Abstract
T cells are crucial for the control of cytomegalovirus (CMV) in infected individuals. Although CMV-specific T cells can be quantified by various methods, clear correlates of protection from CMV disease have not been defined. However, responses to the pp65 protein are believed to play an important role. Here, the proportions of interferon γ–producing T cells following ex vivo activation with pools of overlapping peptides representing the pp65 and immediate early (IE)-1 proteins were determined at multiple time points and related to the development of CMV disease in 27 heart and lung transplant recipients. Frequencies of IE-1–specific CD8 T cells above 0.2 and 0.4% at day 0 and 2 wk, respectively, or 0.4% at any time during the first months discriminated patients who did not develop CMV disease from patients at risk, 50–60% of whom developed CMV disease. No similar distinction between risk groups was possible based on pp65-specific CD8 or CD4 T cell responses. Remarkably, CMV disease developed exclusively in patients with a dominant pp65-specific CD8 T cell response. In conclusion, high frequencies of IE-1 but not pp65-specific CD8 T cells correlate with protection from CMV disease. These results have important implications for monitoring T cell responses, adoptive cell therapy, and vaccine design.
CMV reactivation is frequent in virus carriers (1); however, CMV disease occurs only if the T cell response is compromised (2, 3). Because T cells are instrumental in graft rejection, drugs suppressing the T cell response are required after transplantation. They do not as such cause CMV disease; however, they facilitate this life-threatening complication (2). Therefore, prophylactic and/or preemptive antiviral treatment is often administered (4). CTL responses have been known to correlate with recovery from CMV disease in bone marrow transplant (BMT) recipients since 1982 (3). In murine CMV infection (BALB/c mice), the immediate early (IE) protein, pp89, was known to be an important CTL target protein already in the late 1980's, and CTL responses against pp89 were shown to mediate protection in several studies (5, 6). IE-1–specific T cell responses in CMV-infected humans were described as early as 1991 (7). However, a study in fully immune reconstituted BMT recipients in 1994 revealed that their CMV-specific CTLs were able to kill CMV-infected fibroblasts even if preincubated with actinomycin D (an inhibitor of CMV replication), and, therefore, these CTLs were believed to be directed at virion proteins such as pp65 (8). Although actinomycin D permits the synthesis of IE gene products (9), IE-1 was no longer considered a relevant target, because reports had suggested that it was not efficiently presented on class I MHC after infection (10). By contrast, virion proteins were known to be presented even in the absence of viral replication (11), and additionally, pp65 itself was found to interfere with IE-1 presentation (12). As a result, research in this field concentrated on pp65 (13, 14). The interest in IE-1 as a T cell target in human CMV infection was not revived until 1999, when IE-1–specific CD8 T cells were described to occur in infected individuals at frequencies at least comparable to those of pp65-specific CD8 T cells (15). To date, both pp65 and IE-1 are considered dominant T cell targets (6). Some recent studies confirmed a positive correlation between immune reconstitution and rising numbers of pp65-specific T cells (16–18). Unfortunately, IE-1–specific CD8 T cells were not included in these investigations, and, after BMT, any parameter indicating the return or advent of CMV-specific T cell immunity would be expected to correlate with a lower incidence of CMV disease to some degree (3). Detection of antigen-specific T cells in this study relies on the intracellular accumulation of the effector cytokine, IFN-γ, in functional but secretion-inhibited T cells. IFN-γ induction was achieved by ex vivo stimulation with peptide pools representing all possible CD4 and CD8 T cell epitopes in pp65 and IE-1 (19, 20). CD4 and CD8 T cell responses to both proteins were measured at multiple time points in 27 heart and lung transplant recipients and correlated with clinical data. Our study clearly shows that dominance and magnitude of the IE-1– but not the pp65-specific CD8 T cell response correlate with protection from CMV disease.
