Abstract
Cytokine receptor signals have been suggested to stimulate cell differentiation during hemato/lymphopoiesis. Such action, however, has not been clearly demonstrated. Here, we show that adult B cell development in IL-7 −/− and IL-7Rα2/− mice is arrested at the pre–pro-B cell stage due to insufficient expression of the B cell–specific transcription factor EBF and its target genes, which form a transcription factor network in determining B lineage specification. EBF expression is restored in IL-7 −/− pre–pro-B cells upon IL-7 stimulation or in IL-7Rα−/− pre–pro-B cells by activation of STAT5, a major signaling molecule downstream of the IL-7R signaling pathway. Furthermore, enforced EBF expression partially rescues B cell development in IL-7Rα−/− mice. Thus, IL-7 receptor signaling is a participant in the formation of the transcription factor network during B lymphopoiesis by up-regulating EBF, allowing stage transition from the pre–pro-B to further maturational stages.
B cell development is regulated by multiple B cell–specific transcription factors (1, 2). Both E2A and EBF are necessary for initiating B cell differentiation through the cooperative regulation of B cell–specific genes, such as RAG1, RAG2, Igα, Igβ, λ5, Vpre-B, and CD19 (1, 2). Another transcription factor, PU.1, as well as E2A can up-regulate the promoter activity of the EBF gene (3, 4). Moreover, Pax5 gene expression is regulated by E2A and EBF (5). Once Pax5 is expressed, B cell progenitors are irreversibly committed to B cell lineage (6). Therefore, hierarchical regulation of these transcription factors may be a key for acquisition of B lineage specificity in B cell progenitors. It remains unclear, however, whether the formation of this transcription factor network occurs in a cell autonomous manner or requires extracellular stimuli provided by stromal cells in bone marrow.
Flt3 ligand, stem cell factor (SCF), and IL-7 are important cytokines for B cell development (7–9). Among these cytokines, only IL-7 can promote stage transition from B220+CD19− pre–pro-B cells to B220+CD19+ pro-B cells, whereas Flt3 ligand or SCF enhances the proliferation of B cell progenitors in in vitro cultures (10). Studies from IL-7Rα−/− mice reported conflicting results of the stage at which B cell development is arrested. One paper suggests a role of IL-7R signaling in regulation of IgH gene rearrangement for transition from pro-B to pre-B stage (11). Other papers indicate that B cell development in IL-7Rα−/− mice is arrested at an earlier point than the pro-B/pre-B transition stage, such as the early pro-B stage (using Hardy's nomenclature, fraction B; references 12, 13) or even earlier at the common lymphoid progenitor (CLP) stage (14). Furthermore, impaired T cell development in IL-7Rα−/− or γc −/− mice is rescued by enforced Bcl-2 expression, but rescue of B cell development is not observed in the same mice (15–17), suggesting that the primary function of IL-7R signaling is not to maintain cell survival but to promote cell differentiation and/or stimulate cell proliferation during B cell development. Together, these observations imply a role of IL-7 in stage transition during early adult B cell development. Nonetheless, the molecular basis of its function remains totally unknown.
In this study, we clarified that B cell development is arrested at the pre–pro-B cell stage in the absence of IL-7R signaling. We investigated the role of IL-7R in expression of B cell–specific factors at this developmental stage. We found a direct connection between IL-7R signaling and up-regulation of EBF expression. This finding demonstrates an indispensable role of IL-7 in the formation of transcription factor networks during adult B cell development.
