2-IRES-US11 and incubated in the presence of CpG. 2 d after infection, cells were pulse-labeled with [35S]methionine for 20 min and chased for up to 80 min in the presence or absence of the proteasome inhibitor ZL3VS. Cells were lysed in 1% SDS; the lysate was diluted to 0.07% SDS with NP-40 lysis mix followed by immunoprecipitation with anti-class I heavy chain serum (αHC) and analysis by SDS-PAGE (12%). US11 was immunoprecipitated sequentially from the zero time point and similarly analyzed. (B) Autoradiograms were quantified as mentioned above and the deglycosylated HC/glycosylated HC ratio was calculated. (C) Similarly to (A), cells were transduced with a retrovirus that encodes TCRα-IRES-YFP. Cells were pulse-labeled with [35S]methionine for 20 min and chased for up to 2 h in the presence or absence of the proteasome inhibitor ZL3VS. Cells were lysed in 1% SDS; the lysate was diluted to 0.07% SDS with NP-40 lysis mix followed by immunoprecipitation with anti-TCRα and analysis by SDS-PAGE (12%). (D) Autoradiograms were quantified as mentioned above and the relative amount of TCRα to the zero time point was calculated.
TCRα, when expressed in the absence of the TCRβ chain, is misfolded and recognized as such by the ER quality control machinery, and directed to proteasome-mediated degradation (19, 20). We transduced the B cells with a TCRα-encoding retrovirus, and followed their degradation by pulse-chase analysis. As expected, inclusion of the proteasome inhibitor ZL3VS inhibited the degradation of TCRα. Degradation rates of TCRα were similar in XBP-1−/− and XBP-1+/+ B cells (Fig. 4 C, quantified in Fig. 4 D). We conclude that XBP-1 is not involved in the degradation of TCRα.
Membrane μ is an endogenous substrate for degradation in secretory μ (μs)−/− B cell blasts
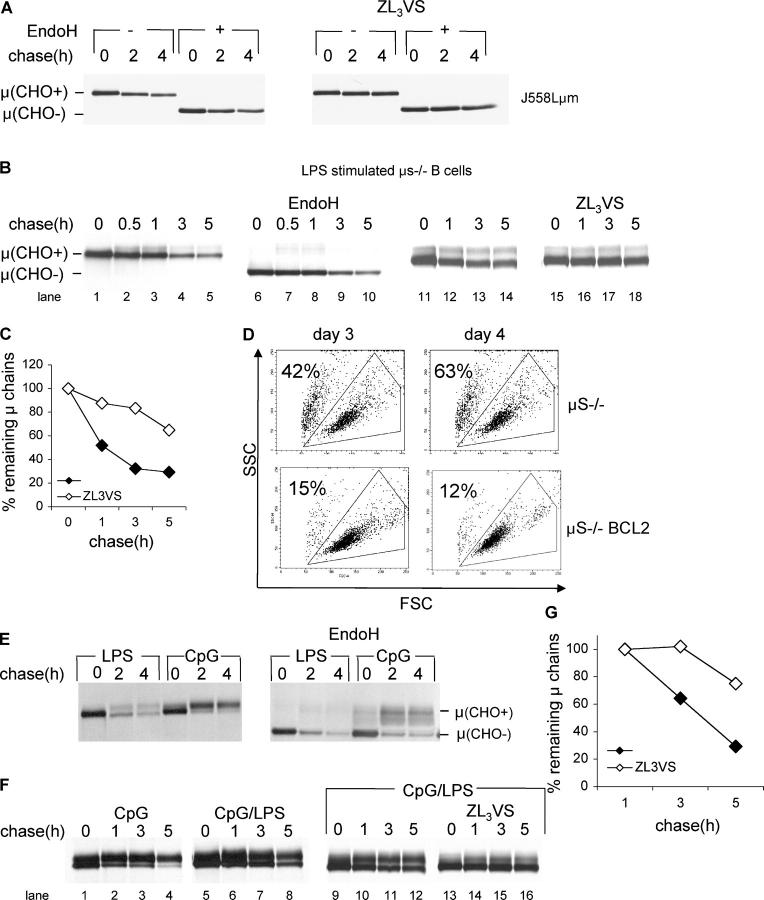
Membrane IgM is presented at the cell surface in a complex with Igα and Igβ (21). Membrane μ chain (μm), in which the authentic transmembrane domain was replaced with that of TCRα, associates normally with the light chain, but undergoes proteasome-mediated degradation (22). Therefore, we assumed that in the absence of Igα or Igβ μm would be retained in the ER and be subjected to dislocation and degradation. For reference purposes, we followed the fate of μm in the Igα-deficient myeloma cell line J558L reconstituted with μm (J558Lμm). As anticipated, μm chains were retained in the ER, as inferred from their EndoH sensitivity, and were degraded by the proteasome, as indicated by their stabilization in the presence of the proteasome inhibitor ZL3VS (Fig. 5 A).
Figure 5.
