Abstract
The mutation pattern of immunoglobulin genes was studied in mice deficient for DNA polymerase η, a translesional polymerase whose inactivation is responsible for the xeroderma pigmentosum variant (XP-V) syndrome in humans. Mutations show an 85% G/C biased pattern, similar to that reported for XP-V patients. Breeding these mice with animals harboring the stop codon mutation of the 129/Olain background in their DNA polymerase ι gene did not alter this pattern further. Although this G/C biased mutation profile resembles that of mice deficient in the MSH2 or MSH6 components of the mismatch repair complex, the residual A/T mutagenesis of polη-deficient mice differs markedly. This suggests that, in the absence of polη, the MSH2–MSH6 complex is able to recruit another DNA polymerase that is more accurate at copying A/T bases, possibly polκ, to assume its function in hypermutation.
Somatic hypermutation of immunoglobulin genes is initiated by activation-induced cytidine deaminase (AID; reference 1). Most experimental evidences suggest that AID initiates this process by deaminating cytidines into uracils in DNA (2). Accordingly, overexpression of AID in fibroblasts is sufficient to induce a G/C-targeted mutagenesis on a highly transcribed reporter gene (3). This mutagenesis is supposed to occur by saturation of the repair capacity of the cell, generating transition mutations by replication over the uracils created by deamination. Transversion mutations are likely generated by one or several translesional polymerases that ensure the replication bypass of abasic sites that failed to be corrected by the classical base excision repair pathway after excision of uracils by uracil glycosylases.
In contrast with this “passive” mutagenic process, hypermutation in activated B cells generates mutations at all four bases at the Ig locus (4). This mutagenesis is entirely dependent on the initial handling of the lesion by only two pathways, one dependent on the uracil glycosylase UNG, the other one upon a subset of the mismatch repair complex, its DNA binding moiety MSH2–MSH6, together with exonuclease 1 (Exo1; references 5–13). The UNG pathway appears necessary for generating most transversion mutations at G/C bases, whereas the MSH2–MSH6 pathway is required for a large part of the mutagenesis at A/T positions.
Translesional DNA polymerases are specific enzymes, recruited at replication forks stalled in front of noninstructional DNA lesions, and able to bypass them with various specificities and efficiencies (for review see reference 14). Moreover, based on in vitro assays, many of them are inherently error prone when copying undamaged DNA, again with various intrinsic misincorporation specificities, which made them prime candidates for the Ig gene mutational process, and several experimental data have indeed documented their involvement. The Ig gene mutation profile of xeroderma pigmentosum variant (XP-V) patients, deficient in DNA polymerase η (15), has suggested that this enzyme is a major contributor of the MSH2–MSH6 driven A/T mutagenesis (16–18). The DNA polymerase ι, which colocalizes with polη at replication foci (19), has been implied in the Ig gene mutagenesis induced in a Burkitt's lymphoma cell line (20). However, inactivation of this enzyme in the mouse, by a stop codon present as a natural mutation of various 129 substrains, failed to show any phenotype on hypermutation (21). DNA polymerase ζ is proposed to contribute to the overall quantitative efficiency of the process, but not to any specific pathway (22, 23).
To look for possible differences in the various DNA polymerases involved between mouse and humans, we inactivated polη in the mouse, and analyzed the Ig mutation pattern in the context of the polι mutation of the 129/Ola strain. This analysis confirms the phenotype observed for polη deficiency in humans, which is not altered further in the presence of the polι mutation of the 129 background.