Results and Discussion
To assess whether the presence and/or magnitude of CMV-specific T cell responses had an influence on the development of CMV-related complications in 23 heart and 4 lung transplant recipients, responses to IE-1 and pp65 were measured beginning at the time of transplantation and ending between 3 mo and 2 yr after it (monitoring period). Clinical data evaluation always included the complete monitoring period but at least 1 yr after transplantation. No significant effects of donor CMV serostatus and drug regimen on the development of pp65 antigenemia and CMV disease were elicited.
In all patients with a CD4 T cell response to pp65 (“CD4/pp65 response”), a CD8 T cell response to pp65 (“CD8/pp65 response”) or a CD8 T cell response to IE-1 (“CD8/IE-1 response”), the reponses were consistently found over time. By contrast, the CD4 T cell response to IE-1 (“CD4/IE-1 response”) was frequently undetectable during the first 100 d and therefore not included in further data analysis. Overall, the response distribution was in keeping with previous results (Table I; references 19 and 21).
Table I.
Global CD4 and CD8 T cell response patterns
| Patients responding to
|
|||
|---|---|---|---|
| Responding T cell subset | pp65 pool | IE-1 pool | |
| CD4 T cells | 23 (85%) | 11 (41%)a | |
| Median frequencies (range) | day 0 | 0.27 (0.02–17.23) | 0.04 (0.01–0.16) |
| In % of total CD4 T cells | 3–4 mo | 0.28 (0.03–16.89) | 0.08 (0.05–0.19) |
| Response was dominant inb | 22 | 0 | |
| CD8 T cells | 27 (100%) | 20 (74%) | |
| Median frequencies (range) | day 0 | 0.44 (0.02–8.06) | 0.97 (0.01–10.81) |
| In % of total CD8 T cells | 3–4 mo | 0.55 (0.04–13.01) | 1.05 (0.03–15.47) |
| Response was dominant inb | 17 | 9 | |
In most donors, this response was not observed at all time points.
In one case, no clear dominance of either response was observed.
Initial data inspection suggested that unlike patients who developed CMV disease (“Disease-Group”), those who did not (“No-Disease-Group”) frequently had high CD8 T cell responses to IE-1 (Fig. 1). Based on clinical considerations, the following approximated time points were chosen for assessing T cell responses: day 0 (median: 0 d, range: 0–4 d, n = 27); 2 wk (median: 14 d, range: 11–19 d, n = 27); 1 mo (median: 29 d, range: 24–37 d, n = 27); and 3–4 mo (median 95 d, range: 89–117 d, n = 26), after which CMV disease is rare. Notably, one out of nine patients (11%) developed CMV disease within 2 wk, four out of nine (44%) within the first month, eight out of nine (88%) within 3–4 mo, and only one patient (11%) later than that. Therefore, the most interesting time points for predicting the risk of CMV disease would have been day 0 and 2 wk (Fig. 2).
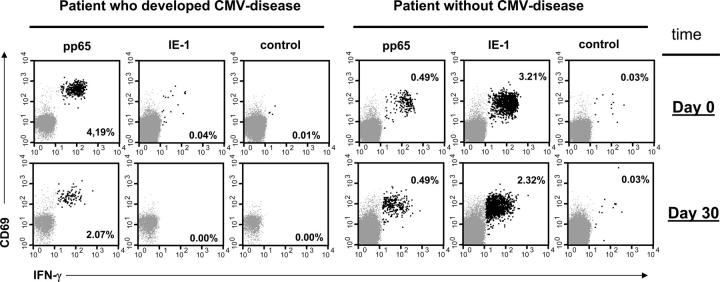
Figure 1.
High frequencies of IE-1–specific CD8 T cells are associated with protection from CMV disease. Dot plots show responses to the CMV pp65 and IE-1 peptide pools in two representative patients, one of whom developed CMV disease on day 39 after transplantation (left). CD8 T cells are shown (IFN-γ+ events highlighted in black). The relative frequencies of IFN-γ1 events are indicated. Axes show log fluorescence.