Membrane μ is an endogenous substrate for degradation in μs−/− B cell blasts. (A) J558Lμm cells were pulse-labeled with [35S]methionine for 30 min and chased for up to 4 h in the presence or absence of ZL3VS. Cells were lysed in 1% SDS; the lysate was diluted to 0.07% SDS with NP-40 lysis mix followed by immunoprecipitation with anti-μ antibodies. Where indicated, immunoprecipitates with EndoH which removes immature N-linked glycans. Immunoprecipitates were analyzed by SDS-PAGE (10%). (B) μs−/− B cells stimulated for 4 d with LPS were pulse-labeled with [35S]methionine for 30 min and chased for up to 5 h. μ-chains were analyzed as above. (C) Autoradiograms were quantified by phosphoimager, and the relative amount of μ to the zero time point was calculated. (D) μs−/− or μs−/− BCL2 B cells were purified from splenocytes by magnetic depletion with anti-CD43. Cells were plated at 106 cells/ml, and stimulated with CpG. Flow cytometry analysis was performed, and live cells were gated based on their forward and side scattering. The percentage of dead cells is indicated. (E) μs−/− BCL2 B cells stimulated for 4 d with LPS or CpG were pulse-labeled with [35S]methionine for 30 min and chased for up to 5 h. μ-chains were analyzed as above. (F) μs−/− BCL2 B cells were stimulated for 4 d with CpG or 2 d with CpG followed by 2 d with LPS (CpG/LPS). Cells were pulse-labeled with [35S]methionine for 30 min and chased for up to 5 h. μ-chains were immunoprecipitated and analyzed as above. (G) Autoradiograms were quantified by phosphoimager, and the relative amount of μ to the 1-h time point was calculated.
Upon mitogenic stimulation of B cells with LPS or CpG, Igα, Igβ, and μm are down-regulated, whereas secretory μ (μs) and J-chain are strongly up-regulated (23, 24). The μs−/− mice, in which the last exon of secreted μ and its poly adenylation sites were deleted, cannot synthesize secretory IgM (25). Therefore, we surmised that μs−/− plasmablasts should reach a stage where, for lack of Igα and Igβ, μm will no longer be assembled correctly, and is subject to ER degradation instead. Pulse-chase experiments conducted at day 4 of LPS stimulation of purified B cells indicate that most of the newly synthesized μm were retained in the ER, followed by proteasome-mediated degradation (Fig. 5 B, quantified in Fig. 5 C). We saw no evidence of class switch recombination to other Ig isotypes as a means of escape from the toxicity of accumulation of unassembled μm. In contrast to J558Lμm, some of the synthesized μm appeared as a more diffuse band of higher apparent molecular weight than the μm seen at time zero. These polypeptides correspond to μ chains that successfully exited the ER and underwent complex-type N-linked glycan modifications in the Golgi complex, as indicated by their EndoH resistance (Fig. 5 B, lanes 7–8).
To investigate a possible role of XBP-1 in the degradation of misassembled μm in primary B cells, we expressed XBP-1s by retroviral transduction in B cells prestimulated for 24 h with CpG. The virus contains an internal ribosome entry site (IRES)-GFP element that allows sorting of GFP-positive cells 24 h after infection. For XBP-1s to exert its effects, we allowed an additional 48 h of expression. Using this protocol, we were left repeatedly with viable cells at numbers too low to allow biochemical analysis. This is probably due to the poor survival of B cells at day 4 after extraction (Fig. 1), and the negative effect of overexpressed XBP-1s on cell viability (unpublished data). Therefore, we reasoned that crossing the μs−/− mice to BCL2 transgenic (BCL2Tg) mice should improve B cell survival, and provide protection from XBP-1s–mediated cell death.
Degradation of μm in μs−/− × BCL2 B cell blasts
We isolated μs−/− × BCL2Tg B cells and stimulated them with LPS or CpG. As expected, the expression of BCL2 protected the B cells from cell death, and resulted in near complete survival after 4 d in culture (Fig. 5 D). We examined the fate of newly synthesized μm by pulse-chase analysis. More μ chains left the ER for the Golgi complex in μs−/− × BCL2Tg B cells than in μs−/− B cells (Fig. 5, B and E, compare top band). The type of stimulation (LPS or CpG) provided to the cells influenced the ratio between immature and mature μ chains, an observation for which we have no satisfactory explanation. In response to CpG stimulation, most of the newly synthesized μ chains acquired EndoH resistance (Fig. 5 E). Regardless of the type of stimulation, the ER-retained μ chains decayed over time, similarly to what was seen for μs−/− B cells and J558Lμm.
We found that stimulation of the μs−/− × BCL2Tg B cells with CpG was a prerequisite to obtain an adequate efficiency of viral transduction. This is probably the result of better induction of proliferation by CpG than by LPS (unpublished data). Therefore, to maximize the amount of ER-retained μ chains available for degradation and to ensure high transduction efficiency, we stimulated B cells with CpG for 48 h, followed by 48 h of stimulation with LPS (Fig. 5 F, lanes 5–8 versus lanes 1–4). Inclusion of proteasome inhibitor stabilized the ER-retained μ chains (Fig. 5 F, lanes 13–16, quantified in Fig. 5 G).