Results and discussion
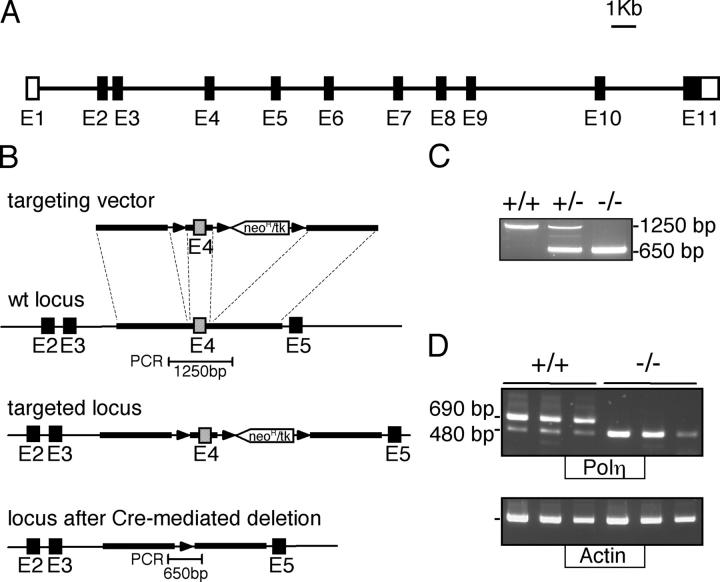
Inactivation of the mouse Polh gene was performed by a conditional knock-out strategy that introduces loxP sites flanking exon 4, an exon containing the DE polymerase motifs conserved in all Y-family polymerases and indispensable for polη catalytic function (Fig. 1 and references 15, 24). This strategy was designed with the aim of crossing the animals obtained with mice harboring other DNA repair defects, allowing the follow-up of the B cell lineage in case of lethality of their combined deficiencies. Deletion of exon 4 was performed here in the targeted embryonic stem (ES) cell clone, leaving in place a single loxP site and, therefore, minimizing disturbance around the gene. The Xpo5 gene, encoding exportin-5 involved in the nuclear export of microRNAs, indeed initiates 200 bp upstream from the first Polh exon in opposite transcriptional orientation; affecting expression of this gene is, therefore, likely to result in an early embryonic lethal phenotype (25).
Figure 1.
Disruption of the mouse Polh gene. (A) Scheme of the mouse Polh gene encoding DNA polymerase η. (Closed boxes) coding exons; (open boxes) noncoding regions. (B) Scheme of the gene targeting strategy. Homologous recombination of the gene-targeting construct results in the introduction of loxP sites on both sides of exon 4. (C) Screening of mouse genotypes: deletion of exon 4 is monitored by a PCR around this exon, using primers shown in B. (D) Polη gene expression in control and gene-targeted mice: amplification of polη transcripts is performed using primers in exons 2 and 6, on twofold consecutive dilutions of RT products from total spleen RNA. The amplified product in polη−/− animals corresponds to the splicing of exons 3–5, which generates an out-of-frame sequence.
The heterozygotes obtained by breeding chimeras with C57Bl/6 mice were crossed together and screened for the presence of the stop codon mutation born by the Poli gene from the 129/Ola genetic background of the ES cells. Wild-type, homozygous polη-deficient, and double polη-polι mutant mice were selected for analysis. Deletion of exon 4 of polη resulted at the transcriptional level in the direct splicing of exons 3–5, an out-of-frame junction that introduces a stop codon 11 amino acids downstream in exon 5 and generates a truncated polη protein of 102 amino acids (Fig. 1 D). Apart from deletion of exon 4, such a truncated protein would lack domains involved in nuclear localization and repair foci formation (19).
Wild-type, polη-deficient, and double polη-polι mutant mice were analyzed for the mutation pattern of their Ig locus. Different types of sequences were analyzed: intronic sequences flanking rearranged JH4 sequences (“JH4 intronic sequences”) and a 560-bp region upstream from the repeats of the Sμ core sequence (hereafter referred to as “pre-switch”), both from Peyer's patch PNAhigh B cells (Table I).
Table I.