Results and Discussion
B cell development is arrested at the pre–pro-B cell stage in IL-7R α −/− and IL-7 −/− mice
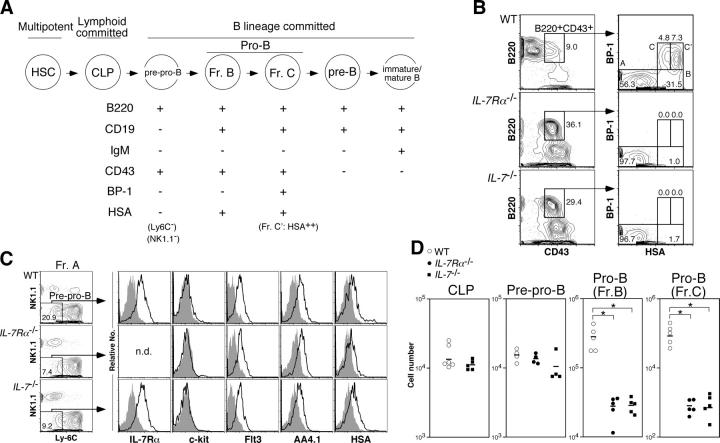
To clarify the role of IL-7R signaling in B cell development, we first confirmed the stage at which B cell development is blocked in IL-7Rα−/− and IL-7 −/− mice (see Fig. 1 A for defined stages of B cell development). Consistent with a previous paper (14), we found that there was no difference in the number of CLPs between IL-7 −/− and WT mice (Fig. 1 D). Next, we analyzed the more mature B220+CD43+ population, which is downstream of CLPs and include the pre–pro-B and pro-B fractions (Fig. 1 A). Total number of B220+CD43+BP-1−HSA−/low subset (fraction A in Hardy's nomenclature [13]) in IL-7Rα−/− and IL-7 −/− mice were also comparable to WT mice (unpublished data). Fraction A contains non-B cell progenitors that express NK1.1 or Ly-6C (18, 19). Therefore, we also compared the number of NK1.1−Ly-6C− cells in fraction A (pre–pro-B cells; Fig. 1 C) and found that the number of these cells were still comparable among WT, IL-7Rα−/−, and IL-7 −/− mice (Fig. 1 D). In contrast, B220+CD43+BP-1−HSAhigh subset (fraction B in Hardy's nomenclature [13]) in IL-7Rα−/− and IL-7 −/− mice was rarely detected (Fig. 1, B and D). Cells in the next maturational stage (B220+CD43+BP-1+HSAhigh subset, fraction C/C′ in Hardy's nomenclature [13]; we simply denote fraction C/C′ as fraction C hereafter in this paper) were almost absent, indicating that IL-7R signaling is required at an earlier stage than the pro-B (fraction B or C) stage in B cell development. The cell surface phenotype of pre–pro-B cells derived from IL-7Rα−/− and IL-7 −/− mice was identical to WT pre–pro-B cells (Fig. 1 C).
Figure 1.
B cell development is arrested at the pre–pro-B stage in the absence of IL-7R signaling. (A) Schematic maturational relationship of B cell populations used in this study. We excluded Ly-6C+ and NK1.1+ cells from Hardy's fraction A (reference 25) except for the experiments shown in Fig. 4. CD19 can be used instead of HSA to distinguish pre–pro-B (fraction A) and pro-B (fractions B and C/C′) cells (reference 13; Fig. S1 D, available at http://www.jem.org/cgi/content/full/jem.20050158/DC1). Hereafter, we used the fraction C/C′ population as fraction C. (B) Bone marrow (BM) cells from 8-wk-old mice were stained with antibodies for lineage markers (Lin: CD3ɛ, Gr-1, Ter119, and Mac-1), B220, CD43, BP-1, and HSA. Propidium iodide+ dead cells as well as Lin+ cells were excluded from this cell surface phenotyping. The numbers in FACS plots are percentages of cells in each cell population for each plot. The representative results of at least three independent experiments are shown. (C) The expression of IL-7Rα, c-Kit, Flt3, AA4.1, and HSA in pre–pro-B cells derived from WT, IL-7Rα−/−, and IL-7 −/− mice. The definition of pre–pro-B cells here is Lin−B220+CD43+CD19−NK1.1−Ly-6C− cells. Removal of CD19+ cells from this analysis is critical to exclude the more mature B cell progenitors from the pre–pro-B cell population (see Fig. S1). Open and filled histograms represent the expression level of various markers and the negative control stained with isotype-matched irrelevant antibodies, respectively. We did not stain pre–pro-B cells from IL-7Rα−/− mice with anti–IL-7Rα antibodies in this analysis. (D) The number of CLP, pre–pro-B, and pro-B cells in bone marrow from bilateral femurs and tibiae derived from WT (open circle), IL-7Rα−/− (closed circle), and IL-7 −/− (closed square) mice. The absolute number of cells was calculated with total bone marrow cell numbers and the percentage of each cell population. The mean is indicated as a horizontal bar. Because IL-7Rα is a marker to define CLPs, we did not determine the number of CLPs in IL-7Rα−/− mice. Asterisks denote significant difference by Student's t test; P < 0.05.
