Abstract
Viral immune evasion strategies target key aspects of the host antiviral response. Recently, it has been recognized that Toll-like receptors (TLRs) have a role in innate defense against viruses. Here, we define the function of the vaccinia virus (VV) protein A46R and show it inhibits intracellular signalling by a range of TLRs. TLR signalling is triggered by homotypic interactions between the Toll-like–interleukin-1 resistance (TIR) domains of the receptors and adaptor molecules. A46R contains a TIR domain and is the only viral TIR domain–containing protein identified to date. We demonstrate that A46R targets the host TIR adaptors myeloid differentiation factor 88 (MyD88), MyD88 adaptor-like, TIR domain–containing adaptor inducing IFN-β (TRIF), and the TRIF-related adaptor molecule and thereby interferes with downstream activation of mitogen-activated protein kinases and nuclear factor κB. TRIF mediates activation of interferon (IFN) regulatory factor 3 (IRF3) and induction of IFN-β by TLR3 and TLR4 and suppresses VV replication in macrophages. Here, A46R disrupted TRIF-induced IRF3 activation and induction of the TRIF-dependent gene regulated on activation, normal T cell expressed and secreted. Furthermore, we show that A46R is functionally distinct from another described VV TLR inhibitor, A52R. Importantly, VV lacking the A46R gene was attenuated in a murine intranasal model, demonstrating the importance of A46R for VV virulence.
The discovery of the Toll-like receptor (TLR) family of proteins has revolutionized the understanding of how the innate immune system recognizes pathogens and initiates an effective and appropriate response (1, 2). TLRs are part of the larger IL-1R/TLR superfamily, which includes IL-1Rs, IL-18Rs, and a group of orphan receptors (3). The family is defined by the presence of a cytoplasmic Toll-like–IL-1 resistance (TIR) domain, which is responsible for mediating downstream signaling. So far, 13 TLRs have been identified; TLRs 1–9 are common to mouse and human, whereas TLR10 is only functional in humans, and TLRs 11, 12, and 13 have been found only in mice (2–5). Many but not all of these receptors have been assigned a role in the initial detection of, and response to, specific pathogen-associated molecules (PAMs). In macrophages and neutrophils, this drives innate immune responses, such as inflammation and induction of microbicidal activity, whereas activation of TLRs expressed on dendritic cells leads to the initiation of adaptive immunity through induction of IL-12 and costimulatory molecules (6, 7).
Although interest focused on the role of TLRs in responding to bacteria initially, there are now three strong lines of evidence that TLRs are also involved in detecting viruses and initiating antiviral responses. First, some TLRs are required for cellular and whole animal responses to certain viruses, viral proteins, and nucleic acids (8, 9). Second, TLRs trigger antiviral signaling pathways leading to the induction of IFN responses (8, 9). Third, viral immune strategies used against TLRs have been identified (8, 9).
Cell surface TLR2 and TLR4 may recognize viral glycoproteins on virions (8, 9). For example, measles virus hemagglutinin activates murine and human cells via TLR2, leading to induction of proinflammatory cytokines and up-regulation of surface expression of CD150, a receptor for measles virus (10). In terms of viral nucleic acids, TLR3 is activated in response to poly(I:C), a synthetic analogue of viral dsRNA (11), whereas TLR7 and TLR8 recognize ssRNA (from influenza, HIV, and vesicular stomatitis virus; references 12–14) and TLR9 responds to dsDNA from herpes simplex virus (15, 16).
That TLR signaling can induce an antiviral state was shown clearly by Doyle et al. (17) in that pretreatment of cells with poly(I:C) or lipid A (the moiety of LPS that is recognized by TLR4) inhibited the replication of a murine herpesvirus in macrophages. Even before the discovery of TLRs, it was known that viral replication and viral pathogenesis often involves NF-κB activation (18). TLRs, like IL-1, mediate downstream signaling mainly through their cytoplasmic TIR domain. This domain mediates homotypic interactions between TLRs, and also recruitment of TIR-containing adaptor proteins, of which myeloid differentiation factor 88 (MyD88) is a prototypical example. Recruitment of MyD88 to TLR complexes leads to activation of IL-1 receptor–associated kinases (IRAKs), which engage with TNF receptor–associated factor (TRAF) 6, leading ultimately to the activation of mitogen-activated protein (MAP) kinases and the transcription factor NF-κB (19, 20). MyD88 is involved in NF-κB activation by every TLR tested thus far, except for TLR3 (21, 22). MyD88 adaptor-like (Mal; reference 23), another TIR adaptor protein, is required specifically for TLR2 and TLR4 signaling (19, 20).
An important MyD88-independent signaling pathway is the activation of the antiviral transcription factor IFN regulatory factor 3 (IRF3) by TLR3 and TLR4. IRF3 activation, together with NF-κB activation, leads to IFN-β induction, which initiates the IFN-based antiviral response. TIR domain-containing adaptor inducing IFN-β (TRIF) is essential for MyD88-independent TLR3 and TLR4 signaling (21, 22, 24). TRIF activates IRF3 via TANK-binding kinase-1 (TBK1), leading to IFN-β expression (25, 26). TRIF probably associates directly with TLR3, but indirectly with TLR4, via a bridging interaction with a fourth adaptor that is unique to the TLR4 signaling pathway, TRIF-related adaptor molecule (TRAM, 27, 28).
Further evidence for the role of TLRs in responding to viruses came from the discovery of viral immune strategies used against TLRs. Vaccinia virus (VV), the poxvirus used to vaccinate against smallpox, encodes proteins that antagonize important components of host antiviral defense. Previously, we showed that VV protein A52R, which has no obvious similarity to host proteins, can block the activation of NF-κB by multiple TLRs, in particular TLR3 (29, 30). A52R associates with both IRAK2 and TRAF6, and disrupts signaling complexes containing these proteins (30). Furthermore, deletion of the A52R gene from VV reduced virus virulence (30).
In contrast with A52R, the VV protein investigated in this work, A46R, has a TIR domain, and as such is the only viral member of the IL-1R/TLR family identified to date (29). Initial studies revealed that A46R could inhibit IL-1, but not TNF-induced NF-κB activation (29). The effect on IL-1, together with the presence of a TIR domain in the protein suggested that A46R may have a role in immune evasion. However, the mechanism of action of A46R, its effect on TLR signaling pathways, and its potential role in virulence were not determined. Here, we show that A46R inhibits TLR-induced signaling, by associating with TIR-domain containing adaptor molecules. This is the first demonstration of direct viral targeting of TIR adaptors. We also demonstrate a role for A46R in VV virulence, and show that A46R and A52R are functionally distinct.
Results
A46R inhibits multiple IL-1–induced signals
Previously, we showed that A46R was capable of suppressing NF-κB activation induced by IL-1, whereas TNF-induced NF-κB was unaffected (29). To further characterize the effects of A46R on cell signaling, first we determined the effect of A46R on other IL-1–dependent signals. The effect of A46R on IL-1–, but not TNF-induced NF-κB–dependent reporter gene activation was confirmed (Fig. 1 a). To measure IL-1–induced transactivation of NF-κB specifically, an assay was performed using an expression plasmid encoding the transactivation domain of the p65 subunit of NF-κB fused to the DNA-binding domain of Gal4, together with a reporter plasmid under the control of a Gal4 upstream activation sequence (31). Reporter gene expression from this plasmid requires p65 transactivation through phosphorylation. Ectopic expression of A46R inhibited IL-1–induced p65 transactivation, but had no effect on basal levels of activity (Fig. 1 b). The effect of A46R on p65 transactivation provided a rationale for the inhibition of the NF-κB–dependent reporter gene. In a similar assay (23), A46R blocked both c-Jun NH2-terminal protein kinase (JNK; Fig. 1 c) and extracellular signal–regulated kinase (ERK; Fig. 1 d) activation induced by IL-1. Thus, A46R inhibited multiple distinct signals emanating from the IL-1 receptor.
Figure 1.