Somatic mutations in JH4 intronic sequences and Sμ core upstream sequences from normal and mutant mice
| JH4 intronic sequence (490 bp)
|
Sμ core upstream sequence (560 bp)
|
|||||
|---|---|---|---|---|---|---|
| Controls | Polη −/− | Polη −/− × polι −/− | Controls | Polη −/− | Polη −/− × polι−/− | |
| Number of sequences | 21 | 71 | 53 | 96 | 122 | 127 |
| Total length sequenced (bp) | 10,290 | 34,790 | 25,970 | 53,760 | 66,320 | 71,120 |
| Unmutated sequences (%) | 14 | 17 | 25 | 42 | 46 | 57 |
| Total number of mutations | 124 | 241 | 129 | 147 | 160 | 122 |
| Number of deletions and insertions | 2 | 4 | 0 | 4 | 3 | 2 |
| Mutation frequency per total sequences (per 100 bp) |
1.2 | 0.7 | 0.5 | 0.3 | 0.2 | 0.2 |
| Mutation frequency per mutated sequences (per 100 bp) |
1.4 | 0.8 | 0.7 | 0.5 | 0.4 | 0.4 |
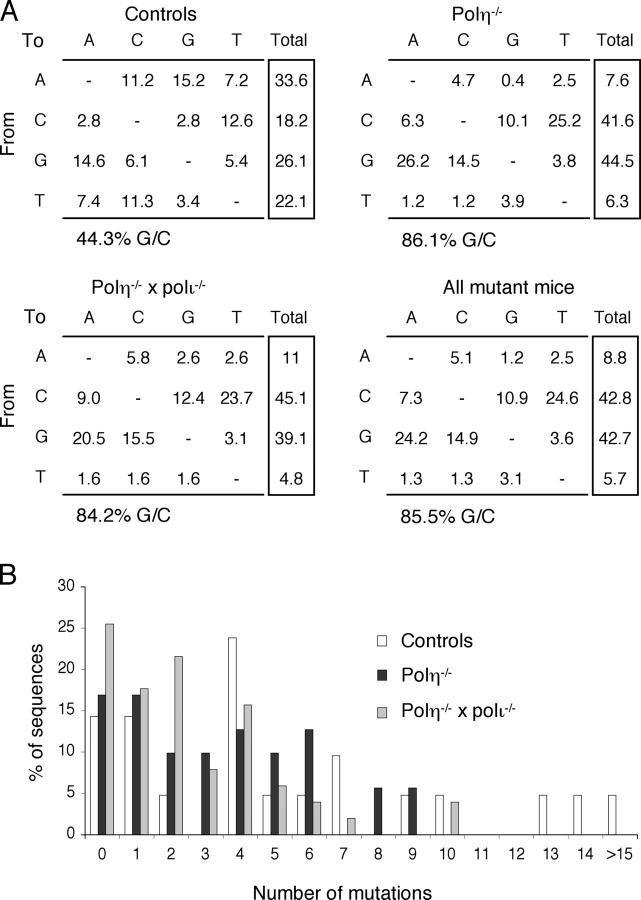
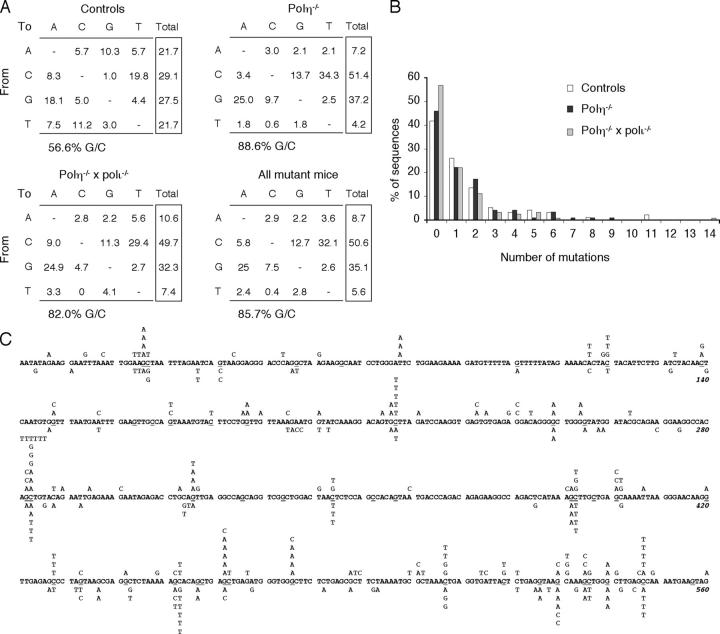
No B cell anomalies were observed in either type of polymerase-deficient animals. Ig sequences from polη−/− PNAhigh B cells displayed the same specific pattern as those obtained from memory B cells of XP-V patients; i.e., a marked reduction of mutations at A/T base pairs: 86.1% mutations at G/C base pairs for the JH4 intron and 88.6% for the pre-switch sequence (vs. 89.3 and 89.8% for the corresponding Ig gene sequences in XP-V patients; Figs. 2 and 3, Table II, and reference 17). Mutation frequencies were somewhat lower for polη-deficient animals compared with wild type when JH4 intronic sequences are considered, but similar for the pre-switch sequences. Such a quantitative dissociation has been described for MSH6- and for UNG/MSH2-deficient animals (5, 13).