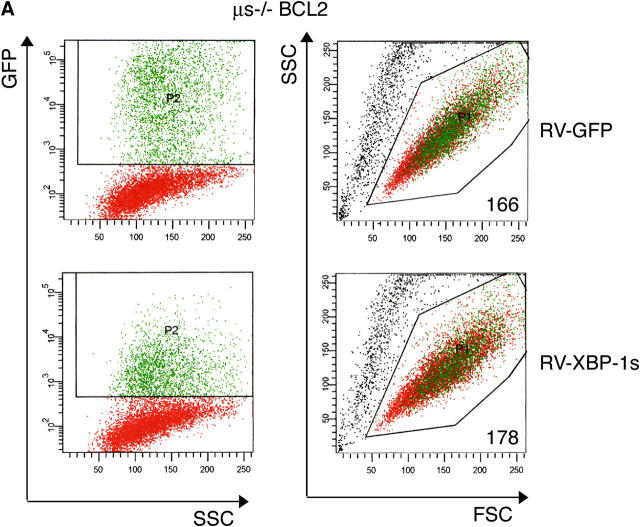
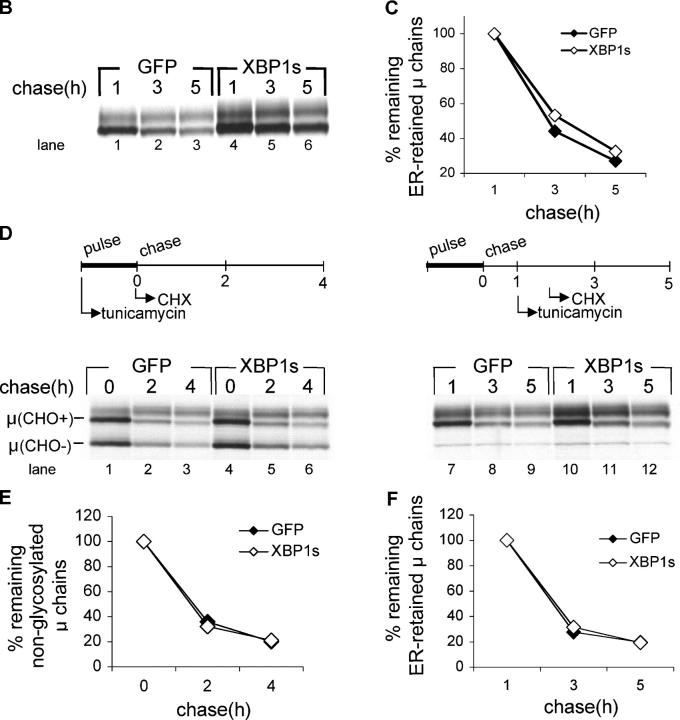
To examine the role of XBP-1 in the degradation of μ, we transduced μs−/− × BCL2Tg B cells with XBP-1s–IRES-GFP or GFP alone. GFP+ cells were sorted ∼18 h after infection (Fig. 6 A, left). At this time, XBP-1s–expressing B cells already showed an increased cell size as indicated by the mean forward scatter of the GFP+ population (Fig. 6 A, right). After 2 d of LPS treatment, equal numbers of live cells were pulse-labeled for 30 min and chased for up to 5 h. The first sample was withdrawn after 1 h of chase—a time point adequate to resolve the ER-retained μ chains from the Golgi-modified chains by SDS-PAGE. Imposition of XBP-1s expression increased the level of μm synthesis when compared with transduction with GFP alone (Fig. 6 B, compare lane 4 with lane 1). However, the rate of μm degradation, as measured relative to the 1-h chase point, was unaffected (Fig. 6 C). We conclude that in accordance with what was seen for secretory IgM, XBP-1s promotes the synthesis of μm, but is not involved in its degradation.
Figure 6.
XBP-1 does not affect degradation of endogenous membrane μ. B cells stimulated for 24 h with CpG and transduced with XBP-1s-IRES-GFP or IRES-GFP encoded retroviruses. 1 d later, the GFP-positive population was sorted, and plated for 2 d in the presence of LPS. (A) Flow cytometry analysis was performed at the time of sorting (∼18 h after infection). GFP-positive B cells are depicted in green. The mean forward scatter of GFP the positive population is indicated. (B) Equal numbers of live cells were pulse-labeled with [35S]methionine for 30 min and chased for up to 5 h. μ-chains were immunoprecipitated and analyzed as above. (C) Autoradiograms were quantified by phosphoimager, and the relative amount of μ to the 1-h time point was calculated. (D) μs −/− BCL2 B cells were stimulated and infected as in (A). Tunicamycin (1 μg/ml) was added during the pulse-labeling period (left), or immediately after removal of the 1-h chase point (right period). Cycloheximide was added at the initiation of the chase (left) or 45 min after tunicamycin addition (right). M-chains were immunoprecipitated and analyzed as above. (E) Autoradiograms were quantified by phosphoimager, and the relative amount of nonglycosylated μ to the zero time point was calculated. (F) Autoradiograms were quantified as above and the relative amount of immature μ to the 1-h time point was calculated.
