Abstract
The pre–T cell receptor (TCR) is expressed early during T cell development and imposes a tight selection for differentiating T cell progenitors. Pre-TCR–expressing cells are selected to survive and differentiate further, whereas pre-TCR− cells are “negatively” selected to die. The mechanisms of pre-TCR–mediated survival are poorly understood. Here, we describe the induction of the antiapoptotic gene BCL2A1 (A1) as a potential mechanism regulating inhibition of pre–T cell death. We characterize in detail the signaling pathway involved in A1 induction and show that A1 expression can induce pre–T cell survival by inhibiting activation of caspase-3. Moreover, we show that in vitro “knockdown” of A1 expression can compromise survival even in the presence of a functional pre-TCR. Finally, we suggest that pre-TCR–induced A1 overexpression can contribute to T cell leukemia in both mice and humans.
The interval between thymic entry of multipotent bone marrow lymphoid progenitors and thymic export of mature αβ CD4+ and CD8+ T lymphocytes requires passage through multiple quality control checkpoints. Only those progenitors successful in producing machinery required for progression will receive survival signals at each checkpoint. Although early T lymphocyte progenitors lack surface expression of CD4 and CD8 (double negative [DN]), their maturation can be tracked by stage-specific changes in surface expression of CD25 and CD44. The population advances from the DN1 (CD44+ CD25−) to DN2 (CD44+ CD25+) to DN3 (CD44− CD25+) and finally to the DN4 (CD44− CD25−) stage (1). Deposition of an intact pre-TCR complex, consisting of a productively rearranged TCRβ chain, the invariant pre-TCRα (pTα) chain, and CD3 signaling chains, on the surface of DN3 thymocytes, is absolutely essential for completion of the αβ T lymphocyte developmental program. αβ T lymphocyte differentiation halts at the DN3 stage in mice lacking pTα, TCRβ, CD3ɛ, or CD3γ (2).
In addition to its role in “selecting” for further development exclusively those progenitors containing a productive TCRβ rearrangement, driving entry into the cell cycle, preventing further rearrangement of TCRβ loci, and down-regulating surface expression of CD25, pre-TCR signaling is essential for survival of αβ T lymphocyte progenitors (2). Analyses of mice unable to generate intact pre-TCR complexes (RAG − / −, CD3γ− / −, SCID) revealed large increases in the proportion of DN thymocytes expressing an apoptotic surface phenotype, thereby linking the pre-TCR to thymocyte survival (3). Complementation with cytoplasmic tail mutant pTα molecules (4) or with a TCRα molecule (5) failed to rectify the survival defect in the DN compartment of pTα2/2mice, implicating specifically the pTα component as crucial for pre-TCR–induced survival signals.
The prevalence of NF-κB consensus sites among cis-elements controlling the expression of multiple antiapoptotic genes (for review see reference 6), in combination with the definitive placement of NF-κB as a distal effector of pre-TCR signaling (7, 8), establishes an indirect association between the pre-TCR and the survival of developing thymocytes. The search for distal mediators of pre-TCR–induced survival has met with little success. The β selection process proceeds normally in progenitors lacking pro-survival BCL-2 family members BCL-2 or BCL-xL (9, 10), and neither BCL-2 nor BCL-xL transgenes restore αβ T lymphocyte development in pre-TCR–deficient mice (11–13). T lymphocyte progenitors lacking the pro-survival Bcl-2 family member MCL-1 fail to progress through the DN2 to DN3 developmental transition, implicating MCL-1 as a crucial mediator of early cytokine-derived, rather than later pre-TCR–derived, survival signals (14). Further refuting a critical role for BCL-2 is the observation that post-β selection thymocytes contain lower levels of Bcl-2 protein than those that have not yet entered β selection (15). However, a recent report presented conflicting evidence, suggesting that BCL-2 expression is induced by the pre-TCR in vitro (16).
Similarly, very little is known about the signaling pathways inducing death in pre-TCR− thymocytes. The initial suggestion that the Fas-induced death pathway is essential for pre–T cell death has been challenged by recent studies of Newton et al. (17), in which the thymocyte profiles of RAG-1−/−lpr mice were found to be similar to those of their RAG-1−/−littermates. Fas is a member of the TNF receptor family, whose members interact via their death domains with cytoplasmic adaptors including FADD and TRADD (18). The same investigators have shown that RAG-1−/−FADD−/−pre–T cells differentiate and reach the double positive (DP) stage (17). These important observations suggested that FADD could be essential for the transduction of death signal within pre–T cells and fueled the search for an active pre-TCR–dependent mechanism of cell death inhibition.
In this report, we direct our search toward antiapoptotic genes potentially targeted by pre-TCR–induced NF-κB transcription factors. We identify the pro-survival A1 gene as a direct transcriptional target of both pre-TCR and NF-κB. Moreover, we connect transcriptional activation of A1 to PLCγ, mobilization of Ca2+, and activation of protein kinase C (PKC) kinases. We also show that A1 expression inhibits pre–T cell death by suppressing the activation of the effector caspase-3. Finally, we show that A1 is up-regulated in both human and mouse pre–T cell leukemias, making it a possible target of the transformation event. Our observations suggest that through NF-κB activation and A1 expression, the pre-TCR ensures the promotion of physiological T cell development.
Results
Pre-TCR–nonexpressing cells die due to the activation of effector caspase-3
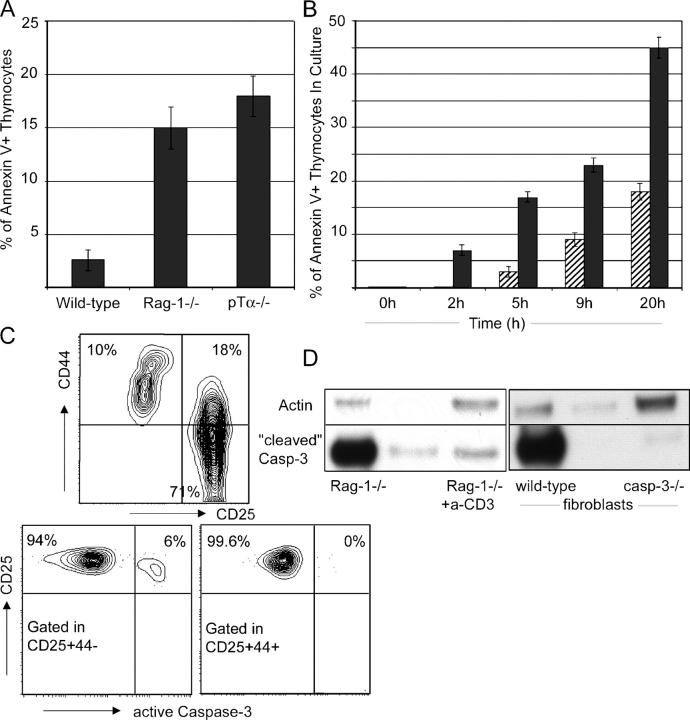
Evidence definitively demonstrating pre-TCR–mediated prevention of imminent apoptosis has not been documented. Here, we addressed this issue both in vivo and in vitro. The percentage of proapoptotic, annexin V+ thymocytes was significantly larger in mice lacking pre-TCR components (pTα2/2, RAG-1−/−) than in their WT counterparts (Fig. 1 A). To demonstrate that pre-TCR expression provides a survival advantage, sorted pre-TCR− DN3 RAG-1−/−and pre-TCR+ DN4 WT thymocytes were cultured in a low concentration of 1 ng/ml IL-7. As evident from Fig. 1 B, pre-TCR–deficient cells die at a significantly higher rate (Fig. 1 B). Similar results were obtained in the absence of IL-7 (not depicted).
Figure 1.
Pre-TCR− thymocytes die by apoptosis due to caspase-3 activation. (A) Percentage of thymocytes binding annexin V in the CD4−8− lineage− compartment of WT, Rag-1−/−, and pTα−/− thymi. (B) In vitro pre–T cell survival. Pre-TCR+ (hatched bars) and pre-TCR− (solid bars) CD4−8− lineage− thymocytes were cultured in the absence of exogenous factors. Survival rates were determined by annexin V staining at the indicated time points. (C) Flow cytometric detection of caspase-3 activation. The left plot illustrates the CD25/CD44 surface profile of Rag-1−/− thymocytes. The right dot plots illustrate caspase-3 activation within indicated cellular compartments. (D) Activation of caspase-3 in Rag-1−/− thymocytes is down-regulated upon anti-CD3ɛ injection. Positive (WT fibroblasts treated with brefeldin A) and negative (caspase-3−/− fibroblasts treated with brefeldin A) controls are included. Blots were reprobed with anti-actin as a loading control.