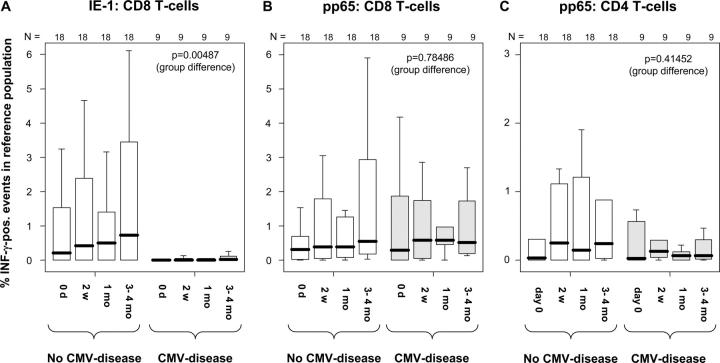
Figure 2.
Median frequencies of IE-1–specific CD8 T cells early after transplantation are higher in patients who do not develop CMV disease. CD4 and CD8 T cell responses to the CMV IE-1 and pp65 peptide pools were measured in 27 transplant recipients at specific time points using intracellular IFN-γ staining. (A) The results for IE-1–specific CD8 T cells, (B) the results for pp65-specific CD8 T cells, and (C) the results for pp65-specific CD4 T cells. In each panel, the results are divided by clinical group (CMV-disease or not). Box plots show the responses at the indicated time points (median, 25th and 75th percentiles, and extreme values). Nonparametric longitudinal analysis was performed using SAS software (p-values are given for differences between the groups). A global significance level of 0.05 is maintained if P ≤ 0.0166 for each of three tested end points.
Apart from their magnitude (relative frequencies), the relationship between the responses was considered. A response was considered dominant in a patient if it was the biggest response (by relative frequencies) at a majority of time points. The CD8/pp65 response was the most dominant overall (56%), followed by the CD8/IE-1 response (26%), and the CD4/pp65 response (19%). Considering the CD8 compartment alone, CD8/pp65 or CD8/IE-1 responses dominant at day 0 remained so for at least 3 mo in 25 out of 27 patients (93%), whereas changes of the dominant response within the CD4 compartment were not observed at any time. Interestingly, 8 out of 15 patients with a dominant CD8/pp65 response at day 0 developed CMV disease but only 1 out of 12 of the remaining patients did (P < 0.05; Fischer's exact test [FET]). Patients with no CD4 response to one of the proteins or both and patients with no CD8/IE-1 response seemed to develop CMV disease more often than the remaining patients (not statistically significant).
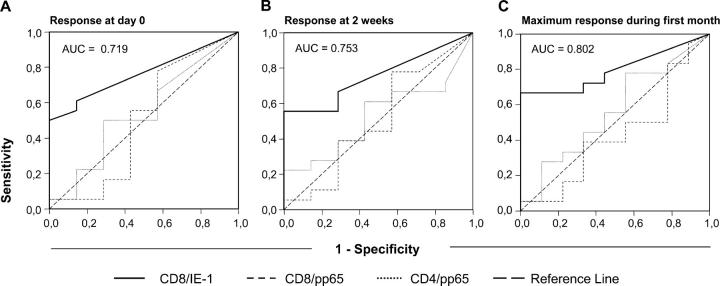
Nonparametric longitudinal analysis (22) of the CD8/pp65, CD8/IE-1, and CD4/pp65 responses between day 0 and 3–4 mo was performed including the factors, time, and group (CMV disease or not). Although time had no significant effect on any parameter, the CD8/IE-1 response (but not the other responses) was significantly different in the two groups (P = 0.00487, with P adjusted for multiple [i.e., three] end points = 0.01461; Fig. 2, A–C), indicating that this was the only relevant parameter for discrimination between the groups. Therefore, receiver operating characteristics (ROC) analysis (23) was used to identify cut-off levels at day 0 or 2 wk, CD8/IE-1 responses above or below which would indicate a decreased or increased risk of CMV disease, respectively (Fig. 3, A and B). Interestingly, 100% (9 out of 9) of patients whose CD8/IE-1 response exceeded 0.2% at day 0 remained free of CMV disease compared with only 50% (9 out of 18) of the patients with lower frequencies (P = 0.012, FET; sensitivity = 50%, specificity = 100%, positive predictive value = 100%, and negative predictive value = 50%). Identical results were obtained for a threshold of 0.4% at 2 wk. Moreover, 100% (12 out of 12) of patients whose CD8/IE-1 response exceeded 0.4% at any time during the first month remained free of CMV disease compared with only 40% (6 out of 15) with a lower maximum response (P = 0.001 FET; sensitivity = 66.7%, specificity = 100%, positive predictive value = 100%; and negative predictive value = 60%; Fig. 3 C). Importantly, IE-1 is the first protein expressed upon CMV reactivation (24), and as a result, IE-1–specific T cells should be the first to be activated and drawn to sites of replication. Notably, in monocytic precursor cells, which are known to harbor latent CMV, replication is arrested before early-phase proteins are made (24, 25). Therefore, they express IE products only and might be recognized (and killed) by IE-1– but not pp65-specific CTL. These cells are known to shuttle CMV to endothelia where they mature, become permissive to CMV replication, and eventually pass on the infectious virus (26). This “feeding” mechanism can probably be intercepted by IE-1–specific but not pp65-specific T cells, which could be one reason why high levels of IE-1–specific CD8 T cells are associated with protection from CMV disease.
