Abstract
Murine Tcrd and Tcra gene segments reside in a single genetic locus and undergo recombination in CD4−CD8− (double negative [DN]) and CD4+CD8+ (double positive [DP]) thymocytes, respectively. TcraTcrd locus variable gene segments are subject to complex regulation. Only a small subset of ∼100 variable gene segments contributes substantially to the adult TCRδ repertoire. Moreover, although most contribute to the TCRα repertoire, variable gene segments that are Jα proximal are preferentially used during primary Tcra recombination. We investigate the role of local chromatin accessibility in determining the developmental pattern of TcraTcrd locus variable gene segment recombination. We find variable gene segments to be heterogeneous with respect to acetylation of histones H3 and H4. Those that dominate the adult TCRδ repertoire are hyperacetylated in DN thymocytes, independent of their position in the locus. Moreover, proximal variable gene segments show dramatic increases in histone acetylation and germline transcription in DP thymocytes, a result of super long-distance regulation by the Tcra enhancer. Our results imply that differences in chromatin accessibility contribute to biases in TcraTcrd locus variable gene segment recombination in DN and DP thymocytes and extend the distance over which the Tcra enhancer can regulate chromatin structure to a remarkable 525 kb.
V(D)J recombination is essential for TCR (Tcra, Tcrb, Tcrg, and Tcrd) gene assembly and for the generation of diverse receptor repertoires on αβ and γδ T lymphocytes (1). A striking feature of Tcra and Tcrd genes is their unique organization, with Tcrd gene segments nested among Tcra gene segments within the complex TcraTcrd locus (2). The murine TcraTcrd locus spans 1.6 megabases (mb) and at its 3′ end includes Dδ, Jδ, and Cδ gene segments, a large array of Jα segments, and Cα. However, the largest portion of the locus (1.5 mb) is devoted to an array of ∼100 V gene segments. Most of the V gene segments are contained within two highly homologous 400-kb segments of DNA resulting from a recent duplication event. Smaller numbers of unique V gene segments flank the duplicated region on each end. A single V segment is located downstream of Cδ in an inverted orientation.
During T lymphocyte development, Tcrd rearrangement occurs in CD4−CD8− double negative (DN) thymocytes, whereas Tcra rearrangement occurs in CD4+CD8+ double positive (DP) thymocytes. These rearrangements are controlled by two developmental stage–specific enhancers: the Tcrd enhancer (Eδ) and the Tcra enhancer (Eα; reference 2). Eδ is thought to regulate Tcrd rearrangement through relatively short-range control of Dδ and Jδ chromatin (3, 4). Eα regulates Tcra rearrangement through long-distance control of chromatin across the 70-kb Jα array and is also known to stimulate transcription of rearranged Tcrd genes at a distance of 100 kb (5, 6).
Little is understood about the regulation of TcraTcrd locus V gene segment usage. Although the V gene segments should theoretically rearrange to either Dδ or Jα, only a subset of V segments dominate the Vδ repertoire. At least 16 V gene segments have been classified as Vδ (7, 8). However, analyses of Tcrd rearrangements in γδ hybridoma panels and analyses of Tcrd transcripts in unbiased panels of Tcrd cDNA clones revealed that TRDV2-2, TRDV5, and the four members of the TRAV15 family (IMGT nomenclature; reference 8) can compose up to 90% of the adult Vδ repertoire (9–11). Biased usage of V gene segments is not the result of selection based on surface TCR expression because the biases are apparent in thymocytes of CD3ɛ−/− mice that cannot assemble surface TCRs (10, 12) and in nonfunctional rearrangements of γδ hybridomas (9, 10). The molecular basis for selectivity in V segment rearrangement is unclear. It cannot be explained as a function of V gene segment proximity to Dδ, because although several Vδ gene segments are found in the proximal 200 kb (TRDV1, TRDV2-2, TRDV4, and TRDV5), members of the prominent TRAV15 family are dispersed across the central duplicated regions as far as 1 mb from Dδ. Selectivity also seems unlikely to be explained by recombination signal sequence (RSS) differences. RSSs of the dominant Vδ gene segments share no obvious features that distinguish them from other Vα and Vδ gene segments. Moreover, an RSS spacer motif shared by many Vα segments is absent not only from the dominant Vδ segments, but from minor Vδ segments and one fifth of Vα segments as well (13).