Enforced expression of XBP-1s does not affect μm turnover
Finally, we examined whether overexpression of XBP-1s would improve the capacity of the primary B cells to handle unphysiologic levels of misfolded proteins. To this end, we designed a pulse-chase experiment that incorporates 45 min of tunicamycin treatment, as a means to generate a pool of nonglycosylated, unfolded proteins in the ER. This was followed by the addition of cycloheximide to block the possibility of de novo synthesis of endogenous XBP-1 in the μs−/− BCL2 cells. Under these conditions, ∼50% of newly synthesized μ is nonglycosylated (Fig. 6 D, lanes 1 and 4); this value indicates the efficacy of tunicamycin treatment. Hence, in these tunicamycin-treated cells, the ER should contain a far greater quantity of misfolded protein than what is present in control cells. We measured the turnover of nonglycosylated μ when tunicamycin was added during the pulse and the entire chase. We found no difference between XBP-1s–transduced cells and GFP controls (Fig. 6 E). Next, we tested the effect of tunicamycin treatment in the course of the chase on the degradation of ER-retained fully glycosylated μ. This protocol was applied to explore whether the turnover of preexisting μ chains would be affected if a UPR were induced later. Again, kinetics of turnover were highly comparable for XBP-1s–transduced cells and GFP controls (Fig. 6 F). μm degradation was affected minimally by tunicamycin treatment (compare Fig. 6, C and F), which suggested that the ER degradation machinery in B cells was not saturated. We conclude that XBP-1 is not required for the degradation of μ chains, even under conditions of nonphysiologic levels of misfolded proteins.
DISCUSSION
The IRE1/XBP-1 arm of the mammalian UPR was suggested to coordinate the protein folding capacity and the degradation machinery of the ER (14). In yeast, the UPR is a key regulator of the protein folding capacity of the ER and also of the ER degradation machinery. When engaged in response to ER stress, the UPR augments the folding capacity of the ER and simultaneously ensures the degradation of misfolded ER proteins (12, 13). Augmentation of the protein-folding capacity in the ER and facilitation of the degradation of misfolded ER cooperate to reduce the levels of misfolded proteins in the ER. Therefore, these two processes are intimately linked and share important molecular components. For example, the ER-resident protein disulfide isomerase assists in protein folding by catalyzing disulfide bond formation, and is reported to participate in ER retention and dislocation of misfolded proteins (26); Bip and its yeast equivalent Kar2p are chaperones necessary for protein translocation into the ER and protein folding, and are also key elements in the process of ER quality control and ER degradation (27).
Thus far, the knowledge obtained for the biologic activity of XBP-1 is based mostly on experiments that provoke extreme nonphysiologic conditions, such as treatment with reagents that massively disrupt protein folding (tunicamycin), or overexpression of XBP-1s. These studies ascribed different roles to XBP-1, including the biogenesis of ER membrane (8), up-regulation of protein folding in the ER, an increase in lysosome and mitochondria content, and playing a central role in the degradation of ER proteins (11, 14). The mere presence of a misfolded protein fails to induce the UPR to the extent seen when tunicamycin is used (12). To our knowledge, there are very few data on glycoprotein quality control in cultures of primary cells, and most evidence has been gathered for permanently established cell lines. We chose differentiating B cells as the model with which to explore this issue for primary nontransformed cells.
We examined the role of XBP-1 under physiologic induction of the UPR, such as differentiation of B lymphocytes into plasma cells (28, 29). Examination of μ chain biosynthesis showed that XBP-1 was not required for the induction of IgM synthesis in response to CpG or LPS stimulation—a response probably linked to the activation of NF-κB through Toll-like receptors—or for the engagement of other signaling pathways. At later times, XBP-1 was required to sustain high levels of IgM synthesis because in the absence of XBP-1, synthesis and secretion of μ chains were attenuated over 3 d of stimulation (Fig. 2); these suggested possible XBP-1 control over assembly and trafficking of IgM. The assembly of IgM monomers into pentamers was normal in XBP-1−/− B cells (unpublished data).
In response to stimulation of splenic B cells with LPS and CpG, splicing of XBP-1 mRNA occurs after 48 h. Using a conditional allele of μ, excision of the μ locus strongly reduced XBP-1 splicing (28). Therefore, XBP-1 splicing requires the expression of μ heavy chains. Furthermore, proteomic analysis of the B cell line, i.29μ+ subjected to LPS stimulation, revealed detectable levels of XBP-1s only after induction of IgM synthesis (9). Accumulation of μ chains in the ER is the major trigger of IRE1 activation and subsequent XBP-1 mRNA splicing in B cells. In contrast, analysis of the time course of IgM synthesis and XBP-1 splicing in the B cell line, CH12 stimulated with LPS, indicated that splicing of XBP-1 preceded IgM up-regulation (29). Our results show that XBP-1 is not required for the initial induction of IgM synthesis, but does plays a role later in sustaining high levels of IgM synthesis. IgM synthesis and XBP-1 activation are part of a positive feedback loop in primary B cells in the course of differentiation into plasma cells.