Figure 2.
Distribution of mutations in the VHDJH4 flanking sequence (“JH4 intronic region”) of Peyer's patches PNAhigh B cells from polη- and polη-polι-deficient mice. (A) Pattern of nucleotide substitution in control mice (two animals), polη-deficient mice (three animals), polη- and polι-deficient mice (two animals), and from all mutant animals together. Data are given as percent substitution after correction for base composition (490 bp starting from the exon/intron border: A, 26.3%; C, 14.9%; G, 27.4%; T, 31.4%). (B) Accumulation of mutations in individual JH4 intronic sequences. The distribution of sequences harboring a given number of mutations relative to the total number of mutated sequences is represented for the three mouse genotypes.
Figure 3.
Distribution of mutations in the Sμ core upstream region (pre-switch) of Peyer's patches PNAhigh B cells from polη- and polη-polι-deficient mice. (A) Pattern of nucleotide substitution from the same control and mutant animals described in Fig. 2. Base composition: 560 bp; A, 33.2%; C, 16.4%; G, 27.5%; T, 22.9%. (B) Accumulation of mutations in individual pre-switch sequences. Same representation as in Fig. 2 B. (C) Distribution of mutations along the pre-switch sequence. Mutations from polη-deficient mice are listed above the sequence, and mutations from double-deficient mice are listed below the sequence. “G” within RGYW motifs and “C” within WRCY motifs are underlined.
Table II.
Pattern of nucleotide changes in JH4 intronic sequences of normal, polη-, and MSH2-deficient mice
| Within G/C
|
Within A/T
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Trans.
|
Transv.
|
Trans.
|
Transv.
|
|||||
| GC:AT | Transitions: transversions |
G/AC/T | G/TC/A | G/CC/G | A/GT/C | A/TT/A | A/CT/G | |
| Mouse | ||||||||
| Controls 1a | 44.3:55.7 | 53.7:46.3 | 61.4 | 18.5 | 20.1 | 47.6 | 26.2 | 26.2 |
| Controls 1b | 49.9:50.1 | 53.3:46.7 | 54.5 | 16.8 | 28.7 | 52.1 | 25.9 | 22.0 |
| Polη−/− c | 85.5:14.5 | 51.3:48.7 | 57.1 | 12.7 | 30.2 | 17.2 | 26.2 | 56.2 |
| Msh2 −/− d | 87.0:13.0 | 71.2:28.8 | 74.0 | 14.0 | 12.0 | 52.7 | 25.7 | 21.6 |
| Human | ||||||||
| Controls 1e | 46.0:54.0 | 51.2:48.8 | 55.4 | 16.5 | 28.1 | 47.6 | 25.0 | 27.4 |
| XP-V e | 89.3:10.7 | 46.6:53.4 | 51.1 | 17.1 | 31.8 | 9.4 | 36.4 | 54.2 |
Values in bold represent significant differences from controls discussed in the text.
This study (122 mutations).
From a larger sample (334 mutations; reference 36).
This study, data from all deficient mice together (366 mutations).
Data from mice deficient in the MSH2-dependent pathway (Msh2 −/−, Exo1 −/−, Msh2 G674A) were pooled (326 mutations obtained by Pfu polymerase; references 8, 11, 12).
From ref. 17 (168 mutations).
We next compared the mutation profile of mice harboring both polη and polι inactivation. These profiles appear extremely similar, either in their distribution along the sequence (shown for the pre-switch sequence in Fig. 3 C), or in their base substitution characteristics (Figs. 2 A and 3 A). The lack of phenotype of polι-deficient mice on Ig gene hypermutation is, therefore, not due to a compensatory role of polη in this specific strain.