Another important and unexplained feature of TcraTcrd locus V gene segment regulation is the orderly progression of Vα to Jα rearrangement events that occur during DP thymocyte development. Essentially all V gene segments, including dominant Vδ segments, can rearrange to Jα (14). It is well established that DP thymocytes can undergo primary, followed by several rounds of secondary, Vα to Jα recombination events, using progressively more 3′ Jα segments and progressively more 5′ Vα segments (14–17). The 5′ to 3′ progression of Jα usage is dictated in part by the ability of 5′ promoters (T early α and Jα49) to target initial rearrangements to the 5′ end of the Jα array (18, 19). This progression in Jα usage occurs in rough coordination with a reciprocal progression in Vα usage (14, 16, 20). Jα-proximal Vα segments rearrange exclusively to 5′ Jα segments, whereas Jα-distal Vα segments rearrange exclusively to more 3′ Jα segments. However, the factors that bias initial Vα to Jα recombination events to use proximal Vα segments are not understood. Without such a bias, the opportunity for multiple rounds of secondary rearrangement would be compromised.
Here we investigate the role of local chromatin accessibility in the regulation of TcraTcrd locus V gene segment usage during T cell development. In the DN compartment we find that V gene segments are heterogeneous with respect to acetylation of histones H3 and H4 and that V gene segments that dominate the adult Vδ repertoire are included in the hyperacetylated subset. In the DP compartment, we find that 3′ V gene segments show dramatic increases in histone acetylation and germline transcription, a consequence of super long–distance regulation by Eα. Our results imply that differences in chromatin accessibility contribute to biases in TcraTcrd locus V gene segment usage in both the DN and DP compartments and extend the distance over which Eα can regulate chromatin structure to a remarkable 525 kb.
Results AND Discussion
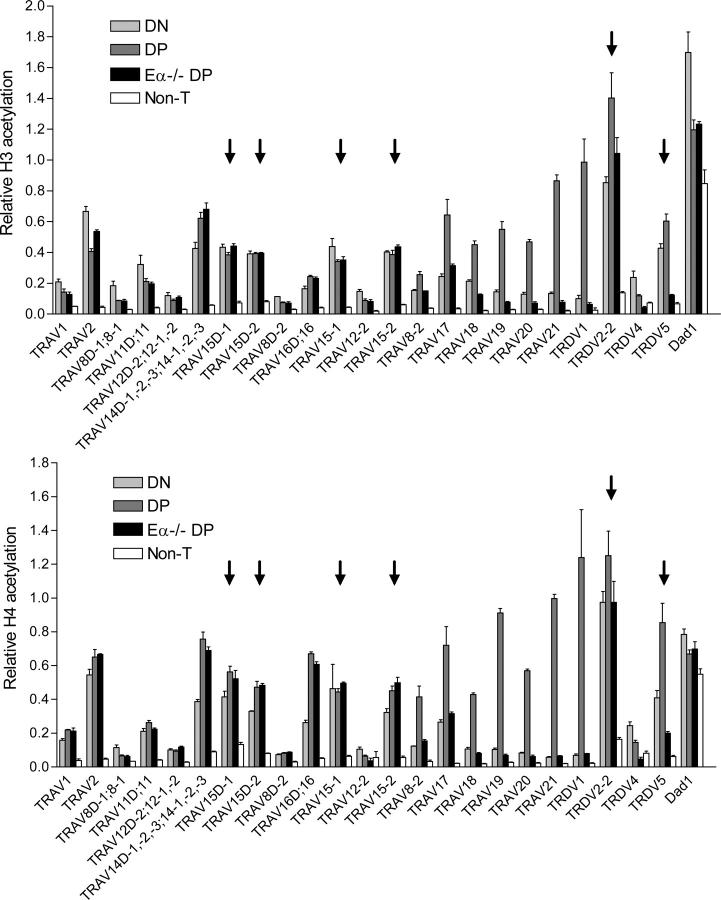
Chromatin that is accessible for V(D)J recombination in vivo typically displays elevated acetylation of histones H3 and H4 (21). These modifications are thought to be functionally important in vivo because forced recruitment of a histone methyltransferase that diminishes H3 and H4 acetylation can block recombinase access (22). Therefore, to evaluate local changes in TcraTcrd locus V gene segment accessibility in DN and DP thymocytes, we used a chromatin immunoprecipitation approach to measure acetylation of histones H3 and H4. We prepared mononucleosomes from thymocytes of Rag2−/− mice and Rag2−/− mice carrying a rearranged Tcrb transgene (Rxβ) and, as a negative control, from T cell–deficient splenocytes of Tcrb−/−Tcrd−/− mice. Acetylated mononucleosomes were immunoprecipitated using antibodies specific for diacetylated histone H3 and tetraacetylated histone H4, and real-time PCR was used to quantify acetylation within the promoter regions of selected V gene segments. In the case of multimember V segment families, it was possible in some instances to design primers that amplified individual family members. In other instances the average behavior of several family members was analyzed in the same PCR reaction. The analyzed V gene segments span 1.5 mb (Fig. 1) and can be segregated into different groups based on the extent to which they contribute to the adult TCRδ repertoire. As noted previously, TRDV2-2, TRDV5, and TRAV15 family members TRAV15D-1, TRAV15D-2, TRAV15-1, and TRAV15-2 constitute the most commonly used Vδ gene segments in adult mice. Additional Vδ segments analyzed are TRDV1 (variable usage in different mouse strains), TRDV4 (usage is restricted to the fetus), the TRAV14 family (usage is low in adults but substantial in neonates), TRAV16 (usage is low in adults), and TRAV21 (usage is low in adults but substantial in neonates; references 9–11). Many of the remaining V gene segments, including TRAV1, TRAV2, the TRAV8, TRAV11, and TRAV12 families, TRAV19, and TRAV20, almost never rearrange to Dδ and are used exclusively by TCRα (23).