Figure 3.
The magnitude of the CD8/IE-1 response identifies patients who will not develop CMV disease. ROC analysis was performed to define optimum thresholds of the CD8/IE-1 response (relative frequency) that would discriminate between patients who did or did not develop CMV disease. Panels show ROC curves for the response at day 0 (A), 2 wk (B), and for the maximum response during the first month (C). The area under the curve (AUC) for the CD8/IE-1 response graph is indicated. Graphs for the CD8/pp65 and CD4/pp65 responses are shown for reference.
There was no statistical association between pp65 antigenemia occurring during the first 3–4 mo and the presence and/or magnitude of any of the measured T cell responses. However, in all patients who had CMV disease, this coincided with their first pp65 antigenemia (tested at least weekly during hospital stays or at each visit). Late pp65 antigenemia (after day 100) occurred in only six patients and was not linked to CMV disease.
Interestingly, in 15 patients the CD8/IE-1 responses were below the above described threshold levels for the response at day 0 or 2 wk, or the maximum response during the first month; however, 6 out of these 15 did not develop CMV disease, indicating that factors apart from the magnitude of the CD8/IE-1 response were related to protection. In this regard, it appeared of particular interest to investigate if patients had a lower risk of developing CMV disease if they recognized a greater number of epitopes. Although all 15 patients had a CD8/pp65 response (13 of which had been mapped), only 12 had a CD4/pp65 response (9 of which had been mapped). The CD8/IE-1 response had been mapped only in patients with a high response, who were not included in this group. In fact, the median number of CD4 epitopes recognized in pp65 was two in patients who did not develop CMV-disease (range 0–3) compared with one in patients who developed CMV disease (range 0–1); however, the difference was statistically not significant (P = 0.232, Mann-Whitney U test [MWU]). It was interesting, however, that four out of six patients who remained free of CMV disease recognized two or more CD4 T cell epitopes in pp65, but none of the patients who developed CMV disease did (P = 0.061, cross-table analysis using FET). At least, this tendency suggested that recognition of several CD4 pp65 epitopes may also be related to protection, which will be investigated in future studies. Meanwhile, there was no difference between the groups with regard to the numbers of CD8 epitopes or the sum of CD4 and CD8 epitopes recognized in pp65 (P = 0.881 and P = 0.624, respectively, MWU). The T cell responses mapped in this patient cohort recognized known dominant epitopes including the CD8 epitopes pp65495–503 (NLVPMVATV, presented by HLA-A2) and IE-1309–317 (CRVLCCYVL, presented by HLA-B7; reference 6), the CD4 epitope pp65365–379 (EHPTFTSQYRIQGKL, presented by HLA-DR11; reference 21), and others (6). Recognition of these determinants indicated that mutations of IE-1 or pp65 in the clinical strains infecting our patients did not play a significant role for this study.