Reimold et al. reported minor changes in transcription of the IgH locus after 4 d of LPS treatment of XBP1-1−/− and XBP-1+/+ B cells, as measured by RT-PCR (1). Gene chip analysis that compared the transcriptome of XBP-1−/− B cells with WT controls in response to LPS showed increased levels of μ transcripts in the WT cells (11). To resolve these differences, we analyzed the μ mRNA levels, and found no more than a twofold reduction of μ transcripts in XBP-1−/− cells (Fig. 2, E and F). This moderate reduction in mRNA levels cannot account for the dramatic reduction in μ synthesis in XBP-1−/− B cells (Fig. 2 B). Our data also are in agreement with the observation that transduction of splenic B cells with a dominant negative allele of ATF6 attenuates IgM secretion with minimal effect on μ transcription (30). Combined, these results suggest that posttranscriptional mechanisms control the rate of IgM synthesis in an XBP-1–dependent manner. The mechanism by which XBP-1 controls IgM synthesis does not result in general modulation of translation and insertion into the ER, because genes introduced by retroviral transduction were synthesized at comparable levels, and the endogenous immunoglobulin light chains, class I MHC, and class II MHC products continued to be synthesized and transported intracellularly at similar rates in both genetic backgrounds (Figs. 3 and 4). These findings rule out the phosphorylation of eIF2α by PERK as a plausible explanation for IgM down-regulation in the XBP-1−/− B cells, because this phosphorylation results in general attenuation of translation initiation.
Cotranslational insertion of nascent proteins into the ER is a process that involves several steps (for review see reference 31). Upon emergence from the ribosome, the signal peptide is recognized instantly by the signal recognition particle (SRP). The SRP-bound signal peptide interacts with the SRP receptor at the cytosolic face of the ER membrane. At the ER, the signal peptide is transferred—by a poorly understood mechanism—into the Sec61 translocon, the entry channel into the ER (32). The fidelity with which proteins enter the ER varies greatly between different signal peptides and between different cell types (17). Therefore, it is possible that XBP-1 in B cells controls specifically the efficiency by which nascent μ chains are targeted into the ER. By avoiding general inhibition of translation, B cells ensure a continuous increase in the flux of μ chains that enter the ER. The accumulation of μ chains in the ER lumen promotes the splicing of XBP-1. Although speculative, this suggestion is supported by fact that XBP-1 in B cells promotes the transcription of several genes whose products participate in cotranslational insertion of proteins into the ER, such as TRAM and components of SRP and Sec61 themselves (4, 11, 33).
Transcriptional profiling of B cell indicates control by XBP-1 over components of the ER degradation pathway. In response to LPS, B cells up-regulate several genes whose products are implicated in ER quality control, such as EDEM and the mouse homologues of the yeast Hrd1 and Hrd3. This response is attenuated severely in XBP-1−/− cells (11). We have obtained compelling evidence for the involvement of human Hrd1 and Hrd3 in the US11-dependent pathway of class I MHC turnover, through direct physical interaction with Derlin-1 (Lilley, B.N. & Ploegh, H.L., unpublished data). However, it is not known whether XBP-1 contributes to the degradation of misfolded ER proteins. Thus, we examined the capacity of XBP-1−/− B cells to degrade two well-characterized substrates for ER degradation. The human CMV–encoded glycoprotein US11 reduces the half-life of class I MHC HCs from hours to minutes by catalyzing their dislocation. When we expressed HLA-A2 and US11 in primary B cells, we observed comparable and robust dislocation of the class I MHC HC in XBP-1−/− and XBP-1+/+ B cells (Fig. 4). B cells are equipped in a manner that does not require XBP-1 with the molecular machinery, such as Derlin-1 and associated factors, to degrade HCs.
Similar results were obtained when we examined the degradation of TCRα. When expressed in primary B cells, TCRα was degraded efficiently at rates superior to those measured in COS-1 cells (19, 20) or U373 cells (34). This degradation machinery operates independently of XBP-1 status (Fig. 4). We conclude that XBP-1 does not play an important role in the degradation of ER proteins in B cells. The UPR paradigm, as invoked for clearing the ER of unwanted proteins, may not apply to B cells. When the UPR is induced by stimulation of CH12 cells with LPS, XBP-1 is spliced; ER chaperones are induced; but the proapoptotic gene CHOP, a UPR indicator, is not activated (29). The UPR in B cells might be unusual, in that it supports induction of genes that facilitate IgM synthesis and secretion, but fails to induce apoptosis.