Expression of B cell–specific genes is impaired in pre–pro-B cells in the absence of IL-7R signaling
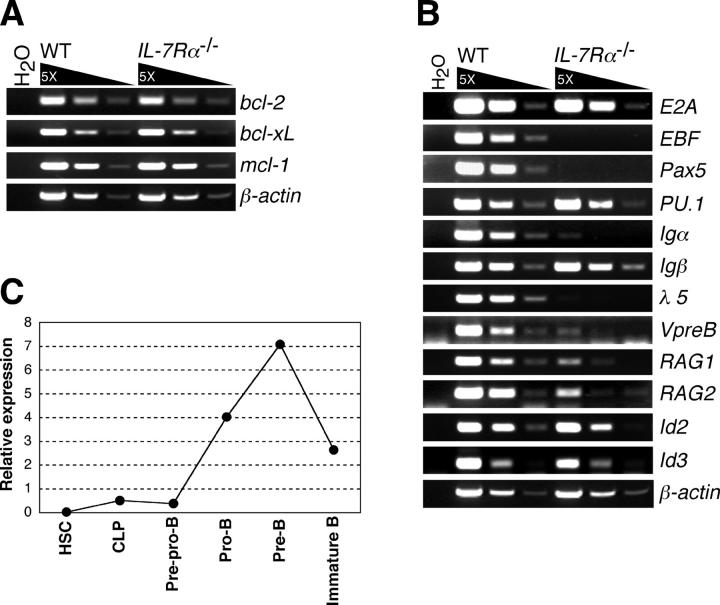
A recent study showed that lack of a cell survival factor, Mcl-1, causes severe B cell developmental arrest at the pro-B cell stage (20), although our previous studies suggest that antiapoptotic signals via IL-7R may not be crucial in B cell development (15, 16). We examined the expression of known survival factor genes bcl-2, bcl-x L, and mcl-1 in WT and IL-7Rα−/− pre–pro-B cells (Fig. 2 A). Expression levels of these survival factors were comparable, indicating the dispensable role of IL-7 for expression of these survival factors at the pre–pro-B cell stage. Because the mcl-1 gene flanked with the loxP sites is not deleted in pre–pro-B cells of CD19-CreMcl-1 f/null mice, it is not clear if Mcl-1 also plays an important role in the pre–pro-B cell stage (20). Regardless, our gene expression profiling suggests that mcl-1 expression is not regulated by IL-7R signaling pathway, at least in pre–pro-B cells.
Figure 2.
Gene expression profiles in IL-7R α −/− pre–pro-B cells. (A) Expression of antiapoptotic genes in WT and IL-7Rα−/− pre–pro-B cells were examined by RT-PCR. RNA was purified from 1.5 × 104 pre–pro-B cells (Lin−B220+CD43+CD19−NK1.1−Ly-6C− cells), which were sorted twice in sequence (double sorted) from WT and IL-7Rα−/− BM cells. After double sorting, the purity of cells was >99% in reanalysis. (B) Expression of genes that are involved in B cell development in WT and IL-7Rα−/− pre–pro-B cells. RT-PCR analyses were done as in A. (C) Expression of EBF gene in various developing B cell populations. The cell populations used here are HSC (Lin−/lowThy-1.1lowc-KithighSca-I+), CLP (Lin−c-KitlowSca-IlowThy-1.1−IL-7Rα+), pre–pro-B (B220+CD43+CD19−NK1.1−Ly-6C−), pro-B (B220+CD43+CD19+HSAhigh), pre-B (B220+CD43−IgM−), and immature B (B220+IgM+IgD−). The lineage cocktail (Lin) used for HSCs and CLPs also contained anti-B220 antibodies. Total RNA was purified from each doubly sorted population and subjected to quantification of EBF mRNAs by real-time PCR after first strand synthesis with reverse transcriptase. The amount of the first strand DNA applied was normalized to the expression level of a reference gene, GAPDH. EBF expression in whole BM was arbitrarily defined as unit one. The mean value of more than three independent samples is shown. Range of error is too small to be displayed in the histogram.
We next examined the expression of various B cell–specific genes critical for B cell development in WT and IL-7Rα−/− pre–pro-B cells. Notably, expression of EBF, Pax5, and λ5 was absent in IL-7Rα−/− pre–pro-B cells but was present in WT pre–pro-B cells (Fig. 2 B). Igα and Vpre-B expressions were also significantly reduced in IL-7Rα−/− pre–pro-B cells. In contrast, E2A and PU.1, which regulate early B cell development (1, 21), were normally expressed (Fig. 2 B). E2A function could be suppressed by direct association with Id2 or Id3 (1), but we did not observe any elevated expression of Id2 or Id3 in IL-7Rα−/− pre–pro-B cells (Fig. 2 B). Because Igα, Igβ, λ5, Vpre-B, RAG1, RAG2, and Pax5 genes are targets of E2A and EBF (1, 2, 5), we hypothesized that the ablated expression of EBF might cause arrested B cell development at the pre–pro-B stage in IL-7Rα−/− mice.