A46R inhibits multiple IL-1–dependent signals. HEK 293 cells were transfected with 100 ng A46R or pcDNA3.1 (EV) and the NF-κB (a), p65 (b), JNK (c), or ERK (d) reporter plasmids as described in Materials and methods. 6 h before harvesting, the cells were stimulated with either 100 ng/ml IL-1 or 100 ng/ml TNF as indicated, and luciferase reporter gene activity was measured.
A46R interacts with MyD88 and antagonizes MyD88-dependent signaling
The results from Fig. 1 suggest that A46R was acting close to the IL-1 receptor complex. This was also likely given that A46R has a TIR domain (29), as illustrated in Fig. 2 a, which shows an alignment of VV and variola virus A46R with several human TIR-containing proteins. Given that all IL-1 signals tested to date are dependent on the TIR domain-containing adaptor MyD88 (32, 33), including p65 transactivation (31) and JNK activation (34), we reasoned that A46R may target MyD88 via a TIR domain interaction. To test this hypothesis, coimmunoprecipitation studies were performed with A46R and MyD88 expressed ectopically. Fig. 2 b shows a clear interaction between A46R and MyD88 (left). As a control, the ability of A46R to interact with TRAF2, an adaptor used by TNF but not IL-1 (35), was tested in parallel. In this case, no interaction was detected (Fig. 2 a, right), consistent with the lack of effect of A46R on TNF signaling (Fig. 1 a). These results were confirmed by GST-pulldown experiments, whereby purified GST-A46R interacted with ectopically expressed MyD88 in a cell lysate, but not with TRAF2 (Fig. 2 c). A46R also failed to interact with TAB1, a signaling molecule downstream of MyD88 (unpublished data). The interaction of A46R with MyD88 was also demonstrated in cells infected by VV and transfected to express MyD88, to use the A46R protein at its physiological concentration (Fig. 2 d). As a control for specificity, this was compared with a VV deletion mutant lacking the A46R gene (vΔA46R), whereupon no band for A46R was detected (Fig. 2 d). A46R also interacted with MyD88 in a yeast two-hybrid pairwise assay (unpublished data), thus demonstrating that the association was direct.
Figure 2.
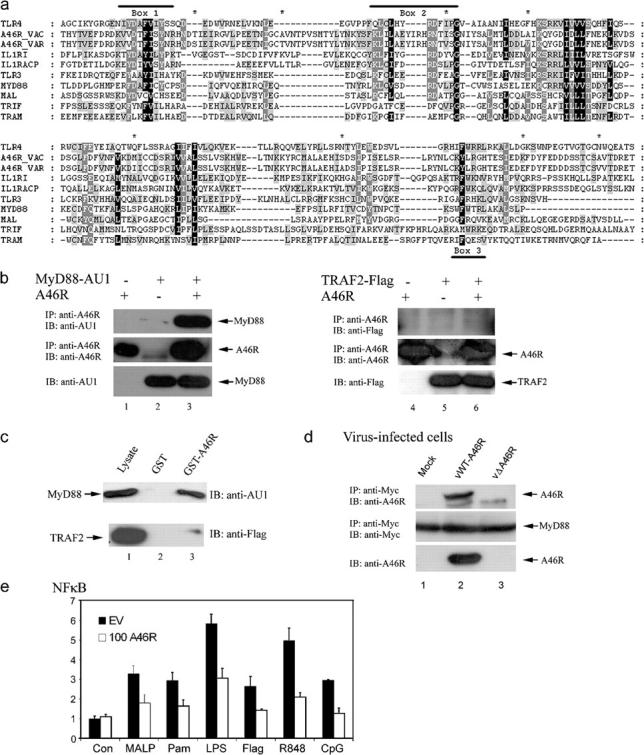
A46R associates with MyD88 and blocks MyD88-dependent signaling. (a) Alignment of A46R with human TIR domains. The conserved motifs Box 1, Box 2, and Box 3 are indicated by a solid line. The eight differences between VV and variola virus A46R sequences are indicated by an asterisk. For A46R, amino acids 35–238 are shown. (b) HEK 293T cells were transfected with A46R and AU1-MyD88 (left) or Flag-TRAF2 (right) as indicated. After 24 h, lysates were subject to immunoprecipitation, SDS-PAGE, and immunoblotting with the indicated antibodies. (c) HEK 293T cells were transfected with 8 μg AU1-MyD88 (top) or Flag-TRAF2 (bottom). After 24 h, lysates were incubated with GST alone (lane 2) or GST-A46R (lane 3), and together with whole cell lysates (lane 1) were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (d) HEK 293 cells were transfected with 4 μg of myc-MyD88. After 24 h, cells were infected with viruses either containing (vWT-A46R) or not (vΔA46R) the A46R gene (MOI = 1) and harvested 24 h after infection. Lysates were subjected to immunoprecipitation, SDS-PAGE, and immunoblotting with the indicated antibodies. Whole cell lysates were analyzed for expression of A46R. (e) Murine macrophage RAW 264.7 cells were transfected with the phRL-TK reporter gene and the NF-κB luciferase construct as described in Materials and methods, together with pcDNA3.1 or 100 ng A46R. Cells were stimulated for 6 h with 10 nM MALP-2 (MALP), 5 μg/ml Pam3Cys (Pam), 1 μg/ml LPS, 250 ng/ml Flagellin (Flag), 1 μM R-848, or 5 μg/ml CpG DNA. Cells were harvested 24 h after transfection and the reporter gene activity was measured.
Apart from its role in IL-1R signaling, MyD88 is also used by murine TLRs 1, 2, 4, 5, 6, 7, and 9 (2). Therefore, we tested the effect of A46R on signaling via these TLRs in RAW264.7 cells, a murine macrophage cell line that expresses most TLRs (36). Stimulation of RAW264.7 cells with MALP-2 (TLR2 and 6), Pam3Cys (TLR2 and 1), LPS (TLR4), flagellin (TLR5), R848 (TLR7), or CpG DNA (TLR9) led to induction of the NF-κB–dependent reporter gene (Fig. 2 e). In each case, the presence of A46R caused strong inhibition of induction, whereas it had no suppressive effect on control levels (Fig. 2 e). Thus, A46R inhibited MyD88-dependent signaling by both IL-1R and TLRs, presumably by interacting with MyD88 via its TIR domain.
A46R targets the TLR4 receptor complex
The effect of A46R on TLR4 signaling was examined in greater detail. A chimeric form of TLR4, comprising the murine CD4 extracellular domain fused to the cytoplasmic domain of human TLR4, which renders TLR4 constitutively active (37), was used. Overexpression of CD4-TLR4 induced NF-κB, and this was inhibited by coexpression of A46R in a dose-dependent manner (Fig. 3 a, left). In fact, the highest concentration of A46R-expressing plasmid almost completely prevented TLR4-induced NF-κB activation, whereas it did not suppress basal levels of reporter gene activity (Fig. 3 a, left, white bars). TLR4-induced activation of the MAP kinases p38 and ERK was also inhibited by A46R expression (Fig. 3, middle and right, respectively).
Figure 3.
A46R inhibits TLR4 signaling and interacts with TLR4, Mal, and TRAM. (a) HEK 293 cells were transfected with 50 ng CD4-TLR4, 25–100 ng A46R, or pcDNA3.1 (EV) and the NF-κB (left), p38 (middle), or ERK (right) reporter plasmids as indicated. Cells were harvested 24 h after transfection, and luciferase reporter gene activity was measured. (b–d) HEK 293T cells were transfected with A46R and Flag-TLR4 (b), Flag-Mal (c), or Flag-TRAM (d) as indicated. After 24 h, lysates were subject to immunoprecipitation, SDS-PAGE, and immunoblotting with the indicated antibodies. (e) HEK 293T cells were transfected with 8 μg of Flag-TLR4 (top), Flag-Mal (middle), or Flag-TRAM (bottom). After 24 h, lysates were incubated with GST-A46R (lane 3) or GST alone (lane 2), and together with whole cell lysate (lane 1), were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. (f) HEK-TLR4 cells were transfected with the indicated amounts (ng) of A46R 24 h before stimulation with 1 μg/ml LPS, and 24 h after stimulation, supernatants were harvested and assayed for IL-8 by ELISA.