Overexpression of XBP-1s in B cell lines induces the expression of genes that participate in protein translocation into the ER, protein folding in the ER, and trafficking in the secretory pathway (4, 11). Expression of XBP-1s in Raji cells also yielded an increase in protein translation and a marked decrease in total degradation of glycosylated proteins; these probably are attributable to an increase in the folding capacity of the ER (11). Because μm is recognized for degradation due to a failure to assemble with Igα and Igβ rather than owing to the misfolding of μ chains themselves, an increase in the folding capacity of the ER as induced by XBP-1s should not rescue μm from degradation. Therefore, we hypothesized that enforced expression of XBP-1s should facilitate the degradation of μm if XBP-1 is involved in this degradation pathway. Expression of XBP-1s increases the synthesis of μm by 20–70%, which supports our previous results that XBP-1 promotes IgM synthesis. Nonetheless, the kinetics of degradation of μm remained unaltered (Fig. 6 C). When we imposed a UPR by treatment with tunicamycin, enforced expression of XBP-1s did not affect the rate of degradation of μm chains (Fig. 6, E and F). Overall, our data indicate a minor role, if any, for XBP-1 in ER quality control in primary B cell blasts.
Why do XBP-1−/− B cells fail to yield plasma cells in vivo? Two alternative mechanisms may apply. Either XBP-1−/− B cells do not differentiate into plasma cells or XBP-1−/− B plasma cells fail to survive. Because XBP-1 has been postulated to be involved in the control of degradation of protein in the ER, a failure of XBP-1−/− B cells to handle those substrates could render them susceptible to apoptosis. Our data argue against this hypothesis and indicate that, in contrast to other cell types, primary plasmablasts are equipped with highly efficient ER degradation machinery that operates independently of XBP-1.
MATERIALS AND METHODS
Mice.
We obtained 129S6 mice from Taconic. BCL2 Tg mice were provided by K. Rajewsky (Harvard Medical School, Boston, MA). μs−/− and μs−/− × BCL2 Tg mice were used at 6–8 wk and maintained in the pathogen-free facilities in accordance with the guidelines of the Committee on Animals of Harvard Medical School.
Cell culture and cell lines.
Mature B cells were purified from mouse splenocytes by magnetic depletion with anti-CD43 (Miltenyi Biotech). Cells were plated at 106 cells/ml in complete medium containing RPMI 1640 supplemented with 10% FBS (Hyclone Laboratories), 2 mM glutamine, 50 U/ml penicillin, 50 μg/ml of streptomycin, 50 μM of β-ME, 25 mM Hepes, 1× nonessential amino acids, and 1 mM sodium pyruvate. Stimulation was performed with 20 μg/ml LPS (Sigma-Aldrich) or 100 nM of CpG (1826-CPG, TIB Molbiol). J558Lμm were provided by J. Haimovich (Tel-Aviv University, Tel Aviv, Israel) and maintained in the same medium.
Metabolic labeling, pulse-chase analysis, and immunoprecipitation.
After starvation in methionine/cysteine-free Dulbecco's modified Eagle's medium for 45 min, cells were labeled metabolically with 500 μCi/ml of [35S]methionine/cysteine (1,200 Ci/mmol, PerkinElmer) at 37°C for the times indicated. Pulse-chase experiments, cell lysis, and immunoprecipitation were performed as described previously (35). The immunoprecipitates were analyzed by SDS-PAGE followed by fluorography. Densitometry was performed by phosphorimager using Imagequant 1.0 software (Molecular Dynamics). Antibodies against HCs, US11, and anti–β-chain of class II MHC have been described (36, 37). Goat anti–mouse I was purchased from Southern Biotechnology. All of the experiments were conducted at least three times with similar outcomes. Each figure shows a representative experiment with the corresponding quantitation.
Northern hybridization and semi-quantitative RT-PCR.
Total RNA was isolated using TriZol (GIBCO BRL) or Qiashedder/Rneasy RNA purification columns (QIAGEN). Northern blotting was performed as described (28). In brief, 7–10 μg RNA were electrophoresed on 1.2% agarose, 6% formaldehyde gels transferred onto Genescreen membrane (NEN) and covalently bound to the membrane using UV Stratalinker (Stratagene). Probe for Cmu (CmuXB) was prepared as described (38). Hybridization was performed with Ultrahyb buffer as recommended by the manufacturer (Ambion). Total RNAs were used for first-strand synthesis with Superscript reverse-transcriptase (Invitrogen). IgM PCR primers were used as described (1). Three-fold dilutions of total cDNA were used for PCR amplification, performed with AmpliTaq Gold polymerase (Applied Biosystem). PCR products were electrophoresed on a 1% agarose gel and visualized by ethidium bromide staining. Primers and probe were provided by J. Manis (Children's Hospital, Boston, MA).
RT-PCR analysis.
Total RNA was isolated using TriZol (GIBCO BRL). RNAs were used for first-strand synthesis with the Superscript reverse transcriptase (Invitrogen). PCR primers 5′-ACACGCTTGGGAATGGACAC-3′ and 5′-CCATGGGAAGATGTTATGGG-3′, encompassing the missing sequences in XBP-1, were used for the PCR amplification with Platinum PCR Supermix (Invitrogen). Cycling conditions were as follows: 95°C for 3 min and 58°C for 40 s, 35 cycles of 72°C for 45 s, and 95°C for 45 s. A PCR for GAPDH was performed to validate cDNA synthesis. We separated PCR products by electrophoresis in 11% PAGE gel and visualized them by ethidium bromide staining.