We further examined the expression of EBF at different maturational stages of developing B cells. As shown in Fig. 2 C, we observed strong up-regulation of EBF expression at the pro-B stage. EBF expression peaked at the small, nondividing pre-B cell stage and declined at the immature-B cell stage (Fig. 2 C). Because B cell development is arrested at the B220+CD43+BP-1−HSA− (fraction A) stage in EBF −/− mice (22), EBF expression in pre–pro-B cells should be required for the stage transition from the pre–pro-B to pro-B cell stage, despite its lower expression level in the pre–pro-B population compared with the more mature B cell progenitors.
EBF is a downstream target of IL-7R signaling at the pre–pro-B cell stage
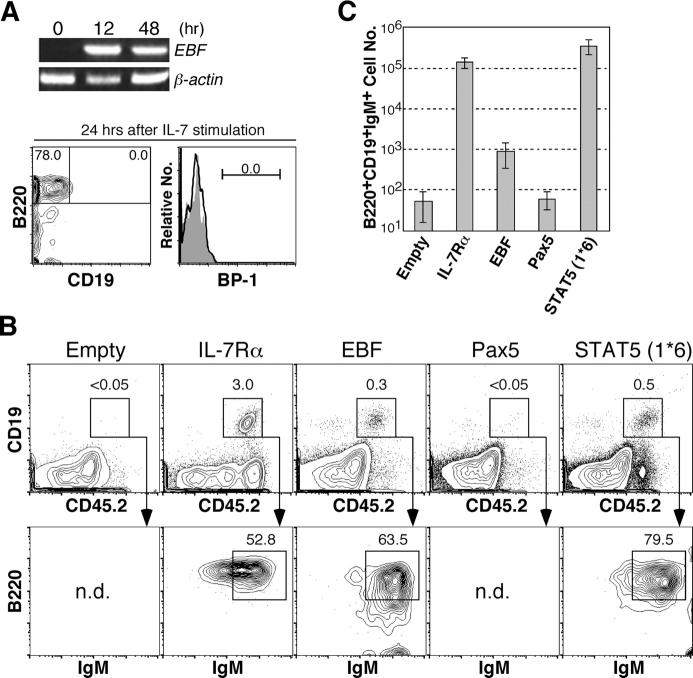
Next, we examined the relationship between IL-7R signaling and EBF expression in pre–pro-B cells. We purified pre–pro-B cells from IL-7 −/− mice and stimulated them with IL-7 in vitro. EBF expression levels were analyzed by RT-PCR. As shown in Fig. 3 A, there was no EBF expression in pre–pro-B cells derived from IL-7 −/− mice, as in the case with IL-7Rα−/− pre–pro-B cells (Fig. 2 B). EBF expression was up-regulated at 12 h after IL-7 stimulation, and its expression was maintained for at least 48 h (Fig. 3 A). We did not observe any change of surface phenotype in these pre–pro-B cells after 24 h of culture in the presence of IL-7 (Fig. 3 A), suggesting that initiation of EBF expression is a direct consequence of IL-7 stimulation in pre–pro-B cells.
Figure 3.
EBF expression is regulated by IL-7R signaling in B cell development. (A) Restoration of EBF expression in IL-7 −/− pre–pro-B cells. 1.2 × 104 pre–pro-B cells (B220+CD43+CD19−NK1.1−Ly-6C−) derived from IL-7 −/− mice were cultured in the presence of 10 ng/ml IL-7 in 96-well plates. The same number of freshly isolated cells was used as a nonstimulated control (time 0). Cells were harvested at the indicated time points and subjected to RT-PCR analysis of EBF expression. We also checked the cell surface phenotype of cells at 24 h after the culture. At this time point, no pro-B cells (CD19+ or BP-1+ cells) were developed from pre–pro-B cells (open histogram). Closed histogram represents background staining with isotype control antibodies. (B) IL-7Rα−/− HSCs (CD45.2+) were infected with empty, IL-7Rα, EBF, Pax5, or STAT5 (1*6) retroviruses by spin infection and injected into sublethally irradiated RAG2 −/− mice (CD45.1+). 5 wk after injection, mice were killed and spleen cell suspensions were stained with antibodies for CD19, B220, and IgM, as well as for CD45.2. CD19+B220+IgM+ cells are mature B cells. Representative data from more than three samples are shown. We did not observe any immature B cells derived from EBF+ IL-7Rα−/− HSCs in bone marrow at this time point but did observe them at an earlier time point (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20050158/DC1). (C) CD19+B220+IgM+ cell numbers of reconstituted mice with IL-7Rα2/− HSCs infected with empty, IL-7Rα, EBF, Pax5, or STAT5 (1*6) retroviruses shown in B were calculated. The cell numbers are expressed as mean ± SD.