Given that TLR4-induced NF-κB, p38, and ERK are only partially MyD88 dependent (38), it was difficult to account for the potent effects of A46R on these signals by an interaction with MyD88 alone. Therefore, we assessed the ability of A46R to target other TIR domain-containing proteins involved in the TLR4 receptor complex. Fig. 3 b shows that A46R could be immunoprecipitated with TLR4 itself, suggesting that A46R can also interact with the TLR4 TIR domain. Next, we tested the ability of A46R to target two important TLR4 TIR adaptors, Mal and TRAM. Mal and TRAM are thought to interact directly with TLR4, and subsequently recruit MyD88 and TRIF, respectively (23, 27, 39). Both Mal and TRAM are essential for robust TLR4-dependent NF-κB activation (19, 23, 27, 28). Consistent with this, A46R immunoprecipitated with both Mal (Fig. 3 c) and TRAM (Fig. 3 d). The interactions with TLR4, Mal, and TRAM were confirmed by GST-pulldown experiments (Fig. 3 e). Furthermore, Mal was shown to coimmunoprecipitate with A46R in infected cells, associate directly with A46R in the yeast two-hybrid pairwise assay, and rMal interacted with GST-A46R in vitro (unpublished data).
Next, we demonstrated that the inhibitory effects of A46R on TLR4 signaling pathways also resulted in a suppression of gene induction. Fig. 3 f shows that LPS stimulation of HEK293 cells expressing TLR4 led to the release of IL-8, which is NF-κB and p38 dependent (not depicted). Transient transfection of these cells with A46R suppressed IL-8 release by ∼50%. Thus, the viral TIR domain of A46R can target multiple human TIR domain-containing proteins important for TLR4 signaling, leading to antagonism of signaling pathways and subsequent suppression of gene induction.
A46R antagonizes TRIF-dependent pathways
TRIF was identified as a TIR adaptor molecule capable of directing both TLR4- and TLR3-induced IRF3 activation, leading to IFN-β induction, which is independent of MyD88 (21, 22, 24). The MyD88-independent late activation of NF-κB by TLR4 was also explained by the discovery of TRIF (21). Given the central role of TRIF in the activation of the antiviral transcription factor IRF3, we wondered whether A46R would also target this TIR adaptor directly. Co-expression of A46R and TRIF, with subsequent coimmunoprecipitation, demonstrated that these two proteins could indeed form a complex (Fig. 4 a). An interaction between A46R and TRIF was confirmed by a GST-pulldown experiment (Fig. 4 b) and also by a yeast two-hybrid pairwise assay (not depicted).
Figure 4.
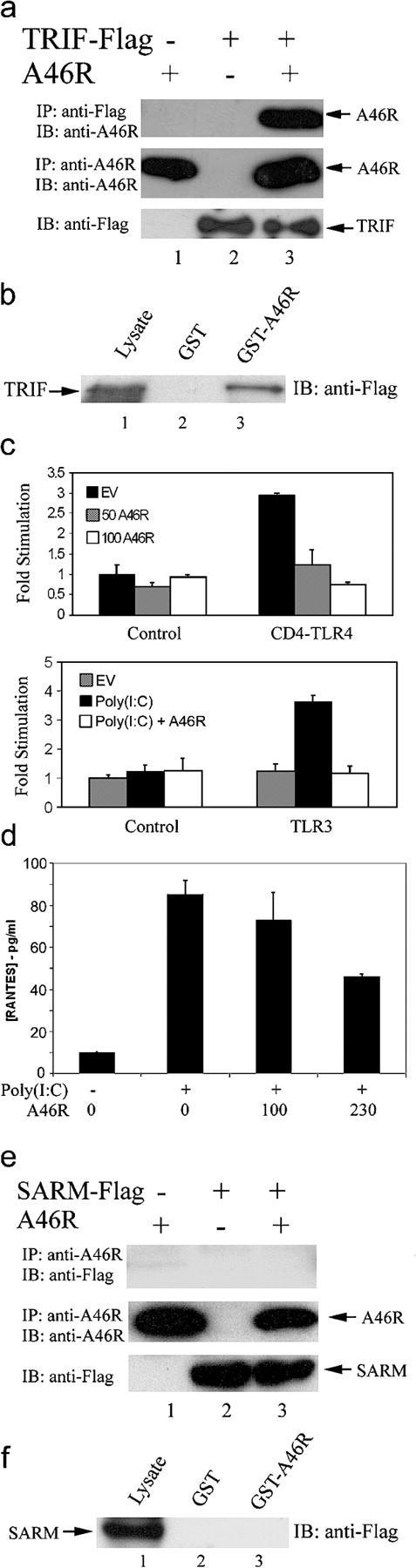
A46R associates with TRIF and inhibits TRIF-dependent signaling and gene induction. (a) HEK 293T cells were transfected with A46R and Flag-TRIF as indicated. After 24 h, lysates were subject to immunoprecipitation, SDS-PAGE, and immunoblotting with the indicated antibodies. (b) HEK 293T cells were transfected with 8 μg Flag-TRIF. After 24 h, lysates were incubated with GST-A46R (lane 3) or GST alone (lane 2), and together with whole cell lysates (lane 1), were analyzed by SDS-PAGE and immunoblotting with anti-Flag Ab. (c) HEK 293 cells were transfected with the IRF3 reporter plasmids (as described in Materials and methods) with either 50 ng CD4-TLR4 (top) or 0.5 ng TLR3 (bottom) and 50–150 ng A46R or pcDNA3.1 (EV) as indicated. (bottom) Cells were stimulated with 25 μg/ml poly(I:C) 6 h before harvesting. Luciferase activity was measured after 24 h. (d) HEK-TLR3 cells were transfected with the indicated amounts (ng) of A46R 24 h before stimulation with 25 μg/ml poly(I:C), and 24 h after stimulation supernatants were harvested and assayed for RANTES by ELISA. (e and f) As in a and b, except Flag-SARM was transfected instead of Flag-TRIF.
To determine the functional relevance of this interaction, the effect of A46R on TRIF-dependent signals was examined. For this, IRF3 activation by TLR4 and TLR3 was assessed by using a Gal4–IRF3 fusion protein together with a Gal4-dependent reporter plasmid, which requires IRF3 transactivation to express the reporter gene (27). Ectopic expression of CD4-TLR4 resulted in a threefold induction of this IRF3-dependent reporter, and this was completely blocked by coexpression of A46R (Fig. 4 c, top). Although the interaction of A46R with TLR4 and TRAM probably contributes to this inhibitory effect, induction of an IFN-β promoter reporter gene by ectopic expression of TRIF was also blocked by A46R (unpublished data), suggesting a direct effect on TRIF, which is downstream of TLR4 and TRAM. Further TLR3-dependent activation of IRF3 by poly(I:C), which is entirely TRIF dependent, was also completely blocked by A46R (Fig. 4 c, bottom). A46R also inhibited LPS and poly(I:C)-mediated induction of an ISRE-dependent reporter (unpublished data). In addition, no interaction between A46R and TLR3 was detected (unpublished data). Furthermore, poly(I:C)-induced regulated on activation, normal T cell expressed and secreted (RANTES) release from HEK293 cells stably expressing TLR3 was inhibited by A46R (Fig. 4 d). This represents inhibition of gene induction that is entirely independent of MyD88 (27). Therefore, A46R blocks TLR-induced IRF3 activation and subsequent gene induction by directly targeting TRIF.