Results from adoptive transfer studies have demonstrated that CD4 T cells are essential for a lasting immune response against CMV (27). Although our data suggest that the breadth of the CD4/pp65 response may play a role in protection from CMV disease, we were unable to define critical threshold levels for the frequencies of pp65-specific CD4 T cells at particular time points. With regard to CD8 T cells, however, our results indicate that observing the frequencies of IE-1–specific CD8 T cells early after transplantation might be useful for identifying risk groups in terms of CMV disease. Our findings clearly challenge the paradigm that pp65 is the most important protein recognized by CD8 T cells in human CMV infection. By contrast, the relevance of the IE-1 protein as a T cell target protein is underscored. Moreover, that CMV disease developed only in individuals with a dominant CD8/pp65 response shows that dominance by frequencies does not necessarily indicate relevance in terms of protection. In conclusion, our study demonstrates that the definition of protective levels based on the frequencies of antigen-specific peripheral blood T cells is a realistic possibility for the future. Looking at the right specificities, however, will be crucial.
Materials and Methods
Patients and medication
After written informed consent, 27 patients were enrolled (21 male and 6 female), including 23 heart and 4 lung transplant recipients, with ages ranging from 32 to 67 yr (median 51). Primary disease included cardiomyopathy (15), coronary heart disease (7), malformation (1), primary pulmonary hypertension (2), and α-1-antitrypsin-deficiency (2). Table II shows a summary of CMV-related parameters. Initial maintenance-immunosuppressive medication included cyclosporin A (CsA), prednisolone (Pred), and azathioprine (Aza) in 16 patients and CsA, Pred, and mycophenolate mofetil (MMF) in 11 patients. 7 patients were switched from Aza to MMF later. Immunosuppressive induction therapy included methylprednisolone (Mpred) and ATG bolus in all patients. Acute graft rejection was treated with high dose intravenous Mpred or ATG. Recipients of CMV-seropositive lung transplants received oral ganciclovir (GCV) during the first 3 mo (n = 4). Patients with CMV reactivation (pp65 antigenemia) were treated with intravenous GCV until pp65 antigen was negative followed by oral GCV for 4 wk. Standard comedication included antibiotics, anti-hypertensive drugs, and diuretics. The study was approved by the Charité Internal Review Board (Ethikkomission Charité Mitte).
Table II.
Patients and CMV manifestation
| pp65 antigenemia
|
|||
|---|---|---|---|
| Group description | No antigenemia |
Asymptomatic | CMV disease |
| All patientsa (total = 27) | 8 (30%) | 10 (37%) | 9 (33%) |
| Recipients of CMV+ graft (n = 13) |
4 (31%) | 6 (46%) | 3 (23%) |
| Oral GCV prophylaxis (n = 4) |
2 (50%) | 0 (0%) | 2 (50%) |
All patients were CMV-seropositive before transplantation.
Monitoring parameters and period
Monitoring began immediately before transplantation (day 0) in four cases within 2–4 d of transplantation and was performed weekly during hospitalization and biweekly or less frequently in out-patients. In 27 patients, pp65- and IE-1–specific responses were monitored for 2 mo or more, in 26 patients for 3 mo or more, and in 18 patients for more than 1 yr. The median number of monitoring time points per patient was 14 (range: 7–21). Other parameters determined at each visit included pp65 antigenemia by immune fluorescence, CsA levels, echocardiography, electrocardiogram, and standard blood chemistry.