To determine the biological importance of EBF up-regulation triggered by IL-7 stimulation, we examined whether or not enforced EBF expression could rescue the impaired B cell development in IL-7Rα−/− mice. IL-7Rα, EBF, and Pax5, as well as MSCV-IRES-GFP empty vector, were retrovirally introduced into IL-7Rα−/− HSCs (CD45.2+) for in vivo reconstitution. 4 × 103 GFP+ HSCs were injected into sublethally irradiated RAG2 −/− mice (CD45.1+). We analyzed spleens in reconstituted mice at 5 wk after injection. The top panels of Fig. 3 B show the frequency of CD19+ B cells in CD45.2+ donor-derived cells. In this experimental system, CD19+ B cells were not detected in reconstituted mice with IL-7Rα−/− HSCs introduced with empty vector, but were clearly present in reconstituted mice with IL-7Rα−/− HSCs infected with IL-7Rα viruses (Fig. 3 B). Although the frequency of B cells was lower than this positive control (IL-7Rα−/− HSCs with ectopic IL-7Rα), introduction of EBF clearly rescued B cell development from IL-7Rα−/− HSCs (Fig. 3 B). All of these CD19+ cells were GFP+ (not depicted) and expressed B cell marker B220 and IgM on the cell surface (Fig. 3 B, bottom). The absolute cell number of the CD19+B220+IgM+ spleen B cells from EBF+ IL-7Rα−/− HSCs (8.8 ± 5.5 × 102) was ∼160 times lower than spleen B cells from IL-7Rα−/− HSCs with ectopic IL-7Rα (1.4 ± 0.4 × 105; Fig. 3 C). No mature B cells were observed from IL-7Rα−/− HSCs introduced with Pax5 as well as introduction of empty vectors (Fig. 3 B). These results demonstrated that the enforced expression of EBF, but not Pax5, could rescue B cell differentiation in the absence of IL-7R signaling. The lower yield of mature B cells from IL-7Rα−/− HSC with ectopic EBF reemphasized the importance of IL-7 as a B cell growth factor.
Activated STAT5 can induce EBF expression in pre–pro-B cells
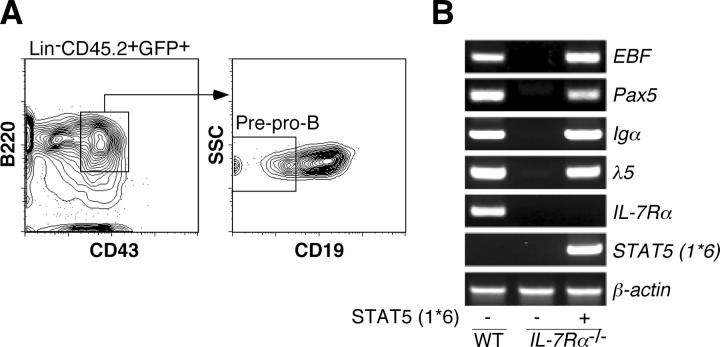
STAT5 is a critical signal molecule downstream of IL-7R (23). In reconstituted mice with IL-7Rα−/− HSCs that were introduced with constitutive active STAT5 (STAT5 (1*6)), we could see both differentiation and expansion of CD19+ B220+IgM+ spleen B cells (3.5 ± 1.4 × 105; Fig. 3, B and C), consistent with a previous observation (24). We isolated pre–pro-B cells from these reconstituted mice (Fig. 4 A) and examined EBF expression. As shown in Fig. 4 B, EBF as well as its downstream target genes, such as Pax5, Igα, and λ5, were expressed in IL-7Rα−/− pre–pro-B cells in the presence of constitutive active STAT5. These data suggest that STAT5 plays a central role in IL-7R signaling pathway to activate EBF expression. In pro-B cells, STAT5 has been shown to induce the expression of cyclin D2, pim-1, and bcl-x L, which are involved in cell proliferation and survival (24). Hence, the lack of induction of proliferation and/or survival factors may be the reason why we could not observe B cell expansion in mice reconstituted with IL-7Rα−/− HSCs introduced with EBF (Fig. 3 C).
Figure 4.
STAT5, a downstream signaling molecule of IL-7R, mediates EBF gene expression. (A) Donor-derived pre–pro-B cells (CD45.2+GFP+B220+CD43+CD19−) were purified from reconstituted mice with IL-7Rα−/− HSCs infected with STAT5 (1*6) retrovirus. (B) 1.0 × 104 pre–pro-B cells derived from WT (left lane), IL-7Rα−/− mice (middle lane), and reconstituted mice with IL-7Rα−/− HSCs expressing STAT5 (1*6) (right lane) were used for analysis of B cell–specific gene expression by RT-PCR. EBF expression as well as expression of its targets, Pax5, Igα, and λ5, were restored in pre–pro-B cells in the presence of constitutive active STAT5. Because IL-7Rα expression was not detected in the right lane, there was no contamination of host-derived cells (RAG2 −/− mice) in the sample.