To gain insight into the mechanism of this cell death, we analyzed activation of the distal effector caspase-3 in apoptotic thymocytes. Using a fluorescently labeled antibody specific for the active form of caspase-3, we detected activation of caspase-3 specifically in the pre-TCR− DN3 subset of RAG-1−/−thymocytes ex vivo (Fig. 1 C). As receptors for prosurvival cytokines IL-7 and stem cell factor are down-regulated at the DN4 stage, these cells do not receive any pre-TCR–independent survival signals. To further support these observations and to increase the sensitivity of detection of active caspase-3, we probed RAG-1−/−thymocyte lysates with a second antibody specific for the activated (cleaved) form of caspase-3. The activation of caspase-3 observed in this system was comparable to that induced by in vitro treatment of mouse embryonic fibroblasts with brefeldin A (Fig. 1 D). Mimicking pre-TCR signaling by CD3ɛ cross-linking relieved the DN3 block of RAG-1−/−thymocytes (7) and decreased caspase-3 activity in these cells, adding one more piece of evidence directly connecting pre-TCR signaling with thymocyte survival (Fig. 1 D). An identical pattern of activation was also seen for the effector caspase 9 (unpublished data).
BCL-2, MCL-1, and BCL-xL are not targets of the pre-TCR
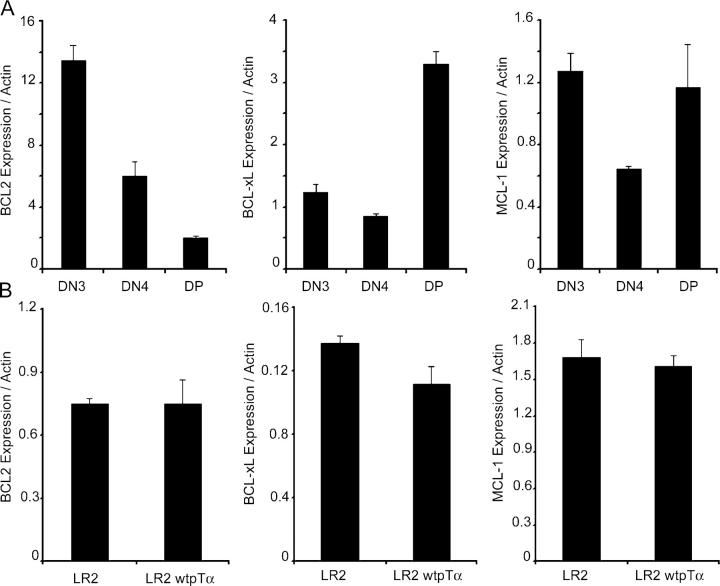
Conflicting reports described the roles of BCL-2 and BCL-xL in pre–T cell death, and recent data indicated that yet another antiapoptotic BCL-2 family member, MCL-1, may regulate survival of thymocytes at an earlier, cytokine-mediated stage (14). To test whether pre-TCR signaling induces expression of these prosurvival factors, we performed real-time RT-PCR analysis using cDNA isolated from sorted DN3, DN4, and DP thymocytes. As evident from Fig. 2, none of these factors were induced by the pre-TCR. MCL-1 expression was high in DN3 cells. In agreement with previous reports, it decreased in DN4 and peaked again at the DP stage. BCL-xL expression was low in both DN3 and DN4 populations, peaking at the DP stage. BCL-2 expression seemed to be actively down-regulated upon pre-TCR expression, reaching a minimum at the DP stage (Fig. 2 A). Similar results showing no up-regulation of BCL-xL and MCL-1 and a down-regulation of BCL-2 were obtained using two distinct pre-TCR− (SCIET27 and LR2) and pre-TCR+ (SCB29 and LR2wtpTα) cell lines (Fig. 2 B and unpublished data; reference 19).
Figure 2.
BCL-2, BCL-xL, and MCL-1 are not induced by pre-TCR signaling. (A) Quantitative real-time RT-PCR analysis of BCL-2, MCL-1, and BCL-xL expression in the indicated thymocyte subsets. (B) Quantitative RT-PCR analysis of gene expression in the LR2 and LR2wtpTα cell lines. Actin expression was used to normalize gene expression.
A1 gene is a direct target of pre-TCR signaling
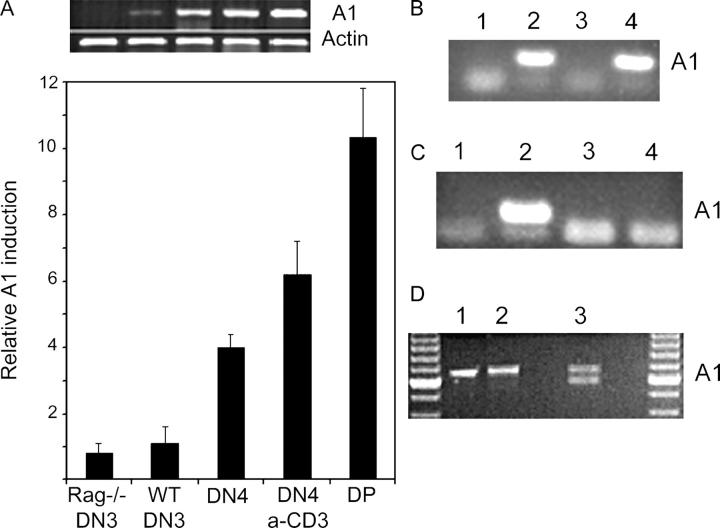
After excluding BCL-2, BCL-xL, and MCL-1 as antiapoptotic pre-TCR transcriptional targets, we focused specifically on antiapoptotic BCL2 family genes whose cis regulatory elements contain NF-κB consensus sites. The BCL2A1 (A1) gene cluster (three A1 genes: A1a, A1b, and A1d; one pseudogene: A1c) fit this description. Expression of this NF-κB–responsive gene cluster is induced by inflammatory stimuli, B cell receptor stimulation, and CD40 signaling (20–22). A1 expression protects cells from various death stimuli including TNF-α stimulation, p53 activation, B cell receptor triggering, and IL-3 deprivation (23–25). Our gene expression analyses identified the A1 gene cluster as a potential pre-TCR target (Fig. 3). Sorted RAG-1−/−pre-TCR− DN3 thymocytes lacked A1 transcripts, whereas sorted preTCR+ WT DN4 thymocytes expressed abundant A1 transcripts. Only after mimicking pre-TCR signaling by injection of anti-CD3ɛ antibody was A1 expression detected in RAG-1−/−thymocytes (Fig. 3 A). These observations were reproduced multiple times using different primer pairs and were confirmed by quantitative real-time PCR (Fig. 3 A). To test whether A1 induction was a direct effect of pre-TCR signaling, we compared A1 expression in pre-TCR− (SCIET27 and LR2) and pre-TCR+ (SCB29 and LR2-WTpTα) pre–T cell lines (7). SCIET27 and LR2 are parental pre-TCR− lines. Introduction of TCRβ (in SCIET) and TCRβ pTα (in LR2) results in the generation of the pre-TCR+ SCB29 and LR2-WTpTα lines (5, 7). As shown in Fig. 3 B, pre-TCR expression induced A1 expression in both cell lines. Moreover, LR2 cell lines engineered to express truncated forms of pTα (mutations of the cytoplasmic tail of the molecule) illustrated the crucial nature of the pTα cytoplasmic tail in pre-TCR–mediated A1 induction (Fig. 3 C). These observations are consistent with our previous report, which showed that thymocytes expressing truncated pTα chains underwent abnormal β selection characterized by defects both in proliferation and in cell survival (4).
Figure 3.
Pre-TCR signaling induces A1 expression both in vivo and in vitro. (A) Semiquantitative and real-time RT-PCR analysis of A1 expression in ex vivo thymocyte subsets. (B) Semiquantitative PCR analysis of A1 expression in pre-TCR+ (lane 2, SCB29; lane 4, LR2-WTpTα) and pre-TCR− (lane 1, SCIET27; lane 3, LR2) cell lines. (C) Pre-TCR complexes containing truncated pTα chains fail to induce A1 expression. Semiquantitative PCR-quantitated A1 expression in LR2 cells expressing (lane 1) no pTα chain, (lane 2) a WT pTα chain, (lane 3) a pTα chain lacking its entire cytoplasmic domain, or (lane 4) a pTα chain lacking its proline-rich domains. (D) Isoform-specific pattern of A1 expression in DN4 pre–T cells. A1 cDNA amplified from DN thymocytes (using A1XhoUp1 and A1BamLow1 primers) was left nondigested (lane 1), digested with NsiI (lane 2), or digested with BglII (3).
The existence of three A1 isoforms, each encoded by a different gene, prompted us to investigate which isoforms of A1 were expressed in pre–T cells. To this end, A1 cDNA amplified from DN4 cells was digested with restriction enzymes whose recognition sites are located in isoform-specific sequence polymorphisms (26). The enzyme NsiI was unable to digest A1 cDNA amplified from pre–T cells (purified DN4), indicating that A1a is either not expressed or expressed at undetectable levels in these cells. In contrast, digestion with the BglI enzyme strongly suggested that A1d is one of the major isoforms expressed in pre–T cells (Fig. 3 D). In addition to verifying these results, sequencing indicated that A1b was also expressed in pre–T cells, albeit at a level lower than that of A1d (not depicted). These observations suggest an isoform-specific pattern of A1 expression in pre–T cells.