Thus, A46R was capable of interacting with four TIR adaptors known to have a key role in IL-1R and TLR-induced signaling. In contrast, A46R did not interact with the fifth human intracellular TIR domain-containing protein, sterile α and HEAT/Armadillo motifs–containing protein (SARM) in a coimmunoprecipitation (Fig. 4 e) or GST-pulldown (Fig. 2 f) assay. This is consistent with the fact that SARM does not lead to NF-κB or IRF3 activation (40), and provided an important specificity control for A46R interactions.
A46R is expressed early during infection and contributes to VV virulence
To determine when A46R was expressed during the virus life cycle, cells were infected for different lengths of time in the presence or absence of cytosine β-D-arabinofuranoside (AraC) (an inhibitor of virus DNA replication and, therefore, of intermediate and late genes) and analyzed by immunoblotting. Fig. 5 a (top) shows that A46R expression was detected in the presence of AraC, whereas a late protein (D8L) was not (Fig. 5 a, bottom), showing early expression. A46R remained in infected cells up to 24 h after infection.
Figure 5.
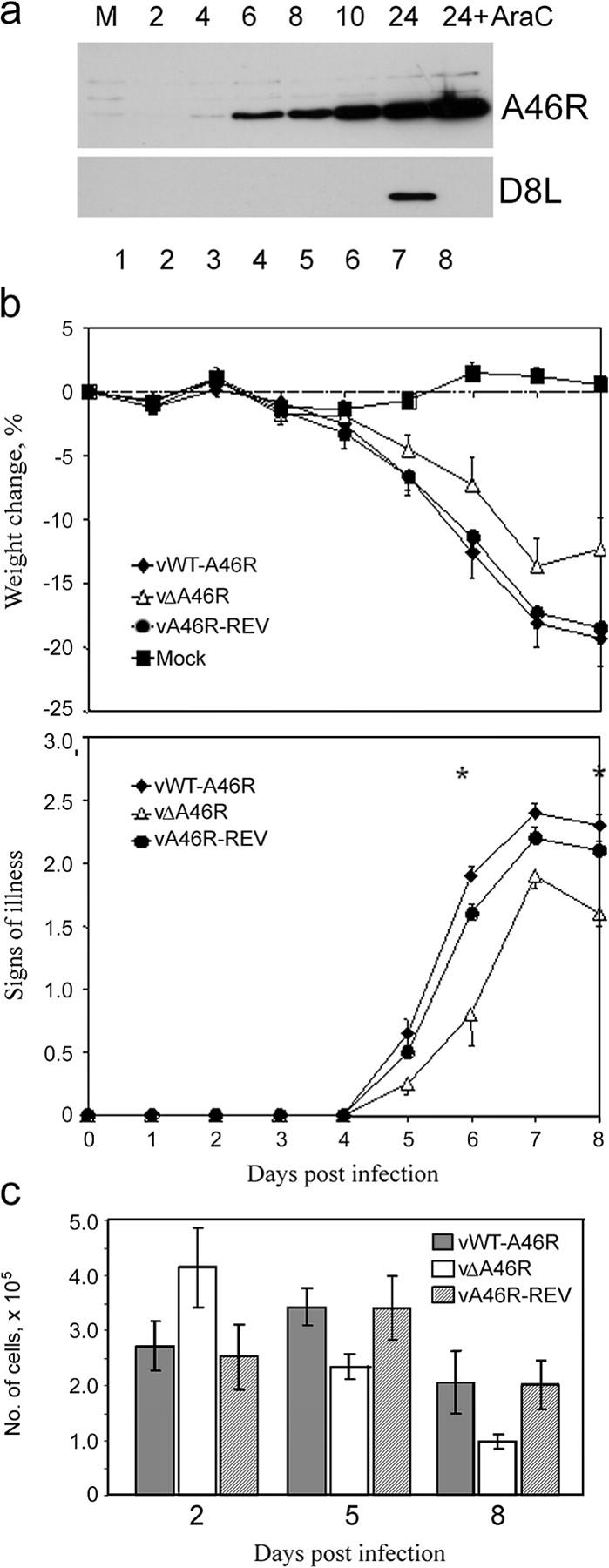
A46R is expressed early during virus infection and contributes to virus virulence. (a) BSC-1 cells were mock infected (M) or infected with VV Western reserve (MOI = 5) in the absence or presence of 40 μg/ml cytosine β-D-arabinofuranoside (AraC). Cells were harvested at the indicated times (h) after infection, and lysates were subjected to SDS-PAGE and immunoblotting, using either anti-A46R (top) or anti-D8L (bottom) Ab. (b) A46R contributes to virus virulence. Groups of 15 female, 6-wk-old Balb/c mice were infected intranasally with 5 × 103 PFU of vWT-A46R, vΔA46R, or vA46R-REV. Each day, animals were weighed and the signs of illness were scored. Data are presented as the mean weight of each group of animals compared with the mean weight of the same group on day 0 (top graph), and the mean signs of illness score (bottom graph). Error bars are SEM. Asterisks represent days on which there was a statistically significant difference (P < 0.05; Student's t test) between the vWT-A46R and both control groups. (c) Number of cells recruited to the lungs of VV-infected mice. On days 2, 5, and 8 after infection, mice were killed, lungs were harvested, and cell suspensions were prepared. The total number of viable lung cells per animal was determined by Trypan blue exclusion. Data are means ± SEM.
To explore the role of A46R in VV virulence, a VV deletion mutant lacking the A46R gene was constructed (vΔA46R). As a control, a revertant virus was also constructed in which the A46R gene was reinserted into the vΔA46R mutant (vA46R-REV). Western blot analysis confirmed expression of A46R in wild-type virus (vWT-A46R) and vA46R-REV, but not in vΔA46R (Fig. 2 d and not depicted). Mice were infected intranasally with vΔA46R, vA46R-REV, or vWT-A46R and were weighed and assessed for signs of illness daily. Fig. 5 b shows that vΔA46R was attenuated relative to both vWT-A46R and vA46R-REV in terms of reduced weight loss (Fig. 5 b, top graph) and milder signs of illness (bottom graph). Assessment of the total number of cells in lungs after infection revealed a difference in the kinetics of the host response to vΔA46R compared with vA46R-REV or vWT-A46R, in that when the virus lacked A46R, the number of cells present on day two was increased, whereas on days 5 and 8, it was reduced. The difference in cell recruitment between the vWT-A46R and vΔA46R on day 5 was statistically significant (P < 0.05). The p-value for the difference between vWT-A46R and vΔA46R on day 8 was 0.09, whereas the value for the difference between vA46R-REV and vΔA46R was 0.04.
A46R and A52R are not functionally redundant
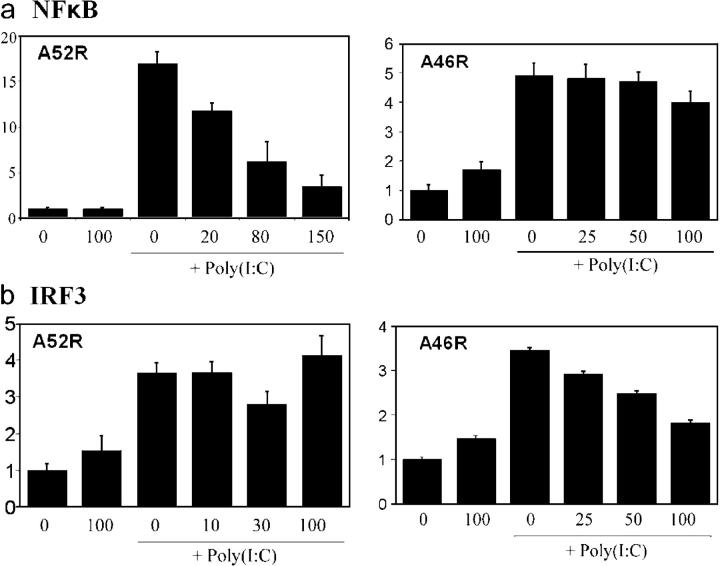
Previously, we showed that the poxviral protein A52R could also inhibit TLR signaling and contribute to virulence (30). A52R does not resemble any host proteins, but does have some similarity to A46R (29, 30). A52R blocked TLR-induced NF-κB activation by targeting TRAF6 and IRAK2, which act downstream of TIR adaptors (30). Therefore, we looked in more detail at the effects of both proteins on signaling by a single TLR, to ascertain whether or not they were functionally redundant. We chose TLR3, given its proposed role in the antiviral response, and also because it signals via a single TIR adaptor, TRIF, thus making the interpretation of the results more definitive. Analysis of NF-κB and IRF3 activation by TLR3 demonstrated a clear difference between A46R and A52R. Fig. 6 a shows that A52R was a potent inhibitor of poly(I:C)/TLR3-induced NF-κB activation, whereas A46R had little effect. In contrast, Fig. 6 b shows that IRF3 activation induced by poly(I:C)/TLR3 was sensitive to A46R, but not A52R. Furthermore, A52R was not capable of inhibiting IL-1 or TLR4-induced MAP kinase activation (not depicted), which were clearly blocked by A46R (Fig. 1, c and d, and Fig. 3, a and b). Hence, A46R and A52R are not functionally redundant in that both are required to effectively shut down TLR3 signaling, whereas only A46R is capable of inhibiting TLR-induced MAP kinase activation.