Figure 1.
Schematic map of the TcraTcrd locus. The map shows the locations of the V gene segments analyzed in this study (approximately one third of the total), as well as Dad1. Eδ and Eα are identified by black circles. The gray bars classify the V gene segments according to location: proximal, central, central duplication, and distal. V gene segment nomenclature is according to IMGT (reference 8), with conversion to the prior nomenclature of Arden (reference 7) provided. The map is drawn to scale with distances indicated on the upper line.
Independent analyses of histone H3 and H4 acetylation yielded remarkably similar profiles (Fig. 2). Relative acetylation levels varied substantially among the different V gene segments. However, the different members of multimember V segment families behaved similarly, independent of their location. We initially focused on the acetylation of V gene segments in the DN compartment. Strikingly, all six dominant Vδ segments (TRDV2-2, TRDV5, TRAV15D-1, TRAV15D-2, TRAV15-1, and TRAV15-2) displayed among the highest levels of relative histone acetylation (values ≥0.4) in DN thymocytes (Fig. 2, arrows). With only a few exceptions, other V segments that are used infrequently or not at all in the adult TCRδ repertoire displayed relative acetylation values ≤0.2. One exception was the TRAV14 family, which displayed acetylation values equivalent to those of the dominant Vδ. Several studies indicate that these V segments contribute minimally to the TCRδ repertoire in adults (9–11). However, the frequency of TRAV14-containing Tcrd transcripts was found to be substantial in one report (23). Moreover, these V segments were shown to make a substantial contribution to the TCRδ repertoire in neonates (11). Neonatal usage could be reflected in our data because we isolated chromatin from Rag2−/− mice as young as 2-wk old. The other exception was TRAV2. This V gene segment was found to be heavily acetylated in DN thymocytes, yet never rearranges to Dδ. However, this V gene segment is situated at the extreme 5′ end of the locus, fully 1.5 mb from Dδ. Recent studies of the Igh locus suggest that without a locus conformational change that promotes synapsis, accessible but widely separated V and D gene segments may be incapable of recombination (24, 25). With the caveat that histone acetylation is likely to provide, at best, an incomplete measure of chromatin accessibility, we conclude that selective chromatin accessibility in the DN compartment is a major factor that defines the dominant Vδ gene segments. Elevated accessibility likely confers on these V segments a competitive advantage for rearrangement in the DN compartment when Dδ, but not Jα, segments are also accessible for recombination. Because Eδ is thought to function over relatively short distances (4), we suspect that (with the possible exception of TRDV5) the acetylation pattern in DN thymocytes is dictated by intrinsic differences among V gene segment promoters. This notion is consistent with the fact that histone acetylation is nearly identical for physically separated V gene segments that are members of the same family because these V gene segments share highly homologous promoters.