IL-7 has been considered to mainly function at the pro-B cell stage in B cell development because pro-B cells proliferate extensively in response to IL-7 (25). In addition to the function of IL-7 as a B cell growth factor, IL-7R signaling regulates the accessibility of distal V segments of the IgH genes, which leads to preferential rearrangement of distal V to DJ in pro-B cells (11, 26). Also, IL-7 may be necessary for pro-B cell survival through Mcl-1 expression (20). In this study, we have demonstrated that IL-7 is necessary for EBF expression in pre–pro-B cells, as well as for transition to more mature stages during adult B cell development. Initiation of EBF expression before the pre–pro-B cell stage might be IL-7 independent, because EBF expression is observed before the IL-7Rα+ CLP stage (27). Our preliminary data also suggests that CLPs derived from IL-7 −/− mice expressed a comparable level of EBF to WT (unpublished data). Therefore, it is possible that IL-7 stimulation is necessary for the maintenance of the EBF expression during the CLP to pre–pro-B cell stage transition. In addition, it is intriguing to know the regulation of EBF gene expression between fetal and adult pre–pro-B cells because fetal B cell development is IL-7 independent (28). Further studies are necessary to elucidate how IL-7 can mediate these different biological processes at different developmental stages of B lymphopoiesis.
Materials and methods
Mice.
IL-7Rα−/− mice were obtained from The Jackson Laboratory. IL-7 −/− mice were provided by I.L. Weissman (Stanford University, Stanford, CA), which were originally provided by R. Murray (DNAX Research Institute, Palo Alto, CA). RAG2 −/− mice were introduced with CD45.1 as described previously (29). All mice were backcrossed onto C57BL/6 background for more than eight generations. Age-matched C57BL/6 mice were used as WT control. The age of mice used in this study was between 4 and 8 wk old. Specifically, we used 8-wk-old mice in the experiment shown in Fig. 1 D. All mice were bred in a specific pathogen-free environment at the mouse facility of Duke University Medical Center. All experimental procedures related to laboratory mice were done according to guidelines specified by the institution.
Cell sorting and flow cytometric analysis.
Antibodies used in this study are listed in the online supplemental material. Preparation of single cell suspension and antibody staining of cells were done as previously described (15). Cell sorting and cell surface phenotyping were performed on a FACSVantage SE with a DiVa option (488 nm argon, 599 nm dye, and 408 nm krypton lasers; BD Biosciences Flow Cytometry Systems), which is available in the FACS facility of Duke University Comprehensive Cancer Center. Data were analyzed with the FlowJo software (Treestar). Dead cells were excluded from analyses and sortings as positively stained cells by propidium iodide.
RT-PCR.
Cells were sorted directly into 1.5-ml microcentrifuge tubes with 1 ml TRIzol reagent (Invitrogen). Total RNA was purified based on the manufactured instruction. First-strand cDNA was synthesized with Superscript III reverse transcriptase and oligo-dT primers (Invitrogen). Verification of the amount of first strand cDNA was done by amplification of β-actin. Genes of interest were amplified by PCR with BD Advantage 2 PCR Enzyme System (BD Biosciences) based on the manufactured protocol. In experiments shown in Fig. 2 C, we quantified EBF expression using the LightCycler system (Roche) after the first strand DNA synthesis. The primers for PCR are listed in the online supplemental material.
Retrovirus production and infection.
Retroviral vectors used in this study are shown in online supplemental material. STAT5A1*6, Mcl-1, Pax5, and Bcl-2 cDNAs were provided by T. Kitamura (University of Tokyo, Tokyo, Japan), S. Korsmeyer (Dana-Farber Cancer Institute, Boston, MA), J. Parnes (Stanford University, Stanford, CA), and K. Sugamura (Tohoku University, Sendai, Japan), respectively. Retroviruses were prepared as described previously (30). For retroviral infection, 2 × 104 IL-7Rα−/− HSCs were placed in 200 μl X-VIVO medium (BIO-WHITTKER) supplemented with 1% BSA, 10 ng/ml IL-11 and TPO, 50 ng/ml SCF (R&D Systems), and 50% of viral supernatants in a well of round bottom 96-well plates. The culture plate was centrifuged at 2,000 rpm at 22°C for 2 h (spin infection). After the spin infection, cells were incubated at 37°C. After 24-h culture, the frequency of GFP+ cells was verified by FACS. The cell numbers for injection were adjusted by the GFP+ cell frequency so that 4 × 103 GFP+ cells were injected into a mouse.