All further analyses were, therefore, performed on pooled data from both types of mutant mice, allowing us to compare larger databases (366 mutations for the JH4 intron and 277 for the pre-switch sequence). The targeting was slightly more pronounced for C than for G, in particular for the pre-switch sequence. Such an increase has been described in switch junctions from XP-V patients (18). However, among the three types of sequences that we have analyzed in similar patients, we could not observe this bias in either switch junctions or JH4 introns, the specific increase in C mutations being only significant in pre-switch sequences from XP-V individuals (17). The general relevance of this observation is, therefore, difficult to assess at this stage. Moreover, among G/C mutations, no difference from wild type is noticeable in the relative proportion of transitions and transversions (Table II).
MSH2 deficiency, together with defects in MSH6 and Exo1, results in a similar bias toward G/C mutagenesis (7–13). We, therefore, wanted to compare the mutation pattern in the JH4 intronic sequence between Msh2 − / − and Polh − / − animals. This pattern was established using data from both MSH2- and Exo1-deficient animals, as well as MSH2-ATPase mutant mice (326 mutations), taking into account only data obtained using Pfu polymerase, to exclude any contribution of less accurate enzymes whose intrinsic error spectrum might specifically impinge on the A/T pattern. At G/C base pairs, these mutant mice show a strong bias toward transitions, which has been interpreted as an increased replicative bypass of uracils, the absence of the mismatch-binding complex leading to an increased fraction of deaminated bases escaping detection and repair. Polh − / − animals, in contrast, show a transition/transversion distribution at G/C bases similar to controls. Strikingly, the mutation pattern at A/T bases pairs shows the reverse situation. MSH2 deficiency shows a distribution similar to wild type, whereas polη deficiency results in a strong transversion bias. This transversion bias is consistent enough to emerge from both types of sequences analyzed (JH4 and pre-switch), and in both mouse and human polη deficiencies (Table II). Moreover, among transversions at A/T bases, most of them are A to C or T to G changes, which represent more than half of all A/T mutations. Such a mutation bias has been described for DNA polymerase κ (26, 27). It is thus possible that, in the absence of polη, the mismatch binding complex is able to recruit a different translesional enzyme, which, due to its specific misincorporation pattern, would be less mutagenic at A/T base pairs, leading to a lower frequency of A/T mutations with a specific transversion bias. Because polκ-deficient animals do not harbor the slightest alteration of their A/T mutation pattern (28), it is probable that this enzyme is not a normal actor of the Ig gene mutation process, being on duty only in case of absence of the regular partner of the MSH2–MSH6 complex (i.e., polη).
Polμ is another enzyme with such a transversion bias (29). However, contrary to polη and polκ, its preferred misincorporation is at copying A, and not T. It is therefore unlikely, in the mouse at least, that polμ could generate the A over T bias of mutations in JH4 sequences, present in control mice and conserved in polη-deficient animals.
UNG and MSH2–MSH6 are the only two repair pathways handling uracils generated by AID during Ig gene hypermutation. The model of Neuberger et al. (5) posits that G/C mutations are introduced by replication over the uracils or the abasic sites generated by uracil glycosylase, both events occurring on the DNA strand opposed to the lesion and without repair. Replication over the abasic site would involve translesional DNA polymerases in their “classical” role of damage bypass. Effectively, in the AID-dependent mutations observed in the chicken cell line DT40 that are almost entirely restricted to G/C bases, Rev1 has been proposed to be the major enzyme involved (30). As the mutation pattern of DT40 is more biased toward G to C and C to G transversions (the hallmark of Rev1) than it is in the mouse, it is so far unclear how many translesional polymerases are involved in the G/C mutation pattern in mouse (and human) B cells. Mutations at A/T bases are more difficult to explain on a strict replication mode within this model.
In another scenario, the MSH2–MSH6 complex would recruit polη for an error-prone repair of the lesion (i.e., on the same DNA strand) that would remove the uracil or the abasic site and create mismatches at nearby A/T bases because this enzyme is inherently more mutagenic at copying Ts. Along this line, specific modifications of AID in hypermutating B cells might also actively recruit UNG at the site of the lesion (31), driving the G/C mutagenesis to proceed differently from a strict saturation of the normal repair of uracils. In such a scheme, the dichotomy between translesion bypass and error-prone repair for the G/C versus A/T mutation pathways might have to be reassessed.