Figure 2.
V gene segment histone modifications. Histone H3 and H4 acetylation was measured in chromatin prepared from DN thymocytes (Rag2−/−), DP thymocytes (Rxβ), Eα−/− DP thymocytes (Eα−/−Rxβ), and non–T cells (T-deficient splenocytes of Tcrb−/−Tcrd−/− mice). Data for all individual gene segments are displayed according to the order in which the gene segments appear in the TcraTcrd locus. In cases where multiple family members are analyzed in a single PCR, the data is displayed according to the locus position of the most distal family member. Arrows identify the dominant adult Vδ gene segments. The data represent the mean ± SEM of triplicate PCR reactions. Analysis of two independent chromatin preparations gave similar results.
We next focused on the acetylation state of V gene segments in the DP compartment (Fig. 2). For most V gene segments in the central, central duplication, and distal portions of the locus, acetylation in the DP compartment was unchanged from the DN compartment. However, all but one V gene segment analyzed in the proximal portion of the locus (TRAV17, TRAV18, TRAV19, TRAV20, TRAV21, TRDV1, TRDV2-2, and TRDV5) displayed increased acetylation in DP as compared with DN thymocytes. Such increases were also apparent for the TRAV14 family, the TRAV16 family (although only for H4 acetylation), and for TRAV8-2, the 3′ member of the TRAV8 family. Because Eα is activated in association with differentiation to the DP stage, we analyzed chromatin prepared from thymocytes of Eα−/−Rxβ mice and Eα−/− mice to determine whether these changes in chromatin structure were dependent on Eα. Thymocyte development is blocked at the DP stage in both Eα−/−Rxβ mice and Eα−/− mice. However, the former retain a completely germline TcraTcrd locus, whereas the latter would have undergone prior Tcrd rearrangement at the DN stage. Nevertheless, results from the two preparations were similar (Fig. 2 and not depicted). With the exception of TRAV14 and TRAV16, deletion of Eα abolished the increases in H3 and H4 acetylation that characterized the transition from DN to DP. The influence of Eα extended as far as TRAV8-2 at the 3′ end of the central region at a distance of 525 kb. That more distal but otherwise homologous TRAV8 family members failed to display Eα-dependent increases in acetylation in DP thymocytes suggests that the defining characteristic of Eα-dependent V gene segments is their position within the locus rather than their promoter structure. Nevertheless, Eα had no effect on the chromatin structure of Dad1, situated only a few kilobases away in the 3′ direction. The highly polarized activity of Eα may reflect the presence of enhancer-blocking elements within the HS2-6 locus control region, which separates Eα from Dad1 (26). The only proximal V gene segment that was insensitive to the influence of Eα was TRDV4. Because TRDV4 is used selectively as a fetal Vδ, we suspect that its promoter may be specifically suppressed in adult thymocytes, rendering it unable to interact with Eα.