Retroviral transduction of B cells.
The XBP-1s vector has been described (28). cDNAs encoding TCRα were inserted into the GFP-RV retroviral vector using double blunt ligation. HLA-A2-IRES-US11 was cloned into the RV retroviral vector. We used Effectene transfection (QIAGEN) to cointroduce the retroviral DNA and packaging vectors ENV and Gag-Pol into 293T HEK cells in a 2:1:1 ratio. Viral supernatants were collected after 72 h and frozen at −80°C for later use. Purified B cells were stimulated for 24 h with CpG and spin-infected with virus-containing supernatants supplemented with 4 μg/ml polybrene. Cells were incubated for 24 h, washed, and replated at 106 cells/ml. Where indicated, GFP-positive cells were sorted by flow cytometry. Pulse-chase experiments were performed 72 h after stimulation or 4 d after stimulation for the BCL2 transgenic cells.
Acknowledgments
B. Tirosh is supported by a Dorot Foundation fellowship. N. Iwakoshi is supported by an Irvington Institute Postdoctoral Fellowship Award. This study was supported by National Institutes of Health grant nos. AI32412 (to L.H. Glimcher) and 5R37-AI33456 (to H.L. Ploegh).
L.H. Glimcher holds equity in Mannkind Corporation, which has licensed the XBP-1 technology. The authors have no other potential conflicting financial interests.
The authors have no conflicting financial interests.
Abbreviations used: μm, membrane μ; μs, secretory μ; ATF, activating transcription factor; EDEM, ER degradation enhancing α-mannosidase-like protein; HC, class I MHC heavy chain; IRES, internal ribosome entry site; PERK, PKR-like endoplasmic reticulum eIF2α kinase; SRP, signal recognition particle; UPR, unfolded protein response; XBP-1, X-box binding protein 1; XBP-1s, spliced XBP-1.
References
- 1.Reimold, A.M., N.N. Iwakoshi, J. Manis, P. Vallabhajosyula, E. Szomolanyi-Tsuda, E.M. Gravallese, D. Friend, M.J. Grusby, F. Alt, and L.H. Glimcher. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 412:300–307. [DOI] [PubMed] [Google Scholar]
- 2.Harding, H.P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575–599. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman, R.J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211–1233. [DOI] [PubMed] [Google Scholar]
- 4.Lee, A.H., N.N. Iwakoshi, and L.H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sidrauski, C., and P. Walter. 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 90:1031–1039. [DOI] [PubMed] [Google Scholar]
- 6.Calfon, M., H. Zeng, F. Urano, J.H. Till, S.R. Hubbard, H.P. Harding, S.G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96. [DOI] [PubMed] [Google Scholar]
- 7.Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R.J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sriburi, R., S. Jackowski, K. Mori, and J.W. Brewer. 2004. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Anken, E., E.P. Romijn, C. Maggioni, A. Mezghrani, R. Sitia, I. Braakman, and A.J. Heck. 2003. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 18:243–253. [DOI] [PubMed] [Google Scholar]
- 10.Iwakoshi, N.N., A.H. Lee, P. Vallabhajosyula, K.L. Otipoby, K. Rajewsky, and L.H. Glimcher. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321–329. [DOI] [PubMed] [Google Scholar]
- 11.Shaffer, A.L., M. Shapiro-Shelef, N.N. Iwakoshi, A.H. Lee, S.B. Qian, H. Zhao, X. Yu, L. Yang, B.K. Tan, A. Rosenwald, et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. [DOI] [PubMed] [Google Scholar]
- 12.Casagrande, R., P. Stern, M. Diehn, C. Shamu, M. Osario, M. Zuniga, P.O. Brown, and H. Ploegh. 2000. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell. 5:729–735. [DOI] [PubMed] [Google Scholar]
- 13.Travers, K.J., C.K. Patil, L. Wodicka, D.J. Lockhart, J.S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida, H., T. Matsui, N. Hosokawa, R.J. Kaufman, K. Nagata, and K. Mori. 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 4:265–271. [DOI] [PubMed] [Google Scholar]
- 15.Krieg, A.M., A.K. Yi, S. Matson, T.J. Waldschmidt, G.A. Bishop, R. Teasdale, G.A. Koretzky, and D.M. Klinman. 1995. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 374:546–549. [DOI] [PubMed] [Google Scholar]
- 16.Stevens, R.H., B.A. Askonas, and J.L. Welstead. 1975. Immunoglobulin heavy chain mRNA in mitogen-stimulated B cells. Eur. J. Immunol. 5:47–53. [DOI] [PubMed] [Google Scholar]