The absence of phenotype of the polι mutation of the 129 strains obviously questions the relevance of the polι-dependent mutagenic process that we described in the BL2 Burkitt's lymphoma cell line (20). In fact, it has been reported that such a polι-dependent mutagenesis can be induced at the Ig locus in B cell lines infected by the hepatitis C virus (32). These mutations would be related to the specific metabolic alterations brought upon infection by this oncogenic virus, alterations that may be shared by many B cell lymphomas (33). Alternatively, inactivation of polι in the 129/Ola mouse strain might show leakiness, in particular in activated B cells, by either read-trough of the stop codon or alternative splicing. A classical inactivation of the Poli gene and the analysis of mice cumulating several deficiencies in the activities involved in hypermutation might contribute to address these issues.
Materials and Methods
Construction of targeting vectors.
Three fragments from the POLH locus were amplified and cloned in the pFlox vector (34): fragment 1, 5′ flank; fragment 2, exon 4; and fragment 3, 3′ flank. Construction of the targeting vector and screening of recombinant clones are described in supplemental Materials and methods (available at http://www.jem.org/cgi/content/full/jem.20050292/DC1). Transfection of E14.1 ES cells was performed as described (35). One recombinant clone was obtained out of 287 ES clones, and subjected to Cre-mediated excision to generate exon 4–deleted clones.
Analysis of gene-targeted mice.
Screening of gene targeted mice was performed by PCR around exon 4 (described in Fig. 1) using the following primers: 5′-screen, 5′-GTCTCCCCAGATGTATCTCC-3′, and 3′-screen, 5′-GCATTGTACACCTGTGGTCC-3′ (45 s at 94°C, 30 s at 59°C, 3 min at 72°C, 40 cycles with Taq polymerase; Biolabs). The polι mutation of the 129/Ola background was screened by direct sequencing of a 540-bp genomic fragment encompassing exon 2 (amplified by iota-exon 2-5′, 5′-TTAAAGCAGGACTGAAGACC-3′; iota-exon 2-3′, 5′-CACATTTACTCTCGGTTGCA-3′, for 15 s at 98°C, 30 s at 60°C, 15 s at 72°C, 40 cycles with Phusion DNA polymerase; Finnzymes). Polη mRNA expression was analyzed by RT-PCR (Stratascript; Stratagene), using splenic RNA extracted by the QIAGEN RNeasy kit and PCR primers located in exons 2 and 6 (exon 2, 5′-GGGCAGAATCGAGTGGTTGC-3′; exon 6 reverse, 5′-GCGGTTGGGCTTATTTAGTCC-3′, 30 s at 95°C, 90 s at 68°C, 40 cycles with Advantage II polymerase mix; CLONTECH Laboratories, Inc.).
Analysis of mutations in the Ig locus.
B220+ PNAhigh B cells from Peyer's patches were isolated from 3–4-mo-old animals as described previously (8). Amplification of the JH4 intronic sequence flanking rearranged VH genes was performed using a mixture of primers amplifying most VH gene families described (compiled from the mouse V gene IMGT database, http://imgt.cines.fr): V1-FR3, 5′-GAGGACTCTGCRGTCTATTWCTGTGC-3′; V5-FR3, 5′-GAGGACACRGCCATGTATTACTGTGC-3′; V3-FR3, 5′-GAGGACACACCCACATATTACTGTGC-3′; V7-FR3, 5′-GAGGACAGTGCCACTTATTACTGTGC-3′; and V9-FR3, 5′-ATGAGGACATGGCTACATATTTCTGT-3′, respectively, in a 6:3:1:1:1 ratio, and JH3′, 5′-TGAGACCGAGGCTAGATGCC-3′. PCR was performed on five aliquots of 500 or 1,000 cells (15 s 98°C, 30 s 64°C, 30 s 72°C for 50 cycles using Phusion DNA polymerase) and 480 bp sequences were determined using the JH3′ primer. A 735-bp fragment upstream from the Sμ core repeat sequence was amplified using primers Sμ-5′, 5′-GTTGAGGTACTGATGCTGTC-3′ and Sμ-3′, 5′-CCAGCCTAGTTTAGCTTAGC-3′ (45 s at 94°C, 30 s at 58°C, 2 min at 72°C, 40 cycles with Pfu Turbo), and a 560-bp sequence was determined using primer 5′-CTATTCTGGCTCTTCTTAAGC-3′. Sequences were obtained with an ABI Prism 3100 Genetic Analyzer after cloning in the Zero Blunt vector (Invitrogen).