Figure 6.
A46R and A52R target different TLR3-mediated signaling pathways. (a and b) HEK 293 cells were transfected with 0.5 ng TLR3 and the indicated amounts (ng) of either A52R or A46R, together with the NF-κB (a) or IRF3 (b) reporter plasmids as described in Materials and methods. Cells were stimulated with 25 μg/ml poly(I:C) 6 h before harvesting where indicated. Luciferase activity was measured after 24 h. Relative stimulation is shown on the y axis.
The results demonstrate that A46R has a viral TIR domain with which it targets TIR adaptor molecules, resulting in inhibition of both MyD88-dependent and TRIF-dependent signaling and gene induction. Furthermore, A46R is functionally distinct from A52R and contributes to VV virulence, most likely due to its inhibitory effects on TIR-dependent signaling.
Discussion
The identification of viral immune evasive strategies and the analysis of the molecular aspects of host–pathogen interactions are crucial to enhancing understanding of microbial pathogenesis and immunity to infection. Given the emerging importance of the TLR system in the antiviral response, understanding how viruses target this receptor family is of particular interest. During a database search for novel TIR domain–containing proteins, A46R from VV was identified (29). This was potentially interesting because many poxviral immunomodulatory proteins, such as cytokine-binding proteins, bear sequence similarity to host factors (41). To date, A46R is the only identified viral TIR domain–containing protein. In this paper, we show that A46R is an intracellular inhibitor of multiple TLR-dependent signaling pathways, define host signaling molecules that it targets, and demonstrate that the protein contributes to VV virulence in vivo.
The observation that A46R blocked all IL-1 signals tested (NF-κB, JNK, and ERK activation) suggested that it was acting close to the IL-1R complex. Furthermore, the presence of a TIR domain within A46R, the knowledge that TIR domains participate in homotypic interactions, and the fact that all the signals blocked by A46R were MyD88 dependent (31–34), suggested that MyD88 may be sequestered by A46R. Coimmunoprecipitation and GST-pulldown experiments demonstrated an interaction between A46R and MyD88. This also suggested that A46R would antagonize TLR signaling, given the central role of MyD88 in many of these pathways. In fact, A46R inhibited every murine TLR pathway to NF-κB activation known to involve MyD88 in a mouse macrophage cell line, together with TLR4-mediated NF-κB, p38, and ERK activation, and IL-8 induction in human 293 cells.
Inhibition of TLR4-dependent NF-κB signaling in human cells was particularly potent, which led us to test the effect of A46R on other TIR domain–containing proteins with a role in this pathway. Altogether, five such proteins are known to be involved in TLR4-mediated NF-κB activation, namely the receptor itself, MyD88, Mal, TRAM, and TRIF (19–23, 28). A46R was found to be capable of associating with all five of these proteins, and hence it probably prevents the formation of the TLR4 receptor complex, thus accounting for its potent inhibition of NF-κB activation. Although these results were based on overexpression, several lines of evidence suggest that the interactions detected are specific, direct, and likely to occur in vivo. First, normally VV-expressed A46R interacted with MyD88 and Mal. Second, the interaction of A46R with MyD88, Mal, and TRIF were confirmed in yeast two-hybrid. Third, rMal interacted directly with GST-A46R in vitro. Finally, A46R displayed specificity for certain TIR domain–containing proteins and did not interact with TLR3 or SARM.
A46R is the first viral protein identified that can target host TIR domain–containing proteins. Given its ability to interact with different and diverse TIR domains (TRIF and TRAM are quite distinct in sequence from MyD88 and Mal; Fig. 1 a and reference 20), elucidation of the crystal structure of A46R should provide important information of general relevance as to how TIR domains interact. Because Box 2 of the TIR domain is particularly important in signaling (42), the extra amino acids surrounding the A46R Box 2 (Fig. 1 a) may represent inhibitory loops that account for the ability of A46R to prevent TIR-dependent signaling. This hypothesis is being explored using mutagenesis studies.
The fact that A46R associated with all four TLR4 adaptors may suggest that TLR4 is a particularly important target for VV immune evasion. Indeed, TLR4 has been proposed to have a role in responding to fusion (F) protein of respiratory syncytial virus (43), although the functional significance of this remains to be clarified (44). TLR4 is also activated by envelope proteins from both mouse mammary tumor virus and Moloney murine leukemia virus, which could also be coimmunoprecipitated with TLR4 (45). VV might interact with other or multiple TLRs that also use these adaptors. Possible VV PAMs detected by TLRs could be proteins on the surface of the intracellular mature virus or extracellular enveloped virus particles (potentially detected by TLR2 or TLR4; references 8, 9), intracellular dsRNA produced from the bidirection transcription of the VV genome (potentially detected by TLR3; reference 11), or the dsDNA genome itself (potentially detected by TLR9, which responds to the dsDNA genome of herpes simplex virus; reference 15, 16). The role of TLRs in responding to VV PAMs is currently being investigated.
Although Mal and TRAM are yet to be directly implicated in responding to viruses, MyD88 (for TLR7; references 8, 9) and TRIF (for TLR3) have been shown to have a role, particularly in relation to type I IFN production (15, 16, 21, 22). TRIF is essential for both TLR4- and TLR3-mediated IRF3 activation and IFN-β production (21, 22) and probably has a key role in the initiation of the IFN-based antiviral response, leading to the inhibition of viral replication and spread. Although the relevance of TLR3 in responding to viruses in vivo has recently been questioned (46, 47), there is evidence that TRIF is important in controlling VV replication. Hoebe et al. (22) showed that macrophages from mice in which trif was disrupted supported the replication of VV to a higher titre than did macrophages from normal mice. Thus, the ability of A46R to inhibit TRIF-mediated IRF3 activation may be of primary importance to VV infection.
Consistent with its proposed role in antagonizing early innate TLR responses, A46R was expressed early during infection. Furthermore, deletion of A46R from VV caused attenuation in a murine intranasal model, and also led to enhanced levels of cells on day 2 in lungs of infected animals. The use of a revertant virus in which the A46R gene was reinserted into the deletion virus confirmed that the attenuation observed in the deletion virus, and the difference in lung cell numbers, were due solely to the absence of A46R. The ability of A46R to target intracellular TIR-dependent signaling most likely accounts for its role in virulence, probably by blocking the induction of immune response genes downstream of TLRs, as has been shown here for the chemokines IL-8 and RANTES. Possibly, inhibition of chemokine induction by A46R might account for the early enhanced levels of cells in the absence of A46R expression.