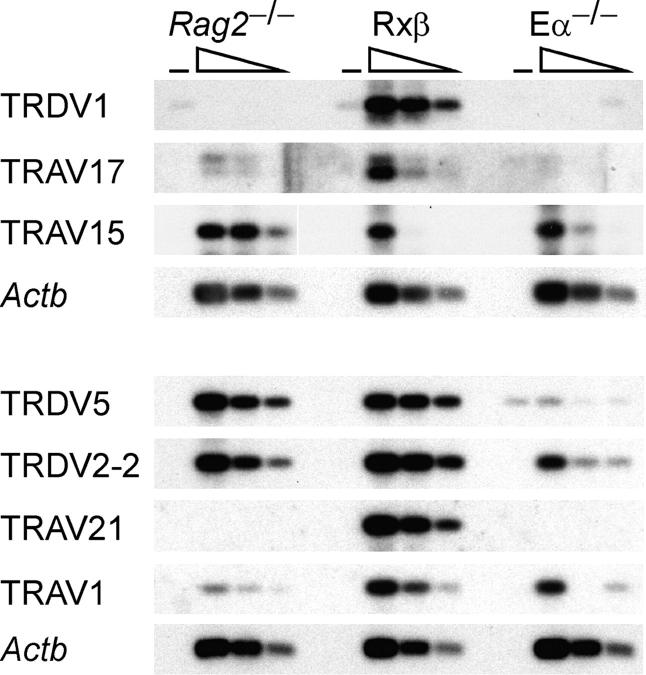
To confirm the results of chromatin analyses, we analyzed germline transcription of selected V gene segments by RT-PCR (Fig. 3). Because of potential differences in transcript stability, PCR efficiency, and detection sensitivity, this approach cannot accurately represent relative rates of transcription at different V segments or relative abundances of different germline transcripts. However, it can provide a good indication of how the abundance of an individual transcript varies in different cell populations. In general, the results closely matched the chromatin data. All of the proximal V gene segments examined (TRDV5, TRDV1, TRDV2-2, TRAV21, and TRAV17) displayed germline transcript levels in DP thymocytes that were highly dependent on Eα. In contrast, Eα had little or no effect on germline transcripts at TRAV15 gene segments in the central and central duplication regions or on germline transcripts at TRAV1 in the distal region. Hence, by these criteria Eα extends its influence at least as far as TRAV17, at a distance of 440 kb. We also note that among the V gene segments examined, only the dominant Vδ gene segments displayed transcript levels in DN thymocytes that were comparable to their levels in DP thymocytes. This supports the idea that chromatin accessibility in DN thymocytes plays a role in defining these Vδ gene segments.
Figure 3.
V gene segment germline transcription. Germline transcription was measured by RT-PCR using serial threefold dilutions of cDNA prepared from DN thymocytes (Rag2−/−), DP thymocytes (Rxβ), and Eα−/− thymocytes (Eα−/−). − identifies control PCR reactions using templates prepared without the addition of reverse transcriptase. Wedges indicate graded amounts of input cDNA. The TRAV15 primers detect all four members of the TRAV15 family. Analysis of two independent cDNA preparations gave similar results. White line indicates that intervening lanes have been spliced out.
Enhancer control of transcription and chromatin structure at a distance of 525 kb is unusual but not unprecedented. Murine Shh gene expression is affected by mutations in an enhancer that is separated from Shh by a distance of 1 mb (27). Complex developmental regulation of V gene segment chromatin structure has been documented for several other antigen receptor loci (21) and in particular for Igh (28, 29). Nevertheless, there is no comparable example of long-distance regulation of V chromatin by the defined enhancers at these loci. Perhaps the clearest description of enhancer control is for Tcrb. Here, in contrast to the TcraTcrd locus, the Tcrb enhancer regulates chromatin structure and transcription within a defined 25-kb region that includes Dβ and Jβ segments but does not extend an influence to Vβ gene segments (30). That the proximal one third of TcraTcrd locus V gene segments are under the influence of Eα may have important implications for how V gene segment usage is regulated in DP thymocytes. First, Eα may predispose initial Vα to Jα rearrangements to involve more accessible, Eα-dependent V gene segments in the proximal portion of the locus. Second, because Tcrd rearrangement and primary Tcra rearrangement result in locus deletion events that may excise several hundred kilobases, V gene segments in the central, central duplication, and distal regions will be brought into proximity of Eα and may therefore come under its regulatory influence. Thus, super long–distance regulation by Eα could promote a sequential activation of V gene segments from proximal to distal across the 1.5-mb V array and could therefore establish a regulated pathway for V gene segment rearrangement in DP thymocytes undergoing primary and secondary Vα to Jα recombination events. Nevertheless, several factors mitigate against highly synchronized Vα usage on different alleles as a consequence of this regulation. First, the region activated by Eα is quite large. Second, even before primary Tcra rearrangement, some alleles will have undergone Tcrd rearrangement involving central V gene segments. This might place central and distal V segments under the influence of Eα at the onset of primary rearrangement. Finally, a subset of centrally located V gene segments should be accessible for primary Tcra rearrangement independent of Eα.