PBMC preparation and ex vivo IFN-γ induction assay
After preparation from citrated blood (Ficoll-Paque; Amersham Biosciences) PBMCs were washed twice in sterile PBS (GIBCO BRL) and resuspended in RPMI 1640 medium (Biochrome) containing 10% (vol/vol) heat-inactivated FCS (Biochrome), 2 mM l-glutamine (Biochrome), and 100 IU of penicillin/streptomycin (Biochrome). Cells were incubated in bulk overnight for 16 h at 37°C in a humidified 5% CO2 atmosphere in loosely covered Falcon 50-ml blue-cap conical tubes (BD Biosciences). Next, 400 μl of the volume-adjusted cell suspension was transferred to Falcon 2025 tubes (BD Biosciences). The maximum number of cells per assay tube was 2 × 106. Immediately, 100 μl RPMI/FCS medium containing single peptides or peptide pool (4 μl of stock solutions) was added. At 2 h, 500 μl RPMI/FCS medium was added containing 20 μg/ml brefeldin A (BFA; Sigma-Aldrich), corresponding to 2 μl BFA stock solution. Final concentrations of (each) peptide and BFA were 1 μg/ml and 10 μg/ml, respectively. Unstimulated control samples were run in parallel (4 μl DMSO instead of peptide stock solution). At 6 h, cells were washed with 3 ml ice-cold PBS (430 g for 8 min at 4°C), resuspended in PBS containing 1 mM EDTA (Sigma-Aldrich), incubated in a 37°C water bath for 10 min to detach adherent cells, spun down (430 g for 8 min at 4°C), and carefully vortexed for 30 s. Cells were then washed with PBS containing 0.5% bovine serum albumin and 0.1% (wt/vol) Na-azide (wash buffer), the supernatant was removed, and tubes were blotted dry. For permeabilization/fixation (one step), the pellets were resuspended in 1 ml of double-concentrated FACS-Lysing Solution (BD Biosciences) containing 0.05% (vol/vol) of Tween 20 (Sigma-Aldrich; reference 21), incubated for 10 min (in the dark at room temperature), washed, resuspended, stained with monoclonal antibodies (100 μl total volume at 4°C for 30 min in the dark), and washed once more before analysis.
Antibodies
FITC-conjugated anti–INF-γ, PE-conjugated anti-CD69, PerCP-conjugated anti-CD3 and allophycocyanin-conjugated anti-CD8 and anti-CD4 were purchased from BD Biosciences.
Peptides
Standard 15–amino acid peptides (11 overlaps) spanning the CMV IE-1 and pp65 proteins (Swiss-Prot Nr. P14332 and P06725, respectively) were purchased (Jerini Biotools). Quality control included mass spectroscopy and HPLC (purity >70%). Stock solutions (8 or 80 mg/ml in DMSO) were stored at −80°C.
Flow cytometry and data analysis
Per sample, 150,000–250,000 events in the FSC/SSC lymphocyte gate were acquired on a FACS Calibur flow cytometer (Becton Dickinson). For data analysis (CELLQuest software; Becton Dickinson), CD3+/CD8+ or CD3−/CD3− events were displayed in a CD69 versus IFN-γ dot plot. CD8+ or CD8−/INF-γ+ cells were expressed as a percent of the respective reference population. CD8− CD3 T cells were used as surrogate for CD4 T cells, which is an acceptable approximation for this type of study (21). Positive events in the corresponding regions in unstimulated samples were subtracted. The assessment of responses was previously described in more detail (19, 21).
Epitope identification
Epitope mapping was performed based on the ex vivo IFN-γ induction assay described above using crossover pools of pp65 and IE-1 peptides. All identified epitopes were confirmed using individual peptides for stimulation as previously described in more detail (15, 21).
Clinical data collection
Complete clinical data analysis covered at least 1 yr and always exceeded the monitoring period. Diagnosis of CMV disease required at least pp65 antigenemia (≥2 positive cells/200,000), neutropenia, and otherwise unexplained fever. Additional criteria included positive chest radiographs, positive organ biopsy, or other relevant laboratory results suggestive of specific organ involvement.
Statistics
SPSS 11.0 (SPSS Inc.) and SAS 9.0 software (SAS Institute) were used. Differences between two unpaired groups were analyzed using the MWU. ROC analysis was used to define cut-off values (22). FET was used for cross-table analysis. Nonparametric, factorial analysis of longitudinal data was performed using the corresponding SAS-module according to Brunner et al. (23). Where applicable, P was adjusted for multiple end-points (Bonferroni adjustment).
Acknowledgments
We would like to thank Dr. Jan Gratama for helpful advice on the manuscript, Dr. B. Wegner for the revision of statistics, and Ms. Christina Nicholson (B.A.) for the English revision.