Previously, we identified another VV TLR antagonist, A52R, which could inhibit TLR-dependent NF-κB activation. There are several lines of evidence that A46R and A52R are not functionally redundant. First, they target distinct TLR signaling molecules (30). Second, although they do have some overlapping effects (such as the inhibition of MyD88-dependent NF-κB activation), their overall effects on TLR signaling are quite distinct. Although A52R is a good NF-κB inhibitor, it has no inhibitory effect on MAP kinase activation (not depicted), nor on TLR3-mediated IRF3 activation (Fig. 6 b). In contrast, A46R blocks both MAP kinase activation and, importantly, TLR3- and TRIF-mediated IRF3 activation. Furthermore, A46R has little effect on TLR3-mediated NF-κB activation, which A52R blocks potently (Fig. 6 a). This was surprising because A46R interacts with TRIF. But, presumably, the interaction of A46R with TRIF has a greater effect on downstream IRF3 activation compared with NF-κB because these two pathways bifurcating from TRIF have been shown to be quite distinct (48, 49). Finally and crucially, the deletion of either A46R or A52R from VV causes attenuation (Fig. 5 b and reference 30) and, thus, both contribute to virulence and are nonredundant.
N1L is another intracellular VV protein that contributes to virulence (50), which has also been shown recently to function by antagonizing TLR signaling, but at the level of IκB kinases and related kinases involved in IRF3 activation (51). Thus, the importance of blocking TLR signaling is demonstrated by the retention by VV of at least three distinct mechanisms of disrupting these pathways. Furthermore, the attenuated phenotypes seen in the absence of A46R, A52R, or N1L provide evidence for a role for TLRs in containing VV infections. The action of these intracellular TLR inhibitory proteins would be expected to be restricted to infected cells, whereas immunomodulatory proteins secreted from VV-infected cells, such as cytokine, chemokine, and IFN-binding proteins, can act on ligands produced from both infected and uninfected cells.
Finally, A46R is also found in variola virus, the causative agent of smallpox. Concern about the threat of the use of variola as a bioweapon has led to a renewed desire to understand this human pathogen. However, little is known about the role of human TLRs in sensing variola virus. Given that VV A46R targets human adaptors, the knowledge that A52R is truncated in variola virus, together with the fact that the VV and variola virus A46R amino acid sequences differ by only eight residues (Fig. 2 a), it is likely that variola virus A46R would have an important role in interactions with the human TLR system.
Materials and Methods
Expression plasmids
Sources of expression plasmids were as follows: AU1-MyD88 and Flag-TLR4 (M. Muzio, Mario Negri Institute, Milan, Italy; references 52, 53), flag-TRAF2 (Tularik Inc.), chimeric receptor CD4-TLR4 (R. Medzhitov, Yale University, New Haven, CT), TLR3 (D. Golenbock, University of Massachusetts Medical School, Worcester, MA), Flag-TRIF (S. Akira, Osaka University, Osaka, Japan), and Myc-MyD88 (L. O'Neill, Trinity College, Dublin, Ireland). Construction of A46R, Flag-A46R, HA-Mal, Flag-TRAM, and Flag-SARM were described previously (23, 27, 29, 40). The glutathione S-transferase (GST) fusion of A46R was synthesized by inserting full-length A46R in the bacterial expression vector GEX4T2.
Antibodies and reagents
Anti-A46R polyclonal Ab was raised against a purified, bacterial-expressed A46R–GST fusion protein. Other antibodies used were anti-Flag M2 mAb, anti-Flag M2–conjugated agarose, anti-myc mAb clone 9E10 (all obtained from Sigma-Aldrich), anti-AU1 mAb (BabCO), anti-HA polyclonal Ab (Y-11), anti-TRAF2 Ab (both obtained from Santa Cruz Biotechnology, Inc.), and anti-D8L mAb (54).
Human rIL-1α was a gift from the National Cancer Institute and human rTNF-α was a gift from S. Foster (Zeneca Pharmaceuticals, Macclesfield, England). TLR agonists used were poly(I:C) (Amersham Biosciences), LPS (Sigma-Aldrich), R-848 (a gift from D. Golenbock, University of Massachusetts Medical School, Worcester, MA), flagellin (a gift from A. Gewirtz, Emory University, Atlanta, GA), phosphothioate CpG DNA (Sigma-Aldrich), synthetic tripalmitoyl lipopeptide Pam3Cys-Ser-(Lys)4 (Pam3Cys; Invivogen), and macrophage-activating lipopeptide 2 kD (MALP-2; Qbiogene).
Reporter gene assays
HEK 293 cells (2 × 104 cells per well) or RAW264.7 cells (4 × 104 cells per well) were seeded into 96-well plates and transfected 24 h later with expression vectors and luciferase reporter genes using GeneJuice (Novagen). In all cases, 20 ng/well of phRL-TK reporter gene (Promega) was cotransfected to normalize data for transfection efficiency. The total amount of DNA per transfection was kept constant at 220 ng (HEK293) or 200 ng (RAW264.7) by addition of pcDNA3.1 (Stratagene). After 24 h, reporter gene activity was measured (30). Data are expressed as the mean fold induction ± SD relative to control levels, for a representative experiment from a minimum of three separate experiments, each performed in triplicate.
For NF-κB assays, 60 ng of a κB-luciferase reporter gene was used (30). For MAP kinase reporter assays, the Pathdetect System (Stratagene) was used, whereby 0.25 ng c-jun–, 2 ng Elk1–, or 0.25 ng CHOP–Gal4 fusion vectors were used in combination with 60 ng pFR-luciferase reporter to measure JNK, ERK1/2, and p38 activation, respectively. For the p65 transactivation assay, 1 ng of a p65 Gal4 fusion vector was used in combination with 60 ng pFR-luciferase reporter (31). For the IRF3 assay, an IRF3-Gal4 fusion vector (3 ng) was used in combination with 60 ng pFR luciferase reporter (27).
Immunoprecipitation and immunoblotting
HEK293 cells were seeded into 10-cm dishes (1.5 × 106 cells) 24 h before transfection with GeneJuice. For coimmunoprecipitations, 4 μg of each construct was transfected. Cells were harvested after 24 h in 850 μl of lysis buffer (50 mM Hepes, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40 containing 0.01% aprotinin, 1 mM sodium orthovanadate, and 1 mM PMSF). For assessment of the A46R–TLR4 interaction, the cells were harvested after 48 h and lysed for 1 h in a different lysis buffer (50 mM Tris/HCl, pH 7.5, 1 mM EDTA, 10% glycerol, 0.05% CHAPS. 0.5% Triton X-100, 250 mM NaCl containing 0.01% aprotinin, 1 mM sodium orthovanadate, and 1 mM PMSF). For assessment of interactions involving VV-expressed A46R, cells were infected (multiplicity of infection [MOI] = 1) with VV Western Reserve 24 h after transfection for 90 min at 37°C. The virus inoculum was aspirated and cell monolayers were overlaid with 2.5% FBS DMEM and harvested 24 h after infection in lysis buffer. For all immunoprecipitations, the appropriate antibodies were precoupled to either protein A or protein G–Sepharose for 1 h at 4°C before incubation with the cell lysates overnight at 4°C. The immune complexes were precipitated, washed, and analyzed by SDS-PAGE and immunoblotting (30).
For analysis of the kinetics of A46R expression, confluent monolayers of BSC-1 cells were infected (MOI = 5) for 90 min at 37°C. After removal of inoculum, cell monolayers were overlaid with 2.5% FBS DMEM in the presence or absence of 40 μg/ml cytosine β-D-arabinofuranoside (Sigma-Aldrich). Cell lysates were analyzed by immunoblotting using anti-A46R or anti-D8L Ab.
GST pulldown assays
Plasmid GEX.4T2-A46R or GEX.4T2 was transformed into Escherichia coli BL21 (DE3) and grown in Terrific Broth. Protein expression was induced with 0.7 mM IPTG. Cells were lysed in NETN and proteins were purified from the insoluble fraction by glutathione sepharose 4B affinity chromatography (Amersham Biosciences).