- 17.Levine, C.G., D. Mitra, A. Sharma, C.L. Smith, and R.S. Hegde. 2005. The efficiency of protein compartmentalization into the secretory pathway. Mol. Biol. Cell. 16:279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tirosh, B., N.N. Iwakoshi, B.N. Lilley, A.H. Lee, L.H. Glimcher, and H.L. Ploegh. 2005. Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J. Virol. 79:2768–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huppa, J.B., and H.L. Ploegh. 1997. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 7:113–122. [DOI] [PubMed] [Google Scholar]
- 20.Yu, H., G. Kaung, S. Kobayashi, and R.R. Kopito. 1997. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J. Biol. Chem. 272:20800–20804. [DOI] [PubMed] [Google Scholar]
- 21.Hombach, J., T. Tsubata, L. Leclercq, H. Stappert, and M. Reth. 1990. Molecular components of the B-cell antigen receptor complex of the IgM class. Nature. 343:760–762. [DOI] [PubMed] [Google Scholar]
- 22.Fagioli, C., A. Mezghrani, and R. Sitia. 2001. Reduction of interchain disulfide bonds precedes the dislocation of Ig-mu chains from the endoplasmic reticulum to the cytosol for proteasomal degradation. J. Biol. Chem. 276:40962–40967. [DOI] [PubMed] [Google Scholar]
- 23.Calame, K.L., K.I. Lin, and C. Tunyaplin. 2003. Regulatory mechanisms that determine the development and function of plasma cells. Annu. Rev. Immunol. 21:205–230. [DOI] [PubMed] [Google Scholar]
- 24.Sakaguchi, N., S. Kashiwamura, M. Kimoto, P. Thalmann, and F. Melchers. 1988. B lymphocyte lineage-restricted expression of mb-1, a gene with CD3-like structural properties. EMBO J. 7:3457–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boes, M., C. Esau, M.B. Fischer, T. Schmidt, M. Carroll, and J. Chen. 1998. Enhanced B-1 cell development, but impaired IgG antibody responses in mice deficient in secreted IgM. J. Immunol. 160:4776–4787. [PubMed] [Google Scholar]
- 26.Tsai, B., Y. Ye, and T.A. Rapoport. 2002. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 3:246–255. [DOI] [PubMed] [Google Scholar]
- 27.Qiu, W., R. Kohen-Avramoglu, S. Mhapsekar, J. Tsai, R.C. Austin, and K. Adeli. 2005. Glucosamine-induced endoplasmic reticulum stress promotes ApoB100 degradation: evidence for Grp78-mediated targeting to proteasomal degradation. Arterioscler Thromb Vasc Biol. 25:571–577. [DOI] [PubMed] [Google Scholar]
- 28.Iwakoshi, N.N., A.H. Lee, and L.H. Glimcher. 2003. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 194:29–38. [DOI] [PubMed] [Google Scholar]
- 29.Gass, J.N., N.M. Gifford, and J.W. Brewer. 2002. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277:49047–49054. [DOI] [PubMed] [Google Scholar]
- 30.Gunn, K.E., N.M. Gifford, K. Mori, and J.W. Brewer. 2004. A role for the unfolded protein response in optimizing antibody secretion. Mol. Immunol. 41:919–927. [DOI] [PubMed] [Google Scholar]
- 31.Walter, P., and A.E. Johnson. 1994. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10:87–119. [DOI] [PubMed] [Google Scholar]
- 32.Song, W., D. Raden, E.C. Mandon, and R. Gilmore. 2000. Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 100:333–343. [DOI] [PubMed] [Google Scholar]
- 33.Voigt, S., B. Jungnickel, E. Hartmann, and T.A. Rapoport. 1996. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 134:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misaghi, S., M.E. Pacold, D. Blom, H.L. Ploegh, and G.A. Korbel. 2004. Using a small molecule inhibitor of peptide: N-glycanase to probe its role in glycoprotein turnover. Chem. Biol. 11:1677–1687. [DOI] [PubMed] [Google Scholar]
- 35.Rehm, A., P. Stern, H.L. Ploegh, and D. Tortorella. 2001. Signal peptide cleavage of a type I membrane protein, HCMV US11, is dependent on its membrane anchor. EMBO J. 20:1573–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tortorella, D., C.M. Story, J.B. Huppa, E.J. Wiertz, T.R. Jones, I. Bacik, J.R. Bennink, J.W. Yewdell, and H.L. Ploegh. 1998. Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol. 142:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villadangos, J.A., R.A. Bryant, J. Deussing, C. Driessen, A.M. Lennon-Dumenil, R.J. Riese, W. Roth, P. Saftig, G.P. Shi, H.A. Chapman, et al. 1999. Proteases involved in MHC class II antigen presentation. Immunol. Rev. 172:109–120. [DOI] [PubMed] [Google Scholar]
- 38.Kuzin, I.I., G.D. Ugine, D. Wu, F. Young, J. Chen, and A. Bottaro. 2000. Normal isotype switching in B cells lacking the I mu exon splice donor site: evidence for multiple I mu-like germline transcripts. J. Immunol. 164:1451–1457. [DOI] [PubMed] [Google Scholar]