How interactions between Eα and V segment promoters change after initial rounds of rearrangement will ultimately depend on what factors limit these interactions in germline configuration. Enhancer–promoter interactions could be limited by distance per se. Alternatively, these interactions could be limited by the positioning of boundary elements within the V array or by additional elements that modulate overall locus conformation. These will be important issues to address in future studies.
MATERIALS AND METHODS
Mice.
Rag2−/− mice were on a 129 background. Rag2−/− mice carrying a rearranged Tcrb transgene (Rxβ) on a C57BL/6 background were crossed with Rag2−/− mice. The progeny were then intercrossed, selecting for the strain 129 TcraTcrd locus using restriction fragment–length polymorphisms. Eα−/− mice, Eα−/− Rxβ mice, and Tcrb−/−Tcrd−/− mice all carry strain 129 TcraTcrd loci. All mice were used in accordance with protocols approved by the Duke University Animal Care and Use Committee.
Chromatin immunoprecipitation.
Mononucleosomes were prepared from thymocytes of 2–6-wk-old mice and from splenocytes of 8–12-wk-old mice as described previously (6). Chromatin immunoprecipitation was performed using rabbit antisera to diacetylated histone H3 (AcH3) and tetraacetylated histone H4 (AcH4; Upstate Biotechnology) and control rabbit IgG (Sigma-Aldrich). Defined fractions of bound and input DNA were analyzed for enrichment of specific sequences by real-time PCR using a LightCycler and a FastStart DNA Master Syber Green I Kit (Roche Diagnostics). Primers and annealing temperatures are provided in Table S1, available at http://www.jem.org/cgi/content/full/jem.20050680/DC1. The PCR program consisted of denaturation for 4 min at 95°C, followed by 50 cycles of 20 s at 95°C, 20 s at annealing temperature, and 20 s at 72°C. To enable accurate comparisons of acetylation levels in different immunoprecipitations, acetylation of mouse β2-microglobulin (B2m) was used as an internal control. Relative acetylation (RA) was then calculated as, RA = (bound/total for experimental)/(bound/total for B2m).
RT-PCR analysis of germline transcription.
Thymocyte RNA was converted to cDNA using oligo dT primers and SuperScript (Invitrogen) according to the manufacturer's instructions. PCR conditions were 94°C for 5 min, followed by either 22 (Actb), 30 (TRDV5, TRDV2-2, TRAV21, and TRAV1), 32 (TRDV1 and TRAV17), or 40 (TRAV15) cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. This was followed by a 10-min extension at 72°C. After agarose gel electrophoresis and transfer to nylon membranes, PCR products were detected by hybridization with 32P-labeled probes. Oligonucleotide primers and probes for amplification and detection of β-actin (Actb) transcripts were as described previously (19). Amplification and detection of germline V transcripts were performed using oligonucleotide primers and probes described in Table S2, available at http://www.jem.org/cgi/content/full/jem.20050680/DC1. PCR from genomic DNA templates indicated that Tcrd rearrangement causes ≤50% loss of proximal V sequences in Eα− / − thymocytes (unpublished data).
Online supplemental material.
Table S1 provides sequences and annealing temperatures for PCR primers used in chromatin immunoprecipitation analysis. Table S2 provides sequences of primers and probes used for RT-PCR analysis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20050680/DC1.