For GST pulldown experiments, HEK 293T cells were transfected and harvested as described for coimmunoprecipitation. 800 μl of cell lysate was added to purified GST-fusion protein coupled to glutathione-sepharose and incubated for 2 h at 4°C. The immune complexes were precipitated, subjected to SDS-PAGE, and were analyzed by immunoblotting.
Determination of cytokine concentrations
HEK293 clonal cell lines expressing either TLR3 (HEK-TLR3) or TLR4 and MD-2 (HEK-TLR4; reference 27) and were used for determination of cytokine production. Cells (2 × 104 cells per well) transfected with the A46R expression plasmid for 24 h were stimulated with 1 μg/ml LPS or 25 μg/ml poly(I:C) 24 h later. Supernatants were harvested 24 h later and IL-8 and RANTES concentrations were determined by ELISA (R&D Systems). Experiments were performed four times in triplicate and data are expressed as the mean ± SD from one representative experiment.
Recombinant VV viruses
A VV mutant (strain WR) lacking 93.5% of the A46R gene (vΔA46R) was constructed by transient dominant selection (55). A plaque-purified wild-type virus (vWT-A46R) and a revertant virus (vA46R-REV) in which the A46R gene was reinserted at its natural locus were also isolated. The virulence of the viruses was investigated in a mouse intranasal model. Female, 6-wk-old BALB/c mice were anesthetized and inoculated with 5 × 103 plaque-forming units of VV in 20 μl of phosphate-buffered saline. A control group was mock-infected with phosphate-buffered saline. Each day, the weights of the animals and signs of illness were measured as described previously (56). On days 2, 5, and 8, single cell suspensions of lung cells were prepared by sieving lungs through a 100-μm nylon mesh followed by hypotonic lysis of erythrocytes. Cell viability was assessed using trypan blue exclusion. Statistical significance was assessed using Student's t test. The animal experiments were conducted under the appropriate licence and regulations stipulated by the Animals (Scientific Procedures) Act 1986, UK government.
Alignment of TIR domains
TIR domains from human proteins with assigned functions were aligned with VV and variola virus A46R using Clustal W. The alignment was viewed and adjusted using GeneDoc (57).
Acknowledgments
This work was supported by the Irish Higher Education Authority, Enterprise Ireland, The Irish Health Research Board, and Science Foundation Ireland. G.L. Smith is a Wellcome Trust Principal Research Fellow. I.R. Haga was a CNPq (Brazil) scholar. K.A. Fitzgerald is supported by a fellowship from the Wellcome Trust, London.
The authors have no conflicting financial interests.
Abbreviations used: ERK, extracellular signal–regulated kinase; IRAK, IL-1 receptor–associated kinase; IRF3, interferon regulatory factor 3; JNK, c-Jun NH2-terminal protein kinase; Mal, MyD88 adaptor-like; MOI, multiplicity of infection; MyD88, myeloid differentiation factor 88; PAM, pathogen-associated molecule; RANTES, regulated on activation, normal T cell expressed and secreted; SARM, sterile α and HEAT/Armadillo motifs-containing protein; TIR, Toll–IL-1 resistance; TLR, Toll-like receptors; TRAF, TNF receptor–associated factor; TRAM, TRIF-related adaptor molecule; TRIF, TIR domain-containing adaptor inducing IFN-β; VV, vaccinia virus.
J. Stack and I.R. Haga contributed equally to this work.
I.R. Haga's present address is Department of Biochemistry, Trinity College, Dublin 2, Ireland.
P.C. Reading's present address is Department of Microbiology and Immunology, University of Melbourne, Victoria, 3010, Australia.
References
- 1.Janeway, C.A. Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197–216. [DOI] [PubMed] [Google Scholar]
- 2.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335–376. [DOI] [PubMed] [Google Scholar]
- 3.Dunne, A., and L.A.J. O'Neill. 2003. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci. STKE. 2003:re3. [DOI] [PubMed]
- 4.Zhang, D., G. Zhang, M.S. Hayden, M.B. Greenblatt, C. Bussey, R.A. Flavell, and S. Ghosh. 2004. A toll-like receptor that prevents infection by uropathogenic bacteria. Science. 303:1522–1526. [DOI] [PubMed] [Google Scholar]
- 5.Tabeta, K., P. Georgel, E. Janssen, X. Du, K. Hoebe, K. Crozat, S. Mudd, L. Shamel, S. Sovath, J. Goode, et al. 2004. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. USA. 101:3516–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reis e Sousa, C. 2004. Toll-like receptors and dendritic cells: for whom the bell tolls. Semin. Immunol. 16:27–34. [DOI] [PubMed] [Google Scholar]
- 7.Pasare, C., and R. Medzhitov. 2004. Toll-like receptors and acquired immunity. Semin. Immunol. 16:23–26. [DOI] [PubMed] [Google Scholar]
- 8.Vaidya, S.A., and G. Cheng. 2003. Toll-like receptors and innate antiviral responses. Curr. Opin. Immunol. 15:402–407. [DOI] [PubMed] [Google Scholar]
- 9.Rassa, J.C., and S.R. Ross. 2003. Viruses and Toll-like receptors. Microbes Infect. 5:961–968. [DOI] [PubMed] [Google Scholar]
- 10.Bieback, K., E. Lien, I.M. Klagge, E. Avota, J. Schneider-Schaulies, W.P. Duprex, H. Wagner, C.J. Kirschning, V. Ter Meulen, and S. Schneider-Schaulies. 2002. Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J. Virol. 76:8729–8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexopoulou, L., A. Czopik-Holt, R. Medzhitov, and R. Flavell. 2001. Recognition of double stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 413:696–712. [DOI] [PubMed] [Google Scholar]
- 12.Diebold, S.S., T. Kaisho, H. Hemmi, S. Akira, and C. Reis e Sousa. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 303:1529–1531. [DOI] [PubMed] [Google Scholar]
- 13.Heil, F., H. Hemmi, H. Hochrein, F. Ampenberger, C. Kirschning, S. Akira, G. Lipford, H. Wagner, and S. Bauer. 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 303:1526–152. [DOI] [PubMed] [Google Scholar]
- 14.Lund, J.M., L. Alexopoulou, A. Sato, M. Karow, N.C. Adams, N.W. Gale, A. Iwasaki, and R.A. Flavell. 2004. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA. 101:5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krug, A., G.D. Luker, W. Barchet, D.A. Leib, S. Akira, and M. Colonna. 2004. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 103:1433–1437. [DOI] [PubMed] [Google Scholar]
- 16.Lund, J., A. Sato, S. Akira, R. Medzhitov, and A. Iwasaki. 2003. Toll-like receptor 9-mediated recognition of herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 198:513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle, S.E., S.A. Vaidya, R. O'Connell, H. Dadgostar, P.W. Dempsey, T.-T. Wu, G. Rao, R. Sun, M.E. Haberland, R.L. Modlin, and G. Cheng. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 17:251–263. [DOI] [PubMed] [Google Scholar]
- 18.Hiscott, J., H. Kwon, and P. Genin. 2001. Hostile takeovers: viral appropriation of the NFκB pathway. J. Clin. Invest. 107:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto, M., K. Takeda, and S. Akira. 2004. TIR domain-containing adaptors define the specificity of TLR signalling. Mol. Immunol. 40:861–868. [DOI] [PubMed] [Google Scholar]
- 20.O'Neill, L.A.J., K. Fitzgerald, and A.G. Bowie. 2003. The Toll-IL-1 receptor adaptor family grows to five members. Trends Immunol. 24:286–289. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signalling pathway. Science. 301:640–643. [DOI] [PubMed] [Google Scholar]
- 22.Hoebe, K., X. Du, P. Georgel, E. Janssen, K. Tabeta, S.O. Kim, J. Goode, P. Lin, N. Mann, S. Mudd, et al. 2003. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 424:743–748. [DOI] [PubMed] [Google Scholar]