This work was partially supported by Charité, Universitätsmedizin Berlin and Immune Tolerance Network (ITN).
The authors have no conflicting financial interests.
T. Bunde and A. Kirchner share senior authorship for this work.
References
- 1.Toro, A.I., and J. Ossa. 1996. PCR activity of CMV in healthy CMV-seropositive individuals: does latency need redefinition? Res. Virol. 147:233–238. [DOI] [PubMed] [Google Scholar]
- 2.Fishman, J.A., and R.H. Rubin. 1998. Infection in organ-transplant recipients. N. Engl. J. Med. 338:1741–1751. [DOI] [PubMed] [Google Scholar]
- 3.Quinnan, G.V., Jr., N. Kirmani, A.H. Rook, J.F. Manischewitz, L. Jackson, G. Moreschi, G.W. Santos, R. Saral, and W.H. Burns. 1982. Cytotoxic T cells in cytomegalovirus infection: HLA-restricted T-lymphocyte and non-T-lymphocyte cytotoxic responses correlate with recovery from cytomegalovirus infection in bone-marrow-transplant recipients. N. Engl. J. Med. 307:7–13. [DOI] [PubMed] [Google Scholar]
- 4.Guiver, M., A.J. Fox, K. Mutton, N. Mogulkoc, and J. Egan. 2001. Evaluation of CMV viral load using TaqMan CMV quantitative PCR and comparison with CMV antigenemia in heart and lung transplant recipients. Transplantation. 71:1609–1615. [DOI] [PubMed] [Google Scholar]
- 5.Jonjic, S., M. del Val, G.M. Keil, M.J. Reddehase, and U.H. Koszinowski. 1988. A nonstructural viral protein expressed by a recombinant vaccinia virus protects against lethal cytomegalovirus infection. J. Virol. 62:1653–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddehase, M.J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2:831–844. [DOI] [PubMed] [Google Scholar]
- 7.Borysiewicz, L.K., J.K. Hickling, S. Graham, J. Sinclair, M.P. Cranage, G.L. Smith, and J.G. Sissons. 1988. Human cytomegalovirus–specific cytotoxic T cells. Relative frequency of stage-specific CTL recognizing the 72-kD immediate early protein and glycoprotein B expressed by recombinant vaccinia viruses. J. Exp. Med. 168:919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, C.R., P.D. Greenberg, M.J. Gilbert, J.M. Goodrich, and S.R. Riddell. 1994. Recovery of HLA-restricted cytomegalovirus (CMV)-specific T-cell responses after allogeneic bone marrow transplant: correlation with CMV disease and effect of ganciclovir prophylaxis. Blood. 83:1971–1979. [PubMed] [Google Scholar]
- 9.Walker, D., and J. Hudson. 1987. Analysis of immediate-early and early proteins of murine cytomegalovirus in permissive and nonpermissive cells. Arch. Virol. 92:103–119. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, M.J., S.R. Riddell, C.R. Li, and P.D. Greenberg. 1993. Selective interference with class I major histocompatibility complex presentation of the major immediate-early protein following infection with human cytomegalovirus. J. Virol. 67:3461–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riddell, S.R., M. Rabin, A.P. Geballe, W.J. Britt, and P.D. Greenberg. 1991. Class I MHC-restricted cytotoxic T lymphocyte recognition of cells infected with human cytomegalovirus does not require endogenous viral gene expression. J. Immunol. 146:2795–2804. [PubMed] [Google Scholar]
- 12.Gilbert, M.J., S.R. Riddell, B. Plachter, and P.D. Greenberg. 1996. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature. 383:720–722. [DOI] [PubMed] [Google Scholar]
- 13.McLaughlin-Taylor, E., H. Pande, S.J. Forman, B. Tanamachi, C.R. Li, J.A. Zaia, P.D. Greenberg, and S.R. Riddell. 1994. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J. Med. Virol. 43:103–110. [DOI] [PubMed] [Google Scholar]