Acknowledgments
The authors thank Y. Zhuang, I. Abarrategui, and B. Beutler for helpful advice and comments on the manuscript.
This work was supported by National Institutes of Health grant GM41052.
The authors have no conflicting financial interests.
References
- 1.Bassing, C.H., W. Swat, and F.W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109:S45–S55. [DOI] [PubMed] [Google Scholar]
- 2.Krangel, M.S., J. Carabana, I. Abbarategui, R. Schlimgen, and A. Hawwari. 2004. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor α/δ (TRA/TRD) locus. Immunol. Rev. 200:224–232. [DOI] [PubMed] [Google Scholar]
- 3.Monroe, R.J., B.P. Sleckman, B.C. Monroe, B. Kohr, S. Claypool, R. Ferrini, L. Davidson, and F.W. Alt. 1999. Developmental regulation of TCR δ locus accessibility and expression by the TCR δ enhancer. Immunity. 10:503–513. [DOI] [PubMed] [Google Scholar]
- 4.Bassing, C.H., R.E. Tillman, B.B. Woodman, D. Canty, R.J. Monroe, B.P. Sleckman, and F.W. Alt. 2003. T cell receptor (TCR) α/δ locus enhancer identity and position are critical for the assembly of TCR δ and α variable region genes. Proc. Natl. Acad. Sci. USA. 100:2598–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sleckman, B.P., C.G. Bardon, R. Ferrini, L. Davidson, and F.W. Alt. 1997. Function of the TCRα enhancer in αβ and γδ T cells. Immunity. 7:505–515. [DOI] [PubMed] [Google Scholar]
- 6.McMurry, M.T., and M.S. Krangel. 2000. A role for histone acetylation in the developmental regulation of V(D)J recombination. Science. 287:495–498. 10642553 [Google Scholar]
- 7.Arden, B., S.P. Clark, D. Kabelitz, and T.W. Mak. 1995. Mouse T-cell receptor variable gene segment families. Immunogenetics. 42:501–530. [DOI] [PubMed] [Google Scholar]
- 8.Bosc, N., and M.-P. Lefranc. 2003. The mouse (Mus musculus) T cell receptor alpha (TRA) and delta (TRD) variable genes. Dev. Comp. Immunol. 27:465–497. [DOI] [PubMed] [Google Scholar]
- 9.Sleckman, B.P., B. Khor, R. Monroe, and F.W. Alt. 1998. Assembly of productive T cell receptor δ variable region genes exhibits allelic inclusion. J. Exp. Med. 188:1465–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pereira, P., V. Hermitte, M.-P. Lembezat, L. Boucontet, V. Azuara, and K. Grigoriadou. 2000. Developmentally regulated and lineage-specific rearrangement of T cell receptor Vα/δ gene segments. Eur. J. Immunol. 30:1988–1997. [DOI] [PubMed] [Google Scholar]
- 11.Weber-Arden, J., O.M. Wilbert, D. Kabelitz, and B. Arden. 2000. γδ repertoire during thymic ontogeny suggests three novel waves of γδ expression. J. Immunol. 164:1002–1012. [DOI] [PubMed] [Google Scholar]
- 12.Gallagher, M., S. Candeias, C. Martinon, E. Borel, M. Malissen, P.N. Marche, and E. Jouvin-Marche. 1998. Use of TCR ADV gene segments by the δ chain is independent of their position and of CD3 expression. Eur. J. Immunol. 28:3878–3885. [DOI] [PubMed] [Google Scholar]
- 13.Probst, J., S.G. Blumenthal, S. Tenzer, T. Weinschenk, J. Dittmer, O. Schoor, A. Six, H.G. Rammensee, and S. Pascolo. 2004. A conserved sequence in the mouse variable T cell receptor α recombination signal sequence 23-bp spacer can affect recombination. Eur. J. Immunol. 34:2179–2190. [DOI] [PubMed] [Google Scholar]