Activation of phospholipase Cγ (PLCγ), Ca2+, and NF-κB are critical for A1 induction
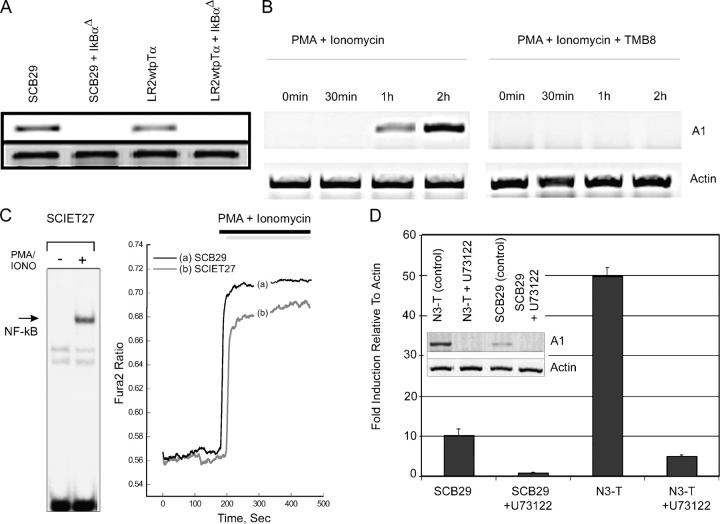
The above observations place A1 expression downstream of the pre-TCR, but the signaling components connecting pre-TCR signals with A1 induction remain unidentified. We initially addressed the role of NF-κB in pre-TCR–induced A1 expression. As mentioned above, pre-TCR+ cell lines (SCB29 and LR2WTpTα), but not their pre-TCR–deficient counterparts (SCIET27 and LR2), contained abundant A1 transcripts. To inhibit NF-κB activation in SCB29 and LR2WTpTα cell lines, we generated a retroviral vector encoding a mutated nondegradable form of the inhibitor of NF-κB α (IKBαΔ; reference 8). When expressed in pre-TCR+ SCB29 and LR2WTpTα cell lines, IKBαΔ completely inhibited pre-TCR–induced A1 expression, suggesting an essential role of NF-kB in pre-TCR–dependent up-regulation of A1 (Fig. 4 A). These observations are consistent with those of Voll et al. (8), who demonstrated that pre–T cell–specific expression of an IKBαΔ transgene (driven by the p56lck promoter) slowed pre-TCR–driven αβ T lymphocyte development.
Figure 4.
A1 is a target of the PLCγ-Ca2+-NF-κB pathway. (A) A1 is a transcriptional target of NF-κB. Semiquantitative RT-PCR analysis of A1 expression in pre-TCR+ cell lines after the inhibition of NF-κB. In the absence of pre-TCR signaling, 50 ng/ml PMA plus 1 μg/ml Iono treatment can induce A1 expression (B), as well as NF-κB nuclear translocation and Ca2+ mobilization (C). (D) Ca2+ mobilization is essential for A1 transcriptional induction. A1 expression was examined by semiquantitative RT-PCR after chelation of intracellular Ca2+ by 1 mM TMB8. (E) PLCγ activation is essential for A1 induction. A1 expression levels were examined by semiquantitative and real-time RT-PCR in pre-TCR+ cell lines (N3T and SCB29) incubated with 10 μM of the specific PLCγ inhibitor U73122.
We have previously shown that activation of PLCγ and Ca2+ mobilization are essential for the transduction of pre-TCR signals (7). To address the role of these two mediators in pre-TCR–induced A1 expression, we used the PMA and ionomycin (Iono), which have been shown to be able to mimic pre-TCR signaling (7). Treatment with PMA and Iono rapidly induced A1 expression in a pre-TCR–deficient line (Fig. 4 B), and Rag-1−/− purified DN3 thymocytes (not depicted). Because this treatment induces both Ca2+ mobilization as well as nuclear translocation of NF-κB (Fig. 4 C), we tested whether A1 expression depends on one or both of these stimuli. Pretreatment of these cell lines with an endoplasmic reticulum Ca2+ chelator, TMB-8, totally abrogated A1 expression (Fig. 4 D) and NF-κB activation (7), highlighting Ca2+ as an essential second messenger. To test whether A1 induction was dependent on PLCγ activation, pre-TCR+ SCB29 and N3-T (a leukemic cell line that has up-regulated A1 expression in response to Notch activation and overexpression of the pre-TCR) cell lines were incubated with the specific PLCγ inhibitor U73122, which inhibits Ca2+ signaling in pre–T cells (7). As shown in Fig. 4 E, inhibition of PLCγ activity totally blocked the ability of the pre-TCR (and of PMA plus Iono treatment) to induce A1 expression in both cell lines. Similar results were obtained using a third pre-TCR+ cell line, LR2-WTpTα (not depicted). The inhibition was complete regardless of the initial level of A1 expression.
PKC activation is essential for A1 induction and survival
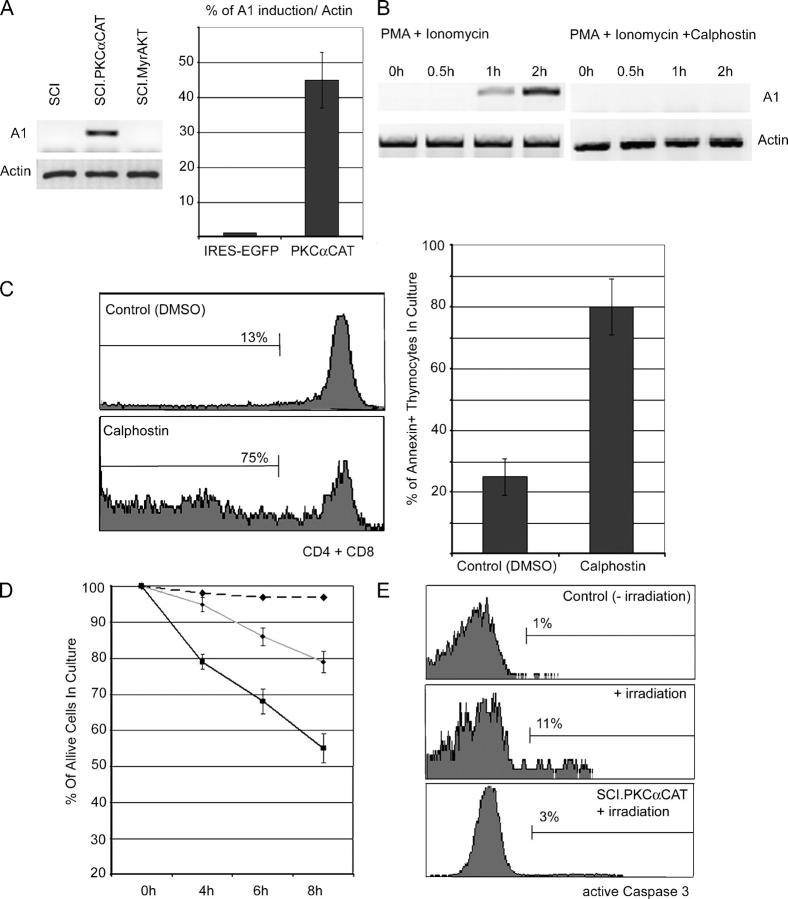
PLCγ activity generates both inositol triphosphate, which initiates Ca2+ mobilization, and diacylglycerol (DAG), which activates downstream kinases such as PKC. To address the potential role of PKC in pre-TCR–induced A1 expression, we expressed a constitutively active form of PKC (PKCαCAT) in pre-TCR–deficient (and thus A1−) SCIET and LR2 cell lines. PKC activation strongly induced A1 expression in the SCIET (Fig. 5 A) and LR2 (not depicted) cell lines. To establish PKC specificity, we expressed an activated (myristoylated) form of PKB/Akt (Myr-Akt), another kinase involved in the transduction of pre-TCR signals (unpublished data). As shown in Fig. 5 A, Myr-Akt failed to up-regulate A1 expression, classifying pre-TCR–induced A1 expression as strictly dependent on PKC.
Figure 5.
PKC is essential for pre-TCR–induced A1 expression. (A) SCIET27 cells were infected with a retrovirus expressing constitutively active forms of PKC (SCI-PKCαCAT) or Akt (SCI-Myr-Akt). A1 expression was compared with uninfected SCIET27 by RT-PCR. (B) In vitro inhibition of PKC activity using 1 μM of the PKC inhibitor calphostin, abrogates A1 expression induced by PMA plus Iono in SCIET27 cells. (C) Inhibition of PKC activity blocks normal T cell development and induces pre–T cell death in an OP-9-DL1 culture. WT DN2 thymocytes were cultured in the presence or absence of calphostin or DMSO control. Differentiation (left) and survival (right) were examined by staining with anti-CD4 and anti-CD8 antibodies and annexin V. Data are representative of at least four independent experiments using 0.1 μM calphhostin. (D and E) SCIET27 cells expressing PKCαCAT are more resistant to irradiation- (480 rads) induced death (left) due to the attenuation of caspase-3 activation (right).