In vivo reconstitution assay.
IL-7Rα−/− HSCs (CD45.2+) infected with retroviruses were injected into retroorbital venous sinus of sublethally irradiated (400 rad) RAG2 −/− mice (CD45.1+). 5 wk after injection, mice were killed and differentiation of donor-derived cells was assessed by FACS.
In vitro stimulation of pre–pro-B cells with IL-7.
pre–pro-B cells derived from IL-7 −/− mice were placed in a well of 96-well plates in IMDM with 10% FCS, 50 μM 2-ME, and 10 ng/ml IL-7 (R&D Systems). Cells were harvested at the time points indicated in the figures and subjected to RT-PCR analyses.
Online supplemental material
A more detailed characterization of pre–pro-B cells is shown in Fig. S1. Rescue of B precursor compartments by ectopic EBF in IL-7Rα2/− HSCs is shown in Fig. S2. Antibodies, retroviral vectors, and PCR primers used in this study are listed in the online supplemental material. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20050158/DC1.
Acknowledgments
STAT5A1*6, Mcl-1, Pax5, and Bcl-2 cDNAs were kindly provided by T. Kitamura, S. Korsmeyer, J. Parnes, and K. Sugamura, respectively. We greatly appreciate the help of L. Martinek and M. Cook with the maintenance of cell sorters and advice in FACS sorting. We thank M. Krangel for access to a real-time PCR machine and W. Zhang and Y. Zhuang for critically reading the manuscript. We also thank H. Singh, C. Klug, B. Kee, J. Hagman, and R. Grosschedl for information on EBF-deficient mice. We wish to acknowledge the help by L. Jerabek and I.L. Weissman to M. Kondo's transition from Stanford to Duke University.
This work was supported in part by the startup fund from Duke University (M. Kondo) and grants from the National Institutes of Health (T32 AI52077 to A.Y. Lai and R01 AI056123 and R01 CA098129 to M. Kondo). K. Kikuchi was a fellow of the Uehara Memorial Foundation. M. Kondo is a scholar of the Sidney Kimmel Foundation for Cancer Research.
The authors have no conflicting financial interests.
References
- 1.Quong, M.W., W.J. Romanow, and C. Murre. 2002. E protein function in lymphocyte development. Annu. Rev. Immunol. 20:301–322. [DOI] [PubMed] [Google Scholar]
- 2.Busslinger, M. 2004. Transcriptional control of early B cell development. Annu. Rev. Immunol. 22:55–79. [DOI] [PubMed] [Google Scholar]
- 3.Kee, B.L., and C. Murre. 1998. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J. Exp. Med. 188:699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medina, K.L., J.M. Pongubala, K.L. Reddy, D.W. Lancki, R. Dekoter, M. Kieslinger, R. Grosschedl, and H. Singh. 2004. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell. 7:607–617. [DOI] [PubMed] [Google Scholar]
- 5.O'Riordan, M., and R. Grosschedl. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 11:21–31. [DOI] [PubMed] [Google Scholar]
- 6.Nutt, S.L., B. Heavey, A.G. Rolink, and M. Busslinger. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401:556–562. [DOI] [PubMed] [Google Scholar]
- 7.Mackarehtschian, K., J.D. Hardin, K.A. Moore, S. Boast, S.P. Goff, and I.R. Lemischka. 1995. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 3:147–161. [DOI] [PubMed] [Google Scholar]
- 8.von Freeden-Jeffry, U., P. Vieira, L.A. Lucian, T. McNeil, S.E. Burdach, and R. Murray. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181:1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sitnicka, E., C. Brakebusch, I.L. Martensson, M. Svensson, W.W. Agace, M. Sigvardsson, N. Buza-Vidas, D. Bryder, C.M. Cilio, H. Ahlenius, et al. 2003. Complementary signaling through flt3 and interleukin-7 receptor α is indispensable for fetal and adult B cell genesis. J. Exp. Med. 198:1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kouro, T., K.L. Medina, K. Oritani, and P.W. Kincade. 2001. Characteristics of early murine B-lymphocyte precursors and their direct sensitivity to negative regulators. Blood. 97:2708–2715. [DOI] [PubMed] [Google Scholar]
- 11.Corcoran, A.E., A. Riddell, D. Krooshoop, and A.R. Venkitaraman. 1998. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 391:904–907. [DOI] [PubMed] [Google Scholar]