- 14.Wills, M.R., A.J. Carmichael, K. Mynard, X. Jin, M.P. Weekes, B. Plachter, and J.G. Sissons. 1996. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T- cell receptor usage of pp65-specific CTL. J. Virol. 70:7569–7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kern, F., I.P. Surel, N. Faulhaber, C. Frommel, J. Schneider-Mergener, C. Schonemann, P. Reinke, and H.D. Volk. 1999. Target structures of the CD8(+)-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J. Virol. 73:8179–8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reusser, P., G. Cathomas, R. Attenhofer, M. Tamm, and G. Thiel. 1999. Cytomegalovirus (CMV)-specific T cell immunity after renal transplantation mediates protection from CMV disease by limiting the systemic virus load. J. Infect. Dis. 180:247–253. [DOI] [PubMed] [Google Scholar]
- 17.Gratama, J.W., J.W. van Esser, C.H. Lamers, C. Tournay, B. Lowenberg, R.L. Bolhuis, and J.J. Cornelissen. 2001. Tetramer-based quantification of cytomegalovirus (CMV)-specific CD8+ T lymphocytes in T-cell-depleted stem cell grafts and after transplantation may identify patients at risk for progressive CMV infection. Blood. 98:1358–1364. [DOI] [PubMed] [Google Scholar]
- 18.Hebart, H., S. Daginik, S. Stevanovic, U. Grigoleit, A. Dobler, M. Baur, G. Rauser, C. Sinzger, G. Jahn, J. Loeffler, et al. 2002. Sensitive detection of human cytomegalovirus peptide-specific cytotoxic T-lymphocyte responses by interferon-gamma-enzyme-linked immunospot assay and flow cytometry in healthy individuals and in patients after allogeneic stem cell transplantation. Blood. 99:3830–3837. [DOI] [PubMed] [Google Scholar]
- 19.Kern, F., N. Faulhaber, C. Frömmel, E. Khatamzas, S. Prösch, C. Schönemann, I. Kretzschmar, R. Volkmer-Engert, H.-D. Volk, and P. Reinke. 2000. Analysis of CD8 T cell reactivity to cytomegalovirus using protein-spanning pools of overlapping pentadecapeptides. Eur. J. Immunol. 30:1676–1682. [DOI] [PubMed] [Google Scholar]
- 20.Gratama, J.W., and F. Kern. 2004. Flow cytometric enumeration of antigen-specific T lymphocytes. Cytometry. 58A:79–86. [DOI] [PubMed] [Google Scholar]
- 21.Kern, F., T. Bunde, N. Faulhaber, F. Kiecker, E. Khatamzas, I.M. Rudawski, A. Pruss, J.W. Gratama, R. Volkmer-Engert, R. Ewert, et al. 2002. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. J. Infect. Dis. 185:1709–1716. [DOI] [PubMed] [Google Scholar]
- 22.Brunner, E., S. Domhoff, and F. Langer. 2002. Nonparametric Analysis of Longitudinal Data in Factorial Experiments. Wiley, New York. 280 pp.
- 23.Greiner, M., D. Pfeiffer, and R.D. Smith. 2000. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev. Vet. Med. 45:23–41. [DOI] [PubMed] [Google Scholar]
- 24.Stinski, M.F. 1978. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J. Virol. 26:686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice, G.P., R.D. Schrier, and M.B. Oldstone. 1984. Cytomegalovirus infects human lymphocytes and monocytes: virus expression is restricted to immediate-early gene products. Proc. Natl. Acad. Sci. USA. 81:6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waldman, W.J., D.A. Knight, E.H. Huang, and D.D. Sedmak. 1995. Bidirectional transmission of infectious cytomegalovirus between monocytes and vascular endothelial cells: an in vitro model. J. Infect. Dis. 171:263–272. [DOI] [PubMed] [Google Scholar]
- 27.Riddell, S.R., and P.D. Greenberg. 1995. Principles for adoptive T cell therapy of human viral diseases. Annu. Rev. Immunol. 13:545–586. [DOI] [PubMed] [Google Scholar]