To further illustrate PKC involvement, pre-TCR− SCIET and LR2 cells were pretreated with calphostin, a potent and specific PKC inhibitor. Calphostin treatment totally blocked PMA plus Iono–induced A1 expression in SCIET (Fig. 5 B) and LR2 (not depicted) cell lines. To demonstrate that PKC inhibition adversely affected pre–T cell survival and differentiation, we used a novel in vitro culture system (27). In this system, OP-9 stromal cells expressing the Notch ligand Delta-1 (OP9-DL1) support thymic-independent development of T cell progenitors. Sorted DN2 cells were cultured with OP9-DL1 cells and supplemented with various concentrations of calphostin or DMSO control. As shown in Fig. 5 C, calphostin, but not DMSO treatment, significantly blocked progression to the DP stage of T cell development. This effect was due to decreased survival, as >85% of the calphostin-treated cells were proapoptotic. Also, the majority (60%) of calphostin-treated cells remained CD4−8−CD25+ (not depicted). To directly connect PKC activation and A1 expression to pre–T cell survival, we compared survival kinetics of irradiated (400 rads) pre-TCR–deficient SCIET27 cells expressing either PKCαCAT-EGFP or EGFP. PKCαCAT expression provided a significant protection from apoptosis (Fig. 5 D), which correlated with the ability of PKC to prevent caspase-3 activation (Fig. 5 E). Identical results were obtained using a second pre-TCR− line (LR2).
A1 as a regulator of pre–T cell survival
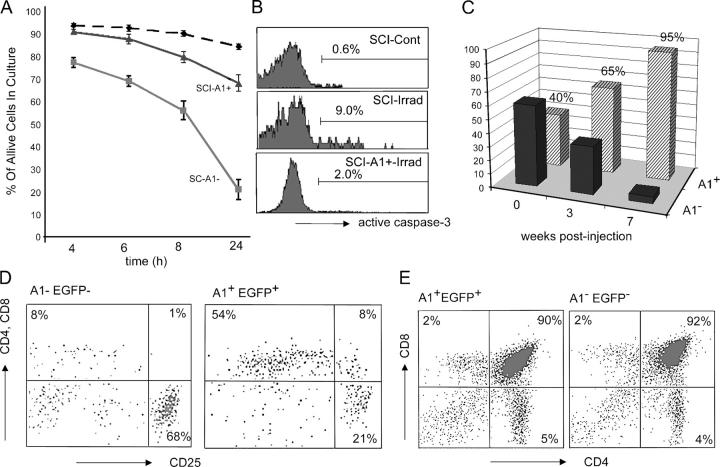
To study the function of A1 in the promotion of pre–T cell survival, we ectopically expressed A1d (the most commonly expressed A1 isoform) in SCIET pre–T cells (4). A1d and EGFP were expressed on a single transcript after infection with a bicistronic A1-IRES-EGFP retrovirus. This retrovirally encoded A1 protein was properly expressed and targeted to the mitochondria (not depicted). As pre–T cells sustain DNA breaks generated during ongoing recombination at their TCR loci, we investigated A1's ability to inhibit death induced by DNA-damaging ultraviolet irradiation. A1d expression efficiently suppressed irradiation-induced cell death in SCIET (Fig. 6 A) and LR2 (not depicted) pre–T cells. Mechanistically, the inhibition of cell death resulted from an A1-mediated reduction in caspase-3 activation (Fig. 6 B). Similar results were obtained with dexamethasone treatment as a death stimulus (not depicted).
Figure 6.
A1 induces pre–T cell survival. SCIET27 cells expressing EGFP (black dashed and gray lines) or A1-EGFP (black line) were irradiated (gray, control; black, A1-expressing) and left in culture for the indicated time periods. Survival was detected using annexin V labeling (A). Caspase-3 activation was measured 4 h after irradiation (B). Retroviral expression of A1 induces survival and differentiation of Rag-1−/− pre-TCR− thymocytes. Ratios of infected (A1+) and uninfected (A1−) RAG-1−/− thymocytes at the indicated time points after injection into Rag-2−/−γc−/− hosts (C). Differentiation of A1+ and A1− thymocytes was analyzed 6 wk after injection by surface staining with antibodies specific for CD4, CD8, and CD25 (D). (E) A1 expression does not alter differentiation of WT thymocytes. CD4/CD8 surface profiles of WT thymocytes infected with A1-IRES-EGFP or IRES-EGFP retroviruses were examined 3 wk after injection into Rag-2−/−γc−/− hosts.
To support these in vitro observations using primary thymocytes, we infected lineage- (CD19, Ter-119, Gr-1, NK1.1, αβ TCR, γδ TCR, Mac-1, CD11c) negative bone marrow progenitors sorted from (Ly5.1+) RAG-1−/−animals with the A1d-IRES-EGFP retrovirus. Alymphoid irradiated (350 rads) Rag − / −γc − / − (Ly5.2+) hosts were injected with the Ly5.1+ A1-infected bone marrow cells. Thymic reconstitution was analyzed at various time intervals after cell transfer. Retroviral A1d bestowed a significant survival advantage upon developing thymocytes (Fig. 6 C). Although injected at a 40:60 ratio of EGFP+A1+/EGFP−A1−, 3 wk's time reversed this ratio to 65:35 (Fig. 6 C). Four additional weeks resulted in a near absence of EGFP− A1− cells in reconstituted thymi, suggesting that EGFP+A1+ thymocytes out-competed their coinjected EGFP− A1− counterparts. Interestingly, despite their lack of a pre-TCR complex, these RAG-1 − / − EGFP+A1+ thymocytes displayed signs of differentiation. More than half of the cells up-regulated CD4 and CD8 coreceptor surface expression (Fig. 6 D). Importantly, annexin V staining revealed little apoptosis among the A1+EGFP+ population and high percentages of apoptotic cells in the EGFP− population (not depicted). No A1+EGFP+ cells were detected in the periphery, due to the inability of the RAG-1−/− cells to express selectable TCRs (not depicted). These results suggest that A1 expression, which is physiologically triggered by pre-TCR signaling, induces pre–T cell survival and ensures differentiation. To illustrate that A1 expression does not affect the physiological progression of T lymphocyte development, we repeated the above reconstitution experiments using bone marrow progenitors sorted from WT mice. No difference was apparent between the differentiation kinetics of WT A1+EGFP+ and WT A1−EGFP− progenitors when injected in age- and sex-matched hosts (Fig. 6 E). These observations are consistent with the recently reported normal T cell development in thymi of mice expressing an A1a transgene driven by a p56Lck promoter (28).
To demonstrate that A1 is necessary for pre–T cell survival, we studied pre–T cell survival in the absence of A1 expression. The existence of three independent A1 isoforms precludes gene-targeting approaches. Multiple consecutive targeting steps would be necessary, as the genes encoding the three A1 isoforms are not all clustered together on chromosome 9. In addition, the significant conservation in both exon and intron sequences shared among the A1 isoforms would make design of gene-targeting vectors extremely difficult. Therefore, we used stable RNA interference to knock down A1 expression in a T lymphocyte–specific manner. To eliminate concern about redundancy among A1 isoforms, we generated short interfering RNAs (siRNAs) capable of knocking down expression of all three A1 isoforms. These siRNAs were expressed using a self-inactivating retrovirus carrying a U6 Pol-II promoter able to drive expression of siRNAs. We tested three different siRNAs, and achieved optimal interference with siRNA-A (Fig. 7 A). Real-time RT-PCR analysis showed silencing efficiency at 75–85% (Fig. 7 A). The effects of A1 deficiency on pre-TCR function were assessed in the pre-TCR+ N3-T cell line (Fig. 8 D). N3-T cells expressing each of the three different siRNAs (EGFP+) were mixed at a ratio of 50:50 with EGFP−N3-T cells. As shown in Fig. 7 B, N3-T cells expressing A1-specific siRNAs exhibited a survival disadvantage, proportional in severity to the efficiency of A1 knockdown. This defect is due to loss of survival, as at any given time of the culture, the proportion of cells binding annexin V was two to four times higher in the population expressing A1-siRNA (siRNA-A) when compared with cells expressing a control EGFP-siRNA (not depicted). No significant differences in cell cycle progression were observed between cells expressing EGFP-siRNA and A1-siRNA constructs (not depicted). When infected with the same siRNAs, neither 3T3, 293T, nor LR2 cells (Fig. 7 C) showed survival defects, suggesting that the effect of A1 knockdown is specific to cells that specifically up-regulate A1 due to pre-TCR overexpression (Fig. 8). Moreover, BCL-xL and BCL-2 expression remained unaltered in N3-T cells that undergo apoptosis due to the A1 deficiency, suggesting that these two genes are not involved in the maintenance of N3-T cell survival (unpublished data).
Figure 7.