- 14.Pasqual, N., M. Gallagher, C. Aude-Garcia, M. Loiodice, F. Thuderoz, J. Demengeot, R. Ceredig, P.N. Marche, and E. Jouvin-Marche. 2002. Quantitative and qualitative changes in V-J α rearrangements during murine thymocytes differentiation: implication for a limited T cell receptor α chain repertoire. J. Exp. Med. 196:1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yannoutsos, N., P. Wilson, W. Yu, H.T. Chen, A. Nussenzweig, H. Petrie, and M.C. Nussenzweig. 2001. The role of recombination activating gene (RAG) reinduction in thymocyte development in vivo. J. Exp. Med. 194:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, C., and O. Kanagawa. 2001. Ordered and coordinated rearrangement of the TCR α locus: role of secondary rearrangement in thymic selection. J. Immunol. 166:2597–2601. [DOI] [PubMed] [Google Scholar]
- 17.Guo, J., A. Hawwari, H. Li, Z. Sun, S.K. Mahanta, D.R. Littman, M.S. Krangel, and Y.W. He. 2002. Regulation of the TCRα repertoire by the survival window of CD4+CD8+ thymocytes. Nat. Immunol. 3:469–476. [DOI] [PubMed] [Google Scholar]
- 18.Villey, I., D. Caillol, F. Selz, P. Ferrier, and J.-P. de Villartay. 1996. Defect in rearrangement of the most 5′ TCR-Jα following targeted deletion of T early α (TEA): implications for TCR α locus accessibility. Immunity 5:331-342. [DOI] [PubMed] [Google Scholar]
- 19.Hawwari, A., C. Bock, and M.S. Krangel. 2005. Regulation of TCRα gene assembly by a complex hierarchy of germline Jα promoters. Nat. Immunol. 6:481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aude-Garcia, C., M. Gallagher, P.N. Marche, and E. Jouvin-Marche. 2001. Preferential ADV-AJ association during recombination in the mouse T-cell receptor alpha/delta locus. Immunogenetics. 52:224–230. [DOI] [PubMed] [Google Scholar]
- 21.Krangel, M.S. 2003. Gene segment selection in V(D)J recombination: accessibility and beyond. Nat. Immunol. 4:624–630. [DOI] [PubMed] [Google Scholar]
- 22.Osipovich, O., R. Milley, A. Meade, M. Tachibana, Y. Shinkai, M.S. Krangel, and E.M. Oltz. 2004. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat. Immunol. 5:309–316. [DOI] [PubMed] [Google Scholar]
- 23.Gallagher, M., P. Obeid, P.N. Marche, and E. Jouvin-Marche. 2001. Both TCRα and TCRδ chain diversity are regulated during thymic ontogeny. J. Immunol. 167:1447–1453. [DOI] [PubMed] [Google Scholar]
- 24.Hesslein, D.G., D.L. Pflugh, D. Chowdhury, A.L. Bothwell, R. Sen, and D.G. Schatz. 2003. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 17:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuxa, M., J. Skok, A. Souabni, G. Salvagiotto, E. Roldan, and M. Busslinger. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18:411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong, X.-P., and M.S. Krangel. 1999. Enhancer-blocking activity within the DNase I hypersensitive site 2 to 6 region between the TCR α and Dad1 genes. J. Immunol. 163:295–300. [PubMed] [Google Scholar]
- 27.Lettice, L.A., S.J. Heaney, L.A. Purdie, L. Li, P. de Beer, B.A. Oostra, D. Goode, G. Elgar, R.E. Hill, and E. de Graaff. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12:1725–1735. [DOI] [PubMed] [Google Scholar]
- 28.Chowdhury, D., and R. Sen. 2001. Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J. 20:6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, K., C. Angelin-Duclos, S. Park, and K.L. Calame. 2003. Changes in histone acetylation are associated with differences in accessibility of VH gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell. Biol. 23:2438–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathieu, N., W.M. Hempel, S. Spicuglia, C. Verthuy, and P. Ferrier. 2000. Chromatin remodeling by the T cell receptor (TCR)–β gene enhancer during early T cell development: implications for the control of TCR-β locus recombination. J. Exp. Med. 192:625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]