Online supplemental material.
Construction of the targeting vector for inactivation of the Polh gene, as well as conditions for analysis of the recombinant ES clones are described online. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20050292/DC1.
Acknowledgments
We thank J. Mégret for performing cell sorting, and the Service d'Expérimentation Animale et de Transgénèse (Villejuif, France) for the generation and handling of gene targeted animals.
This work was supported by a grant from the Ministère de la Recherche (A.C.I. Biologie du Développement et Physiologie Intégrative), the Ligue Nationale contre le Cancer (Equipe Labellisée), and the Fondation Princesse Grace de Monaco.
The authors have no conflicting financial interests.
J.-C. Weill and C.-A. Reynaud share senior authorship for this work.
References
- 1.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 2.Barreto, V.M., A.R. Ramiro, and M.C. Nussenzweig. 2005. Activation-induced deaminase: controversies and open questions. Trends Immunol. 26:90–96. [DOI] [PubMed] [Google Scholar]
- 3.Yoshikawa, K., I.M. Okazaki, T. Eto, K. Kinoshita, M. Muramatsu, H. Nagaoka, and T. Honjo. 2002. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 296:2033–2036. [DOI] [PubMed] [Google Scholar]
- 4.Reynaud, C.-A., S. Aoufouchi, A. Faili, and J.-C. Weill. 2003. What role for AID: mutator, or assembler of the immunoglobulin mutasome? Nat. Immunol. 4:631–638. [DOI] [PubMed] [Google Scholar]
- 5.Rada, C., J.M. Di Noia, and M.S. Neuberger. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T focused phase of somatic mutation. Mol. Cell. 16:163–171. [DOI] [PubMed] [Google Scholar]
- 6.Rada, C., G.T. Williams, H. Nilsen, D.E. Barnes, T. Lindahl, and M.S. Neuberger. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 7.Rada, C., M.R. Ehrenstein, M.S. Neuberger, and C. Milstein. 1998. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 9:135–141. [DOI] [PubMed] [Google Scholar]
- 8.Frey, S., B. Bertocci, F. Delbos, L. Quint, J.C. Weill, and C.A. Reynaud. 1998. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 9:127–134. [DOI] [PubMed] [Google Scholar]
- 9.Phung, Q.H., D.B. Winter, A. Cranston, R.E. Tarone, V.A. Bohr, R. Fishel, and P.J. Gearhart. 1998. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J. Exp. Med. 187:1745–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiesendanger, M., B. Kneitz, W. Edelmann, and M.D. Scharff. 2000. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J. Exp. Med. 191:579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin, A., Z. Li, D.P. Lin, P.D. Bardwell, M.D. Iglesias-Ussel, W. Edelman, and M.D. Scharff. 2003. Msh2 ATPase activity is essential for somatic hypermutation at A-T base pairs and for efficient class switching. J. Exp. Med. 198:1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bardwell, P.D., C.J. Woo, K. Wei, Z. Li, A. Martin, S.Z. Sack, T. Parris, W. Edelman, and M.D. Scharff. 2004. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 5:224–229. [DOI] [PubMed] [Google Scholar]
- 13.Martomo, S.A., W.W. Yang, and P.J. Gearhart. 2004. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J. Exp. Med. 200:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkel, T.A., Y.I. Pavlov, and K. Bebenek. 2003. Functions of human DNA polymerases eta, kappa and iota suggested by their properties, including fidelity with undamaged DNA templates. DNA Repair (Amst.). 2:135–149. [DOI] [PubMed] [Google Scholar]
- 15.Masutani, C., R. Kusumoto, A. Yamada, N. Dohmae, M. Yokoi, M. Yuasa, M. Araki, S. Iwai, K. Takio, and F. Hanaoka. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 399:700–704. [DOI] [PubMed] [Google Scholar]
- 16.Zeng, X., D.B. Winter, C. Kasmer, K.H. Kraemer, A.R. Lehmann, and P.J. Gearhart. 2001. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2:537–541. [DOI] [PubMed] [Google Scholar]