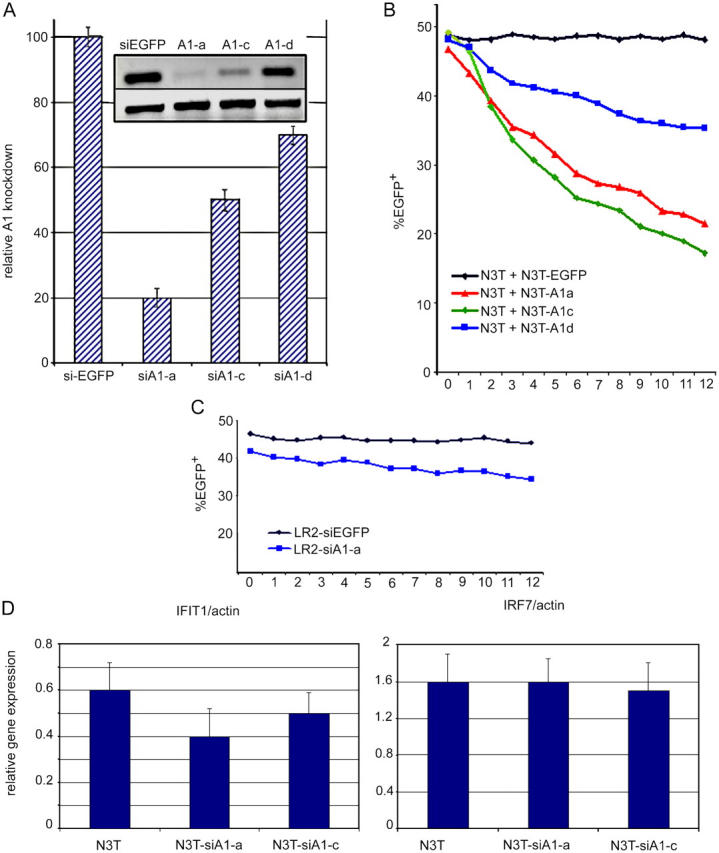
A1 knockdown impairs pre–T cell survival in vitro. (A) Effective A1 silencing in pre–T cells. Pre-TCR+ N3T cells were infected with the Banshee-EGFP retroviral vector alone (siEGFP) or Banshee-EGFP retroviruses encoding one of three independent siRNA complexes specific for A1 (A1-A, A1-C, and A1-D). Semiquantitative and real-time RT-PCR analyses illustrate the efficiency of A1 silencing using each siRNA. (B and C) A1 silencing affects survival of pre-TCR+ (N3T), but not pre-TCR− (LR2), cells. Uninfected N3T or LR2 cells (EGFP−) were mixed at the indicated ratio with N3T or LR2 cells infected with Banshee-EGFP (EGFP) or Banshee-EGFP vectors encoding siA1-A, siA1-C, or siA1-D complexes. Alterations in the ratio of uninfected (EGFP−) to infected (EGFP+) cells were recorded at the indicated time points. This experiment is a representative of four independent experiments. (D) Retrovirally expressed siRNA complexes do not induce IFN response genes. Real-time PCR expression analysis of IFIT-1 and IRF-7 IFN response genes in cell lines infected with the indicated siRNA complexes.
Figure 8.
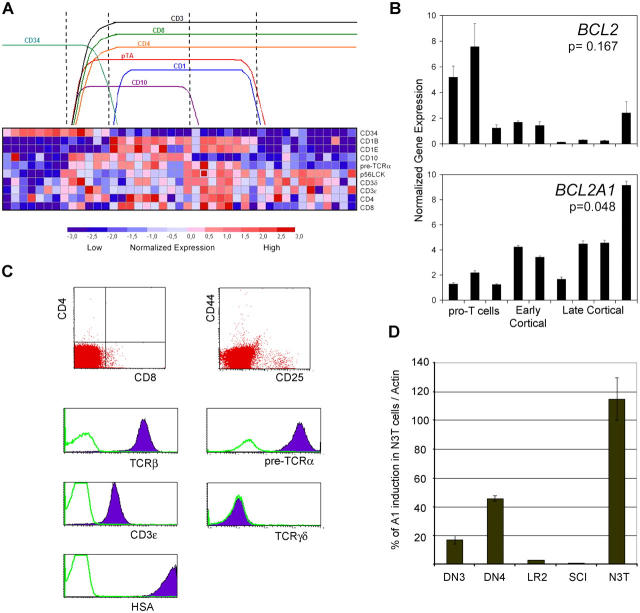
A1 is a target of T cell transformation both in mouse and man. (A) Relative expression levels of pTα and specific T cell differentiation marker genes (rows) across 43 human T cell leukemia samples (columns). Normalized gene expression levels are represented in colors. Groups of samples were identified (top) according to the expression of the indicated T cell differentiation markers. Expression of pTα clearly correlates with a group of genes including p56lck, CD3ɛ, CD4, and CD8. pTα expression is completely absent in early CD34+ pro–T cell leukemias. This is a pictorial representation of the expression levels with no intention to represent specific gene expression levels. (B) A1 and BCL-2 expression was quantitated by real-time RT-PCR analysis in selected representative T-ALL samples. BCL2 and A1 expression were compared in pro–T cell and cortical T-ALL samples using a Wilcoxon rank sum test, showing a significant correlation of BCL2 expression with pro–T T-ALL samples and of A1 expression with cortical T-ALL samples. (C) Surface phenotype characterization of a murine T cell leukemia line (N3T). (D) Expression of A1 in N3T cells. Real-time RT-PCR comparison between N3T, DN3, DN4, SCIET27 (our expression base), and LR2 cells.
To exclude nonspecific siRNA effects due to potential IFN responses, we have selected two well-characterized IFN-responsive genes and studied their expression in pre–T cells expressing our siRNAs (29). There was no specific up-regulation of either IFN-induced protein 1 or IFN regulatory factor 7 in our siRNA-expressing pre–T cells (Fig. 7 D).
A1 expression is a molecular target of pre–T cell transformation
Deregulated pre-TCR signaling triggers aggressive pre–T cell leukemia in mice (30, 31). Genes encoding molecules involved in pre-TCR signaling (p56Lck, ZAP-70, TCRβ, CD3ɛ, and cyclin D3) are overexpressed in human T cell acute leukemias (T-ALLs) arising from early to late cortical thymocytes, which are characterized by the expression of the oncogenes HOX-11+ and TAL-1+. Thus, it is conceivable that the survival advantage conferred by pre-TCR–induced antiapoptotic genes like A1 may contribute to the transformation process (not as a bona fide oncogene but as a target of the transformation event). To connect pre-TCR and A1 expression to a precise window of thymocyte differentiation, we used the patient expression profile data presented by Ferrando et al. (32) to analyze the expression of pTα within the three different clusters of human T-ALL cases. As shown in Fig. 8 A, pTα expression is seen primarily within Hox-11+ early cortical thymocytes, rather than within LYL1+ CD34+ progenitors or TAL-1+ mature T cells. To link A1 expression with induction of T cell leukemia, we selected representative cases from each cluster of human T-ALL cases and performed quantitative real-time RT-PCR. As evident from Fig. 8 B, A1 expression correlated only with HOX-11 or TAL-1–expressing samples. This is consistent with the notion that A1 is induced only after pre-TCR signaling and peaks at the DP stage of development. Interestingly, BCL-2 expression peaked early in development (in CD3− CD34+ samples) and declined after the appearance of the pre-TCR and A1 (Fig. 8 B). This observation is consistent with our previous expression analysis of A1 and BCL-2 during physiological T cell development (Figs. 2 and 3).
To complement these studies, we analyzed A1 expression in a mouse model of T cell leukemia. In this model, introduced by Bellavia et al. (33), T cell–specific expression of a constitutively active form of Notch-3 induces malignant transformation of thymocytes. We have chosen Notch-3 because “nearest neighbor” analysis of the T-ALL database used above had shown that pTα and Notch-3 share a strikingly identical expression profile (unpublished data). The Notch transformation is strictly dependent on pre-TCR signaling (31). N3-T, an immature HSAhi CD25lo CD4− CD8− cell line derived from the thymus of a Notch-3 transgenic animal, most likely represents the pre-TCR+ DN3 to DN4 transitional stage of thymocyte development (Fig. 8 C; reference 33). Surface pre-TCR levels on N3-T cells are even higher than those on WT DN4 cells (Fig. 8 C). In line with our human T-ALL studies, N3-T cells expressed A1 at levels significantly higher that those in WT DN4 thymocytes or in DN cell lines (Fig. 8 D). As shown in Fig. 7 B, A1 is crucial for in vitro survival of N3-T cells, as A1 “knockdown” conferred a significant survival disadvantage.
Discussion
The identification of NF-κB as a pre-TCR target (7, 8) and the phenotype of transgenic mice expressing an inhibitor of the NF-κB pathway (IκBαΔ) specifically in pre–T cells (8) initiated our search for NF-κB–responsive downstream effectors regulating pre–T cell survival. Together with important reports directly connecting A1 transcription to NF-κB activation (34, 35), the recent observation of endogenous c-Rel binding directly to cis elements regulating A1 gene expression (36) directed our search toward A1. Here, we show that A1 expression is directly induced by the pre-TCR. Moreover, A1 appears to be the only antiapoptotic gene induced during β selection, as neither members of the BCL2 family nor members of the IAP family are induced by the pre-TCR (Fig. 2 and unpublished data).