- 12.Peschon, J.J., P.J. Morrissey, K.H. Grabstein, F.J. Ramsdell, E. Maraskovsky, B.C. Gliniak, L.S. Park, S.F. Ziegler, D.E. Williams, C.B. Ware, et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 180:1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy, R.R. 2003. B-cell commitment: deciding on the players. Curr. Opin. Immunol. 15:158–165. [DOI] [PubMed] [Google Scholar]
- 14.Miller, J.P., D. Izon, W. DeMuth, R. Gerstein, A. Bhandoola, and D. Allman. 2002. The earliest step in B lineage differentiation from common lymphoid progenitors is critically dependent upon interleukin 7. J. Exp. Med. 196:705–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo, M., K. Akashi, J. Domen, K. Sugamura, and I.L. Weissman. 1997. Bcl-2 rescues T lymphopoiesis, but not B or NK cell development, in common gamma chain-deficient mice. Immunity. 7:155–162. [DOI] [PubMed] [Google Scholar]
- 16.Akashi, K., M. Kondo, U. von Freeden-Jeffry, R. Murray, and I.L. Weissman. 1997. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 89:1033–1041. [DOI] [PubMed] [Google Scholar]
- 17.Maraskovsky, E., J.J. Peschon, H. McKenna, M. Teepe, and A. Strasser. 1998. Overexpression of Bcl-2 does not rescue impaired B lymphopoiesis in IL-7 receptor-deficient mice but can enhance survival of mature B cells. Int. Immunol. 10:1367–1375. [DOI] [PubMed] [Google Scholar]
- 18.Rolink, A., E. ten Boekel, F. Melchers, D.T. Fearon, I. Krop, and J. Andersson. 1996. A subpopulation of B220+ cells in murine bone marrow does not express CD19 and contains natural killer cell progenitors. J. Exp. Med. 183:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tudor, K.S., K.J. Payne, Y. Yamashita, and P.W. Kincade. 2000. Functional assessment of precursors from murine bone marrow suggests a sequence of early B lineage differentiation events. Immunity. 12:335–345. [DOI] [PubMed] [Google Scholar]
- 20.Opferman, J.T., A. Letai, C. Beard, M.D. Sorcinelli, C.C. Ong, and S.J. Korsmeyer. 2003. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 426:671–676. [DOI] [PubMed] [Google Scholar]
- 21.Singh, H., R.P. DeKoter, and J.C. Walsh. 1999. PU.1, a shared transcriptional regulator of lymphoid and myeloid cell fates. Cold Spring Harb. Symp. Quant. Biol. 64:13–20. [DOI] [PubMed] [Google Scholar]
- 22.Lin, H., and R. Grosschedl. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 376:263–267. [DOI] [PubMed] [Google Scholar]
- 23.Lin, J.X., T.S. Migone, M. Tsang, M. Friedmann, J.A. Weatherbee, L. Zhou, A. Yamauchi, E.T. Bloom, J. Mietz, S. John, and W.J. Leonard. 1995. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 2:331–339. [DOI] [PubMed] [Google Scholar]
- 24.Goetz, C.A., I.R. Harmon, J.J. O'Neil, M.A. Burchill, and M.A. Farrar. 2004. STAT5 activation underlies IL7 receptor-dependent B cell development. J. Immunol. 172:4770–4778. [DOI] [PubMed] [Google Scholar]
- 25.Hardy, R.R., and K. Hayakawa. 2001. B cell development pathways. Annu. Rev. Immunol. 19:595–621. [DOI] [PubMed] [Google Scholar]
- 26.Chowdhury, D., and R. Sen. 2003. Transient IL-7/IL-7R signaling provides a mechanism for feedback inhibition of immunoglobulin heavy chain gene rearrangements. Immunity. 18:229–241. [DOI] [PubMed] [Google Scholar]
- 27.Igarashi, H., S.C. Gregory, T. Yokota, N. Sakaguchi, and P.W. Kincade. 2002. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 17:117–130. [DOI] [PubMed] [Google Scholar]
- 28.Vosshenrich, C.A., A. Cumano, W. Muller, J.P. Di Santo, and P. Vieira. 2003. Thymic stromal-derived lymphopoietin distinguishes fetal from adult B cell development. Nat. Immunol. 4:773–779. [DOI] [PubMed] [Google Scholar]
- 29.Kondo, M., I.L. Weissman, and K. Akashi. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. [DOI] [PubMed] [Google Scholar]
- 30.Kondo, M., D.C. Scherer, T. Miyamoto, A.G. King, K. Akashi, K. Sugamura, and I.L. Weissman. 2000. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 407:383–386. [DOI] [PubMed] [Google Scholar]