- 23.Fitzgerald, K.A., E.M. Palsson-McDermott, A.G. Bowie, C.A. Jefferies, A.S. Mansell, G. Brady, E. Brint, A. Dunne, P. Gray, M.T. Harte, et al. 2001. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 413:78–83. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668–6672. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald, K.A., S.M. McWhirter, K.L. Faia, D.C. Rowe, E. Latz, D.T. Golenbock, A.J. Coyle, S.M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496. [DOI] [PubMed] [Google Scholar]
- 26.McWhirter, S.M., K.A. Fitzgerald, J. Rosains, D.C. Rowe, D.T. Golenbock, and T. Maniatis. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA. 101:233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzgerald, K.A., D.C. Rowe, B.J. Barnes, D.R. Caffrey, A. Visintin, E. Latz, B. Monks, P.M. Pitha, and D.T. Golenbock. 2003. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J. Exp. Med. 198:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto, M., S. Sato, H. Hemmi, S. Uematsu, K. Hoshino, T. Kaisho, O. Takeuchi, K. Takeda, and S. Akira. 2003. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signalling pathway. Nat. Immunol. 4:1144–1150. [DOI] [PubMed] [Google Scholar]
- 29.Bowie, A., E. Kiss-Toth, J.A. Symons, G.L. Smith, S.K. Dower, and L.A.J. O'Neill. 2000. A46R and A52R from vaccinia virus are antagonists of host IL-1 and Toll-like receptor signaling. Proc. Natl. Acad. Sci. USA. 97:10162–10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harte, M.T., I.R. Haga, G. Maloney, P. Gray, P.C. Reading, N.W. Bartlett, G.L. Smith, A. Bowie, and L.A.J. O'Neill. 2003. The poxvirus protein A52R targets Toll-like receptor signalling complexes to suppress host defence. J. Exp. Med. 197:343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jefferies, C., A. Bowie, G. Brady, X. Li, and L.A.J. O'Neill. 2001. Transactivation by the p65 Subunit of NF-kappaB in response to interleukin-1 (IL-1) involves MyD88, IL-1 receptor-associated kinase 1, TRAF-6, and Rac1. Mol. Cell. Biol. 21:4544–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adachi, O., T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi, and S. Akira. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1 and IL-18-mediated function. Immunity. 9:143–150. [DOI] [PubMed] [Google Scholar]
- 33.Burns, K., F. Martinon, C. Esslinger, H. Pahl, P. Schneider, J.-L. Bodmer, F. Di Marco, L. French, and J. Tschopp. 1998. MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 273:12203–12209. [DOI] [PubMed] [Google Scholar]
- 34.Janssens, S., and R. Beyaert. 2002. A universal role for MyD88 in TLR/IL-1R-mediated signaling. Trends Biochem. Sci. 27:474–482. [DOI] [PubMed] [Google Scholar]
- 35.Hsu, H., H.-B. Shu, M.-G. Pan, and D.V. Goeddel. 1996. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84:299–308. [DOI] [PubMed] [Google Scholar]
- 36.Applequist, S.E., R.P.A. Wallin, and H.-G. Ljunggren. 2002. Variable expression of Toll-like receptor in murine innate and adaptive immune cell lines. Int. Immunol. 14:1065–1074. [DOI] [PubMed] [Google Scholar]
- 37.Medzhitov, R., P. Preston-Hurlburt, and C.A. Janeway. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 388:394–397. [DOI] [PubMed] [Google Scholar]
- 38.Kawai, T., O. Adachi, T. Ogawa, K. Takeda, and S. Akira. 1999. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 11:115–122. [DOI] [PubMed] [Google Scholar]
- 39.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-β induction. Nat. Immunol. 4:161–167. [DOI] [PubMed] [Google Scholar]
- 40.Liberati, N.T., K.A. Fitzgerald, D.H. Kim, R. Feinbaum, D.T. Golenbock, and F.M. Ausubel. 2004. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. USA. 101:6593–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alcami, A., and U.H. Koszinowski. 2000. Viral mechanisms of immune evasion. Immunol. Today. 21:447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, Y., X. Tao, B. Shen, T. Horng, R. Medzhitov, J.L. Manley, and L. Tong. 2000. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 408:111–115. [DOI] [PubMed] [Google Scholar]
- 43.Kurt-Jones, E.A., L. Popova, L. Kwinn, L.M. Haynes, L.P. Jones, R.A. Tripp, E.E. Walsh, M.W. Freeman, D.T. Golenbock, L.J. Anderson, and R.W. Finberg. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398–401. [DOI] [PubMed] [Google Scholar]
- 44.Ehl, S., R. Bischoff, T. Ostler, S. Vallbracht, J. Schulte-Monting, A. Poltorak, and M. Freudenberg. 2004. The role of Toll-like receptor 4 versus interleukin-12 in immunity to respiratory syncytial virus. Eur. J. Immunol. 34:1146–1153. [DOI] [PubMed] [Google Scholar]
- 45.Rassa, J.C., J.L. Meyers, Y. Zhang, R. Kudaravalli, and S.R. Ross. 2002. Murine retroviruses activate B cells via interaction with toll-like receptor 4. Proc. Natl. Acad. Sci. USA. 99:2281–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edelmann, K.H., S. Richardson-Burns, L. Alexopoulou, K.L. Tyler, R.A. Flavell, and M.B.A. Oldstone. 2004. Does Toll-like receptor 3 play a biological role in virus infections? Virology. 322:231–238. [DOI] [PubMed] [Google Scholar]
- 47.Levy, D.E., and I.J. Marié. 2004. RIGging an antiviral defense–it's in the CARDs. Nat. Immunol. 5:699–701. [DOI] [PubMed] [Google Scholar]
- 48.Sato, S., M. Sugiyama, M. Yamamoto, Y. Watanabe, T. Kawai, K. Takeda, and S. Akira. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171:4304–4310. [DOI] [PubMed] [Google Scholar]
- 49.Meylan, E., K. Burns, K. Hofmann, V. Blancheteau, F. Martinon, M. Kelliher, and J. Tschopp. 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-κB activation. Nat. Immunol. 5:503–507. [DOI] [PubMed] [Google Scholar]
- 50.Bartlett, N., J.A. Symons, D.C. Tscharke, and G.L. Smith. 2002. The vaccinia virus N1L protein is an intracellular homodimer that promotes virulence. J. Gen. Virol. 83:1965–1976. [DOI] [PubMed] [Google Scholar]
- 51.DiPerna, G., J. Stack, A.G. Bowie, A. Boyd, G. Kotwal, Z. Zhang, S. Arvikar, E. Latz, K.A. Fitzgerald, and W.L. Marshall. 2004. Poxvirus protein N1L targets the I-κB kinase complex, inhibits signaling to NF-κB by the tumor necrosis factor superfamily of receptors, and inhibits NF-κB and IRF3 signaling by Toll-like receptors. J. Biol. Chem. 279:36570–36578. [DOI] [PubMed] [Google Scholar]
- 52.Muzio, M., J. Ni, P. Feng, and V.M. Dixit. 1997. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 278:1612–1615. [DOI] [PubMed] [Google Scholar]
- 53.Muzio, M., G. Natoli, S. Saccani, M. Levrero, and A. Mantovani. 1998. The human toll signaling pathway: divergence of nuclear factor κB and JNK/SAPK activation upstream of tumor necrosis factor receptor-associated factor 6 (TRAF6). J. Exp. Med. 187:2097–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parkinson, J.E., and G.L. Smith. 1994. Vaccinia virus gene A36R encodes a M(r) 43-50 K protein on the surface of extracellular enveloped virus. Virology. 204:376–390. [DOI] [PubMed] [Google Scholar]
- 55.Falkner, F.G., and B. Moss. 1990. Transient dominant selection of recombinant vaccinia viruses. J. Virol. 64:3108–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alcami, A., and G.L. Smith. 1992. A soluble receptor for interleukin-1β encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell. 71:153–167. [DOI] [PubMed] [Google Scholar]
- 57.Nicholas, K.B., H.B. Nicholas Jr., and D.W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW. News. 4:14. [Google Scholar]