One can argue, however, that the data presented are equally consistent with the notion that A1 is a Notch target, as Notch can also activate the NF-kB pathway. This is an intriguing possibility that could provide insights in the mechanism of cooperation between Notch and the pre-TCR (37). However, multiple lines of evidence argue against this scenario and highlight the unique ability of the pre-TCR to regulate A1 expression. Initially, expression of activated forms of Notch-1 (N1-IC) in pre-TCR− lines (SCIET27 and LR2) failed to up-regulate A1 expression (unpublished data). Moreover, specific mutation of the pre-TCR (cytoplasmic truncation) blocked any A1 expression without affecting Notch expression (Fig. 3). Finally, DN thymocytes from pTα−/− mice that overexpress active Notch-3 show activation neither of PKC nor of NF-κB, suggesting that the pre-TCR, rather than Notch, regulates the activity of this pathway in developing pre–T cells (unpublished data).
Why the pre-TCR uses A1 rather than the prototypical antiapoptotic molecules BCL-2 or BCL-xL is not yet understood. However, overexpression of BCL-2 is unable to restore normal developmental progression in pre-TCR–deficient pTα2/2 (unpublished data) or RAG-1−/−mice (11). The preferential use of A1 might be due to the antiproliferative properties ascribed to BCL-2. Introduction of a BCL-2 transgene into WT mice actually impaired the proliferation of TCRβ-selected DN4 thymocytes (38). It was suggested that this antiproliferative effect depends on the ability of BCL-2 to bind to and sequester the calcium-dependent calcineurin phosphatase, thereby inhibiting NF-AT nuclear translocation and activation (39, 40). This interaction between BCL-2 and calcineurin depends on conserved regions in the BH4 domain of BCL-2, which are also present in BCL-xL and MCL-1. In contrast, all three A1 genes lack these conserved calcineurin interaction sites (unpublished data). Thus, BCL-2, BCL-xL, and MCL-1, but not A1, may prevent NF-AT activation, which is important for pre-TCR function (7). Consistent with this notion, Gonzalez et al. (28) recently have shown that overexpression of A1, but not BCL-2, allowed normal proliferation of mature activated T lymphocytes.
Our studies underline the role of PKC as an intermediate transducer of pre-TCR signals. PKC isoenzymes are grouped into two major subfamilies. The activation of conventional PKCs (cPKCs) as well as novel PKCs (nPKCs) typically involves recruitment to membranes and interaction with or allosteric activation by DAG. Ca2+ binding is an essential requirement for cPKCs, but not nPKCs (41). PKCα, cPKC, PKCθ, and nPKC, are the major PKCs expressed in thymocytes (41). In our experiments, PMA plus Iono treatment induced A1 expression in pre-TCR− cells, whereas Iono alone did not. Also, PMA plus Iono in the presence of the ER Ca2+ chelator, TMB-8, induced neither A1 expression nor NF-κB activation. As induction of NF-κB activation and A1 expression required both PMA and Iono, it is tempting to suggest that pre-TCR signaling activates PLCγ, which mobilizes Ca2+, which, along with DAG, activates only cPKC isoenzymes (i.e., PKCα). Consistent with this view is the lack of a pre-TCR–dependent phenotype in mice lacking the PKCθ isoenzyme (42). However, we cannot exclude that PKCθ, although not absolutely essential, plays a secondary role amplifying PKCα-mediated signals to the nucleus.
A recent study illustrated PKC's importance in pre-TCR–mediated allelic exclusion at the TCRβ locus (43). Separate work demonstrated that although activation of PKC can partially restore DN to DP transition in RAG-1 − / − mice, it is not able to drive thymocyte expansion (a phenotype identical to those of mice reconstituted with A1+ RAG-1 − / − progenitors, presented here; reference 37). Perhaps the bifurcation in pre-TCR signaling that occurs downstream of the LAT/SLP-76 complex leads in one direction toward PKC-independent signals inducing thymocyte expansion (Raf/Ras, Akt) and in the other direction toward PKC-dependent signals establishing TCRβ allelic exclusion and ensuring survival.
The critical role of A1 in transmitting pre-TCR–induced antiapoptotic signals and the previous correlation of deregulated pre-TCR signaling with T cell leukemia, prompted us to analyze the patterns of A1 expression in human T-ALL cases and in a Notch-induced model of mouse leukemia. We found a unique correlation between A1 expression and receipt of pre-TCR signals and have shown that A1 knockdown of A1 expression can affect survival of a pre–T cell leukemic line. We recently identified cyclin D3 as an essential mediator of pre-TCR–induced proliferative signals and suggested that cyclin D3 expression was essential for transformation of pre–T cells (44). A1 overexpression may cooperate with cyclin D3 to provide a survival signal rendering transformed pre–T cells resistant to apoptotic signals. These findings could have clinical importance because TAL1+ leukemias are resistant to chemotherapy potentially due to the overexpression of the antiapoptotic A1 (32).
The pre-TCR possesses the unique ability to override cell intrinsic (i.e., DNA damage-induced) and extrinsic (i.e., death receptor–induced) apoptotic stimuli. We describe a novel mechanism by which the pre-TCR meets this responsibility, namely transcriptional induction of A1. Our data do not exclude the possibility that the pre-TCR may use additional mechanisms of cell death inhibition or that non-pre-TCR–induced antiapoptotic factors (like BCL-xL or BCL-2) could assist A1 in the regulation of cell survival. Indeed, the pre-TCR could directly down-regulate the transcription of proapoptotic genes or it could regulate the stability and degradation of proapoptotic molecules at a posttranslational level, via phoshporylation and/or ubiquitination. These potential alternative regulation mechanisms as well as the extent of functional cooperation of A1 with other antiapoptotic genes during β selection are currently under investigation.
Materials and Methods
Mice and cell lines
C57BL/6, pTα2/2, and RAG-2 − / − mice were kept in the sterile Carlson Barrier animal facility. All animal experiments were performed in accordance to the guidelines of the Institutional Animal Care and Use Committee of the university. For pre-TCR mimicking experiments, RAG-2−/−mice were injected with anti-CD3 as described previously (7). All cell lines were cultured in RPMI containing 10% fetal bovine serum (Sigma-Aldrich), 1% penicillin/streptomycin, n and 0.1% β-mercaptoethanol (GIBCO BRL). The caspase-3−/− fibroblasts were as described previously (45).
Patient material
Samples of cryopreserved lymphoblasts from children and young adults treated in Total Therapy studies XI–XIII at St. Jude Children's Research Hospital were obtained with informed consent at the time of diagnosis before any chemotherapy was given. All protocols and consent forms were approved by the hospital's institutional review board, and informed consent was obtained from parents, guardians, or patients (as appropriate).
RT-PCR and quantitative RT-PCR
Total cellular RNA was isolated by using an RNeasy kit (QIAGEN). Each cDNA template was made from total RNA using the Superscript II-RT kit (Invitrogen). PCR reactions were performed with RedTaq (Sigma-Aldrich). Transcripts of all murine A1 isoforms were detected using two primer pairs: forward-1, 5′-cattaactggggaaggattgtgac-3′, reverse-2 5′-gcagaaaagtcagccagccagatt-3′, and forward-2, 5′-tcatgcatatccactccctggctgagc- 3′, reverse-2, 5′-gtcctgtcatctgcagaaaagtcagcc-3′. Murine β actin transcripts were detected using 5′-tggaatcctgtggcatccatgaaac-3′ as the forward primer and 5′-taaaacgcagctcagtaacagtccg-3′ as the reverse primer. For detection of antiapoptotic genes we used the following: BCL2F, 5′-atcttctccttccagcct-3′; BCL2R, 5′-tcattcaaccagacatgc-3′; MCL-1F, 5′-agagcgctggagaccctg-3′; MCL-1R, 5′-ctatcttattagatatgccagacc-3′; BCL-xL forward, 5′-tggagtcagtttagtgatgtc-3′; and BCL-xL reverse, 5′-gctcgattgttcccgtagag-3′.
Quantitative analysis of cDNA amplification was assessed by incorporation of SYBR Green into dsDNA. PCR reactions containing 1 ug cDNA template, 0.5 μM each of the primers, and SYBR Green PCR Master Mix (Applied Biosystems) were performed in a total volume of 25 μl. Gene expression was analyzed using the ABI PRISM 7700 Sequence Detector (Applied Biosystems) and ABI Prism Sequence Detection Software version 1.9.1 (Applied Biosystems). Normalization of samples was performed by dividing the value of the unknown gene by the value of the endogenous reference genes (β actin). Quadruplicate reactions were performed using all cDNA samples. For the real-time PCR analysis using human leukemic samples, total RNA from cryopreserved lymphoblasts was extracted using the RNAqueous kit (Ambion).
The cDNA was analyzed by quantitative PCR using the SYBR Green RT-PCR Core Reagents (Applied Biosystems). Quantitative PCR analyses were performed with an ABI PRISM 7700 Sequence Detection System instrument (Applied Biosystems) in a total volume of 25 μl. Relative expression was calculated for each gene using the ΔCT method. Expression levels were normalized according to expression of GAPDH housekeeping gene. We used the following primers: GAPDH forward, 5′-gaaggtgaaggtcggagt-3′; GAPDH reverse, 5′-gaagaggtgatgggatttct-3′; BCL2 forward, 5′-gattgtggccttctttgagttcg-3′; BCL2 reverse, 5′-cgtacagttccacaaaggcatcc-3′; BCL2-A1 forward, 5′-ccccggatgtggatacctataagg-3′; and BCL2-A1 reverse 5′-attttcccagcctccgttttgc-3′.