- 17.Faili, A., S. Aoufouchi, S. Weller, F. Vuillier, A. Stary, A. Sarasin, C.A. Reynaud, and J.C. Weill. 2004. DNA polymerase ε is involved in hypermutation occurring during immunoglobulin class switch recombination. J. Exp. Med. 199:265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng, X., G.A. Negrete, C. Kasmer, W.W. Yang, and P.J. Gearhart. 2004. Absence of DNA polymerase ε reveals targeting of C mutations on the nontranscribed strand in immunoglobulin switch regions. J. Exp. Med. 199:917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kannouche, P., A.R. Fernandez de Henestrosa, B. Coull, A.E. Vidal, C. Gray, D. Zicha, R. Woodgate, and A.R. Lehmann. 2002. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 21:6246–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faili, A., S. Aoufouchi, E. Flatter, Q. Guéranger, C.-A. Reynaud, and J.-C. Weill. 2002. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature. 419:944–947. [DOI] [PubMed] [Google Scholar]
- 21.McDonald, J.P., E.G. Frank, B.S. Plosky, I.B. Rogozin, C. Masutani, F. Hanaoka, R. Woodgate, and P.J. Gearhart. 2003. 129-derived strains of mice are deficient in DNA polymerase iota and have normal immunoglobulin hypermutation. J. Exp. Med. 198:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz, M., L.K. Verkoczy, M.F. Flajnik, and N.R. Klinman. 2001. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase zeta. J. Immunol. 167:327–335. [DOI] [PubMed] [Google Scholar]
- 23.Zan, H., A. Komori, Z. Li, A. Cerutti, A. Schaffer, M.F. Flajnik, M. Diaz, and P. Casali. 2001. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 14:643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondratick, C.M., M.T. Washington, S. Prakash, and L. Prakash. 2001. Acidic residues critical for the activity and biological function of yeast DNA polymerase eta. Mol. Cell. Biol. 21:2018–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kannouche, P., and A. Stary. 2003. Xeroderma pigmentosum variant and error-prone DNA polymerases. Biochimie. 85:1123–1132. [DOI] [PubMed] [Google Scholar]
- 26.Ohmori, and T.A. Kunkel. 2000. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem. 275:39678–3984. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, Y., F. Yuan, H. Xin, X. Wu, D.K. Rajpal, D. Yang, and Z. Wang. 2000. Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 28:4147–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schenten, D., V.L. Gerlach, C. Guo, S. Velasco-Miguel, C.L. Hladik, C.L. White, E.C. Friedberg, K. Rajewsky, and G. Esposito. 2002. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur. J. Immunol. 32:3152–3160. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Y., X. Wu, F. Yuan, Z. Xie, and Z. Wang. 2001. Highly frequent frameshift synthesis by human DNA polymerase mu. Mol. Cell. Biol. 21:7995–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson, L.J., and J.E. Sale. 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 22:1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhuri, J., C. Khuong, and F.W. Alt. 2004. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 430:992–998. [DOI] [PubMed] [Google Scholar]
- 32.Machida, K., K.T. Cheng, V.M. Sung, S. Shimodaira, K.L. Lindsay, A.M. Levine, M.Y. Lai, and M.M. Lai. 2004. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc. Natl. Acad. Sci. USA. 101:4262–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machida, K., K.T. Cheng, V.M. Sung, K.J. Lee, A.M. Levine, and M.M. Lai. 2004. Hepatitis C virus infection activates the immunologic (type II) isoform of nitric oxide synthase and thereby enhances DNA damage and mutations of cellular genes. J. Virol. 78:8835–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu, H., J.D. Marth, P.C. Orban, H. Mossmann, and K. Rajewsky. 1994. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 265:103–106. [DOI] [PubMed] [Google Scholar]
- 35.Torres, R., and R. Kühn. 1997. Laboratory Protocols for Conditional Gene Targeting. Oxford University Press, Oxford, UK. 167 pp.
- 36.Bertocci, B., A. De Smet, E. Flatter, A. Dahan, J.C. Bories, C. Landreau, J.C. Weill, and C.A. Reynaud. 2002. DNA polymerases mu and lambda are dispensable for Ig gene hypermutation. J. Immunol. 168:3702–3706. [DOI] [PubMed] [Google Scholar]