Microarray analysis.
Expression data from 43 T cell lymphoblastic leukemia samples reported by Yeoh et al. (46). Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling was also according to Yeoh et al. (46). Normalization of global fluorescence differences across arrays and calculation of gene expression values were preformed using dChip (http://www.dchip.org). Normalization of relative expression levels across all 43 samples and visualization of selected T cell differentiation marker genes (TCR-related genes) were performed using Gene Cluster (http://www.broad.mit.edu/cancer/software/genecluster2/gc2.html). Additional information concerning the samples and the microarray analysis can be found at: http://www.stjuderesearch. org/data/ALL1.
Flow cytometric analysis and cell sorting.
Anti-CD4 (L3T4), CD8 (53-6.7), CD25 (3C7), CD44 (IM7), B220 (RA3-6B2), CD11c (HL3), NK1.1 (PK136), TCRβ (H57-597), TER-119 (TER-119), Mac-1 (M1/70), Gr-1 (RB6-8C5), and heat stable antigen (M1/69) mAbs were from BD Biosciences. These mAbs were directly coupled to FITC, PE, CYC, APC, or biotin. Surface marker expression on thymocytes and peripheral T cells was visualized using a FACSCalibur (Becton Dickinson) and analyzed with Flow-Jo and CELLQuest software. Cell sorting was performed using a Mo-Flo (DakoCytomation). DN thymocytes were isolated as described by Aifantis et al. (7).
Retroviral constructs and progenitor infections.
MigR1 (30) and a previously described moloney murine leukemia virus–based retroviral vector (4) were used for retroviral transduction experiments. PKCαCAT (a constitutively active form of PKC) constructs were cloned upstream of the MigR1 IRES-GFP cassette (37). Mouse A1d cDNA was amplified from WT thymocyte cDNA using the following primers: A1XhoUp1, 5′-gcctcgagcaacagcctccagatatgattaggg-3′ and A1BamLow1, 5′-cggatcctaaccattctcctgggagccaagg-3′. After amplification, the PCR product was sequenced and cloned in the XhoI–BamHI sites of the moloney murine leukemia virus–based vector upstream of an IRES-EGFP cassette (4). Retroviral supernatants were generated as described previously (47).
Retrovirally expressed A1 siRNA.
We have used the Banshee-EGFP (48) retroviral vector (provided by J. Alberola-Ila, California Institute of Technology, Pasadena, CA) to express small interfering RNAs in pre–T cells. The cloning procedure has been described by Hernandez-Hoyos et al. (48). The following siRNA sequences were used: siA1-A, 5′-ggaaatgctctttctcctcaa-3′; siA1-C, 5′-gagcagattgccctggatgta-3′; and siA1-D, 5′-ggagaatggatacggcggaat-3′. After cloning, sequencing was used to ensure the insertion of the correct sequence. Concentrated retrovirus was generated as described above.
T cell differentiation in OP9-DL1 culture
2 × 103 sorted DN2 or DN3 thymocytes were placed in each well of a 24-well tissue culture plate containing a confluent monolayer of OP9-DL1 cells in DMEM (Life Technologies) supplemented with 15% FCS, penicillin/streptomycin, l-glutamine, and 5 ng/ml rIL-7 and rFlt3L (PeproTech) as described previously (27). The PKC inhibitor calphostin C (or DMSO) was added (100 nM) in wells. Differentiation was analyzed after 8 d, and viability was assessed by propidium iodide staining.
Active caspase-3 assay
SCIET27, SCIET27 transfected with PKCαCAT, or SCIET27 transfected with A1 were irradiated (480 rads). At various time points after irradiation, 106 cells were harvested, washed, and fixed/ permeabilized. Active caspase-3 was detected using a PE-conjugated active caspase-3 mAb (BD Biosciences).
Western blot analysis
Rag-2−/− sorted CD25+ or CD25−44− (after anti-CD3 injections) cells were used. As controls for the caspase-3 activation, we have used embryonic fibroblasts from either WT or caspase-3−/− mice. Total protein lysates were prepared as described by Aifantis et al. (7). The protein blots were probed with anti-cleaved caspase-3 (Cell Signaling) and anti-actin (Santa Cruz Biotechnology, Inc.) as an internal loading control.
Cytosolic Ca2+ measurements
For Ca2+ measurements, we have used 2 × 106 SCIET and SCB29 cells loaded with 1 μM Fura2-AM (Molecular Probes) dissolved in IMDM containing 1% FBS for 30 min at 37°C in the dark. The exact protocol followed is described elsewhere (7).
Nuclear extracts and electrophoretic mobility shift assay
5–10 × 106 SCIET cells were used for the preparation of nuclear protein extracts. The details of the exact protocol used can be found elsewhere (7).
Acknowledgments
We would like to thank G. Founari and L. Scorrano for experimental help and stimulating discussions. We are grateful to J. Alberola-Ila and G. Hernandez-Hoyos for the Banshee vector, B. Reizis for the LR2 cells, and R. Flavell for the Casp-3−/− fibroblasts.
We would like to acknowledge the Saint Jude Cancer Center Support grant (CA-21765). This work was supported by the Leukemia Research Foundation, the Cancer Research Foundation, the Cancer Research Institute, and the V Foundation For Cancer Research (all to I. Aifantis). J.-C. Zuniga-Pflucker is supported by an Investigator award from the Canadian Institutes of Health Research (CIHR). M. Ciofani is supported by a Studentship award from the CIHR. I. Screpanti was supported by Associazione Italiana per la Ricerca sul Cancro. A. Ruiz-Vela is supported by a fellowship from the European Molecular Biology Organization. T. Palomero is a recipient of a fellowship from the Lady Tata Memorial Trust. C. Borowski was supported by a National Science Foundation Graduate Research Fellowship.
The authors have no conflicting financial interests.
Abbreviations used: cPKC, conventional protein kinase C; DAG, diacylglycerol; DN, double negative; Iono, ionomycin; DP, double positive; nPKC, novel protein kinase C; PKC, protein kinase C; PLCγ, phospholipase Cγ; pTα, pre-TCRα; siRNA, short interfering RNA; T-ALL, T cell acute leukemia.
M. Mandal and C. Borowski contributed equally to this work.
References
- 1.Godfrey, D.I., J. Kennedy, T. Suda, and A. Zlotnik. 1993. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8− triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 150:4244–4252. [PubMed] [Google Scholar]
- 2.von Boehmer, H., I. Aifantis, J. Feinberg, O. Lechner, C. SaintRuf, U. Walter, J. Buer, and O. Azogui. 1999. Pleiotropic changes controlled by the pre-T cell receptor. Curr. Opin. Immunol. 11:135–142. [DOI] [PubMed] [Google Scholar]
- 3.Haks, M.C., P. Krimpenfort, J.H. van den Brakel, and A.M. Kruisbeek. 1999. Pre-TCR signaling and inactivation of p53 induces crucial cell survival pathways in pre-T cells. Immunity. 11:91–101. [DOI] [PubMed] [Google Scholar]
- 4.Aifantis, I., C. Borowski, F. Gounari, H.D. Lacorazza, J. Nikolich-Zugich, and H. von Boehmer. 2002. A critical role for the cytoplasmic tail of pTalpha in T lymphocyte development. Nat. Immunol. 3:483–488. [DOI] [PubMed] [Google Scholar]
- 5.Borowski, C., X. Li, I. Aifantis, F. Gounari, and H. Von Boehmer. 2004. Pre-TCRα and TCRα are not interchangeable partners of TCRβ during T lymphocyte development. J. Exp. Med. 199:607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karin, M., and A. Lin. 2002. NF-kappaB at the crossroads of life and death. Nat. Immunol. 3:221–227. [DOI] [PubMed] [Google Scholar]
- 7.Aifantis, I., F. Gounari, L. Scorrano, C. Borowski, and H. von Boehmer. 2001. Constitutive pre-TCR signaling promotes differentiation through Ca2+ mobilization and activation of NF-kappaB and NFAT. Nat. Immunol. 2:403–409. [DOI] [PubMed] [Google Scholar]
- 8.Voll, R.E., E. Jimi, R.J. Phillips, D.F. Barber, M. Rincon, A.C. Hayday, R.A. Flavell, and S. Ghosh. 2000. NF-kappaB activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity. 13:677–689. [DOI] [PubMed] [Google Scholar]
- 9.Veis, D.J., C.M. Sorenson, J.R. Shutter, and S.J. Korsmeyer. 1993. Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell. 75:229–240. [DOI] [PubMed] [Google Scholar]
- 10.Motoyama, N., F. Wang, K.A. Roth, H. Sawa, K. Nakayama, I. Negishi, S. Senju, Q. Zhang, and S. Fujii. 1995. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science. 267:1506–1510. [DOI] [PubMed] [Google Scholar]
- 11.Maraskovsky, E., L.A. O'Reilly, M. Teepe, L.M. Corcoran, J.J. Peschon, and A. Strasser. 1997. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1−/− mice. Cell. 89:1011–1019. [DOI] [PubMed] [Google Scholar]
- 12.Strasser, A., A.W. Harris, L.M. Corcoran, and S. Cory. 1994. Bcl-2 expression promotes B- but not T-lymphoid development in scid mice. Nature. 368:457–460. [DOI] [PubMed] [Google Scholar]
- 13.Chao, D.T., and S.J. Korsmeyer. 1997. BCL-XL-regulated apoptosis in T cell development. Int. Immunol. 9:1375–1384. [DOI] [PubMed] [Google Scholar]
- 14.Opferman, J.T., A. Letai, C. Beard, M.D. Sorcinelli, C.C. Ong, and S.J. Korsmeyer. 2003. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 426:671–676. [DOI] [PubMed] [Google Scholar]
- 15.Voll, R.E., E. Jimi, R.J. Phillips, D.F. Barber, M. Rincon, A.C. Hayday, R.A. Flavell, and S. Ghosh. 2000. NF-kappa B activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity. 13:677–689. [DOI] [PubMed] [Google Scholar]
- 16.Murga, C., and D.F. Barber. 2002. Molecular mechanisms of pre-T cell receptor-induced survival. J. Biol. Chem. 277:39156–39162. [DOI] [PubMed] [Google Scholar]
- 17.Newton, K., A.W. Harris, and A. Strasser. 2000. FADD/MORT1 regulates the pre-TCR checkpoint and can function as a tumour suppressor. EMBO J. 19:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu, H., J. Huang, H.B. Shu, V. Baichwal, and D.V. Goeddel. 1996. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 4:387–396. [DOI] [PubMed] [Google Scholar]
- 19.Groettrup, M., K. Ungewiss, O. Azogui, R. Palacios, M.J. Owen, A.C. Hayday, and H. von Boehmer. 1993. A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor beta chain and a 33 kd glycoprotein. Cell. 75:283–294. [DOI] [PubMed] [Google Scholar]
- 20.Hinz, M., P. Loser, S. Mathas, D. Krappmann, B. Dorken, and C. Scheidereit. 2001. Constitutive NF-kappaB maintains high expression of a characteristic gene network, including CD40, CD86, and a set of antiapoptotic genes in Hodgkin/Reed-Sternberg cells. Blood. 97:2798–2807. [DOI] [PubMed] [Google Scholar]
- 21.Bernal, A., R.D. Pastore, Z. Asgary, S.A. Keller, E. Cesarman, H.C. Liou, and E.J. Schattner. 2001. Survival of leukemic B cells promoted by engagement of the antigen receptor. Blood. 98:3050–3057. [DOI] [PubMed] [Google Scholar]
- 22.Chen, C., L.C. Edelstein, and C. Gelinas. 2000. The Rel/NF-kappaB family directly activates expression of the apoptosis inhibitor Bcl-x(L). Mol. Cell. Biol. 20:2687–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duriez, P.J., F. Wong, K. Dorovini-Zis, R. Shahidi, and A. Karsan. 2000. A1 functions at the mitochondria to delay endothelial apoptosis in response to tumor necrosis factor. J. Biol. Chem. 275:18099–18107. [DOI] [PubMed] [Google Scholar]
- 24.D'Sa-Eipper, C., and G. Chinnadurai. 1998. Functional dissection of Bfl-1, a Bcl-2 homolog: anti-apoptosis, oncogene-cooperation and cell proliferation activities. Oncogene. 16:3105–3114. [DOI] [PubMed] [Google Scholar]
- 25.Craxton, A., P.I. Chuang, G. Shu, J.M. Harlan, and E.A. Clark. 2000. The CD40-inducible Bcl-2 family member A1 protects B cells from antigen receptor-mediated apoptosis. Cell. Immunol. 200:56–62. [DOI] [PubMed] [Google Scholar]
- 26.Hatakeyama, S., A. Hamasaki, I. Negishi, D.Y. Loh, F. Sendo, and K. Nakayama. 1998. Multiple gene duplication and expression of mouse bcl-2-related genes, A1. Int. Immunol. 10:631–637. [DOI] [PubMed] [Google Scholar]
- 27.Schmitt, T.M., and J.C. Zuniga-Pflucker. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17:749–756. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez, J., A. Orlofsky, and M.B. Prystowsky. 2003. A1 is a growth-permissive antiapoptotic factor mediating postactivation survival in T cells. Blood. 101:2679–2685. [DOI] [PubMed] [Google Scholar]
- 29.Sledz, C.A., M. Holko, M.J. de Veer, R.H. Silverman, and B.R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834–839. [DOI] [PubMed] [Google Scholar]
- 30.Allman, D., F.G. Karnell, J.A. Punt, S. Bakkour, L. Xu, P. Myung, G.A. Koretzky, J.C. Pui, J.C. Aster, and W.S. Pear. 2001. Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J. Exp. Med. 194:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellavia, D., A.F. Campese, S. Checquolo, A. Balestri, A. Biondi, G. Cazzaniga, U. Lendahl, H.J. Fehling, A.C. Hayday, L. Frati, et al. 2002. Combined expression of pTalpha and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc. Natl. Acad. Sci. USA. 99:3788–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrando, A.A., D.S. Neuberg, J. Staunton, M.L. Loh, C. Huard, S.C. Raimondi, F.G. Behm, C.H. Pui, J.R. Downing, D.G. Gilliland, et al. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 1:75–87. [DOI] [PubMed] [Google Scholar]
- 33.Bellavia, D., A.F. Campese, E. Alesse, A. Vacca, M.P. Felli, A. Balestri, A. Stoppacciaro, C. Tiveron, L. Tatangelo, M. Giovarelli, et al. 2000. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 19:3337–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zong, W.X., L.C. Edelstein, C. Chen, J. Bash, and C. Gelinas. 1999. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev. 13:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Antwerp, D.J., S.J. Martin, I.M. Verma, and D.R. Green. 1998. Inhibition of TNF-induced apoptosis by NF-kappa B. Trends Cell Biol. 8:107–111. [DOI] [PubMed] [Google Scholar]
- 36.Edelstein, L.C., L. Lagos, M. Simmons, H. Tirumalai, and C. Gelinas. 2003. NF-kappa B-dependent assembly of an enhanceosome-like complex on the promoter region of apoptosis inhibitor Bfl-1/A1. Mol. Cell. Biol. 23:2749–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciofani, M., T.M. Schmitt, A. Ciofani, A.M. Michie, N. Cuburu, A. Aublin, J.L. Maryanski, and J.C. Zuniga-Pflucker. 2004. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J. Immunol. 172:5230–5239. [DOI] [PubMed] [Google Scholar]
- 38.O'Reilly, L.A., A.W. Harris, and A. Strasser. 1997. bcl-2 transgene expression promotes survival and reduces proliferation of CD3− CD4−CD8− T cell progenitors. Int. Immunol. 9:1291–1301. [DOI] [PubMed] [Google Scholar]
- 39.Linette, G.P., Y. Li, K. Roth, and S.J. Korsmeyer. 1996. Cross talk between cell death and cell cycle progression: BCL-2 regulates NFAT-mediated activation. Proc. Natl. Acad. Sci. USA. 93:9545–9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibasaki, F., E. Kondo, T. Akagi, and F. McKeon. 1997. Suppression of signalling through transcription factor NF-AT by interactions between calcineurin and Bcl-2. Nature. 386:728–731. [DOI] [PubMed] [Google Scholar]
- 41.Tan, S.L., and P.J. Parker. 2003. Emerging and diverse roles of protein kinase C in immune cell signalling. Biochem. J. 376:545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, Z., C.W. Arendt, W. Ellmeier, E.M. Schaeffer, M.J. Sunshine, L. Gandhi, J. Annes, D. Petrzilka, A. Kupfer, P.L. Schwartzberg, and D.R. Littman. 2000. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature. 404:402–407. [DOI] [PubMed] [Google Scholar]
- 43.Michie, A.M., J.W. Soh, R.G. Hawley, I.B. Weinstein, and J.C. Zuniga-Pflucker. 2001. Allelic exclusion and differentiation by protein kinase C-mediated signals in immature thymocytes. Proc. Natl. Acad. Sci. USA. 98:609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sicinska, E., I. Aifantis, L. Le Cam, W. Swat, C. Borowski, Q. Yu, A.A. Ferrando, S.D. Levin, Y. Geng, H. von Boehmer, and P. Sicinski. 2003. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 4:451–461. [DOI] [PubMed] [Google Scholar]
- 45.Cheng, E.H., M.C. Wei, S. Weiler, R.A. Flavell, T.W. Mak, T. Lindsten, and S.J. Korsmeyer. 2001. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell. 8:705–711. [DOI] [PubMed] [Google Scholar]
- 46.Yeoh, E.J., M.E. Ross, S.A. Shurtleff, W.K. Williams, D. Patel, R. Mahfouz, F.G. Behm, S.C. Raimondi, M.V. Relling, A. Patel, et al. 2002. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 1:133–143. [DOI] [PubMed] [Google Scholar]
- 47.Ory, D.S., B.A. Neugeboren, and R.C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA. 93:11400–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernandez-Hoyos, G., M.K. Anderson, C. Wang, E.V. Rothenberg, and J. Alberola-Ila. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 19:83–94. [DOI] [PubMed] [Google Scholar]