Abstract
We analyzed the structure of antigen receptors of a comprehensive panel of mature B non-Hodgkin's lymphomas (B-NHLs) by comparing, at the amino acid level, their immunoglobulin (Ig)VH-CDR3s with CDR3 sequences present in GenBank. Follicular lymphomas, diffuse large B cell lymphomas, Burkitt's lymphomas, and myelomas expressed a CDR3 repertoire comparable to that of normal B cells. Mantle cell lymphomas and B cell chronic lymphocytic leukemias (B-CLLs) expressed clearly restricted albeit different CDR3 repertoires. Lymphomas of mucosa-associated lymphoid tissues (MALTs) were unique as 8 out of 45 (18%) of gastric- and 13 out of 32 (41%) of salivary gland-MALT lymphomas expressed B cell antigen receptors with strong CDR3 homology to rheumatoid factors (RFs). Of note, the RF-CDR3 homology without exception included N-region–encoded residues in the hypermutated IgV H genes, indicating that they were stringently selected for reactivity with auto-IgG. By in vitro binding studies with 10 MALT lymphoma–derived antibodies, we showed that seven of these cases, of which four with RF-CDR3 homology, indeed possessed strong RF reactivity. Of one MALT lymphoma, functional proof for selection of subclones with high RF affinity was obtained. Interestingly, RF-CDR3 homology and t(11;18) appeared to be mutually exclusive features and RF-CDR3 homology was not encountered in any of the 19 pulmonary MALT lymphomas studied.
B cell non-Hodgkin's lymphomas (B-NHLs) comprise >85% of malignant lymphomas worldwide. They are in majority of germinal center (GC) or post-GC phenotype and often harbor chromosomal translocations typically involving immunoglobulin (Ig) loci (1). In spite of their genetic defects, most B-NHLs do not replicate spontaneously in vitro, indicating that they still depend on environmental stimuli for their growth. To date, these external factors are ill defined. Evidence exists that B cell antigen receptor (BCR) ligands have, as in normal B cell development, a pivotal role in the pathogenesis of at least some B-NHLs. For example, the architecture and cellular composition of follicular lymphomas (FLs) is highly reminiscent of normal GCs. Furthermore, extranodal marginal zone B cell lymphomas (MZBCLs) of mucosa-associated lymphoid tissue (MALT) arise at sites of antigenic stimulation due to organ-specific autoimmunity; e.g., Sjögren's sialadenitis (2) and Hashimoto's thyroiditis (3), or infection like Helicobacter pylori gastritis (4, 5) and Borrelia burgdorferi dermatitis (6, 7), respectively. Similarly, a role of hepatitis C virus (HCV) infection has been inferred in the development of malignant B cell proliferation, including splenic MZBCL and MALT lymphoma (8, 9). Most recently, it has been claimed that ocular adnexal MALT lymphoma and immunoproliferative small intestinal disease (also known as α-heavy chain disease) are associated with Chlamydia psittaci and Campylobacter jejuni infections, respectively (10, 11). The low tendency of MALT lymphomas to spread beyond the environment in which they evolve may be related to the expression of certain homing and chemokine receptors, such as α4β7 and CXCR3 (12, 13). In addition, it has been proposed that even during the tumor stage, antigen (Ag) plays a growth-sustaining role. This notion is strongly supported by the observation that a proportion of low grade gastric MALT lymphomas (14, 15) and skin MZBCLs (7) are curable by bacterial eradication alone, while interferon α-2b treatment can cause regression of HCV-associated MZBCLs (16, 17).
Analysis of the Ig variable (IgV) genes supported the concept of Ag-driven lymphomagenesis in FL and MALT lymphoma. The IgV heavy (IgV H) and IgV light (IgV L) chain genes of these malignancies are heavily mutated, compatible with a GC or post-GC derivation (18–22). The mutation patterns unequivocally indicate that Ag-based selection occurs at some stage of their development; despite high mutation loads, the overall structure of the Ig is generally being preserved in these lymphomas often during years of disease. Apparently, selective forces prevent the outgrowth of BCR− lymphoma mutants.
Although many studies on FL and MALT lymphomas allude to a role for Ag in the pathogenesis of these lymphomas, only sporadic data exist on the exact ligands that these B cell neoplasms might recognize. Although the obvious candidate ligand for gastric MALT lymphoma was H. pylori, in vitro cultures revealed that the tumor B cells were not directly stimulated by H. pylori, but indirectly by CD40/CD40L-mediated help of intratumoral, H. pylori–specific T cells (23). Hussell et al. (24) and Greiner et al. (25) observed reactivity of MALT lymphoma–derived Abs with follicular DCs (FDCs), various epithelia, or postcapillary venules of Peyer's patches, but no specific Ags were molecularly defined. It is well documented that a significant proportion of B cell chronic lymphocytic leukemias (B-CLLs) express Ig (poly) reactive with a diversity of autoantigens (26–31). In contrast, with other B-NHL entities, autoreactivity has only sporadically been reported (32–34). Finally, viral antigens of HTLV-I and HCV have been implicated as BCR ligands of individual cases of B-CLL (35) and diffuse large B cell lymphoma (DLBCL; reference 36), respectively.
To address the issue of antigen-receptor specificity of B-NHL, we performed a systematic analysis of the antigen binding sites of 132 extranodal MZBCL (24 from our laboratory and 108 from literature) and, for comparison, from a comprehensive panel of 478 other B-NHL. We provide evidence that among the various B cell neoplasms, gastric- and salivary gland-MALT lymphomas express a distinctive Ig repertoire and frequently originate from precursor B cells clonally selected for auto-IgG binding capacity. The fact that B-NHL entities express qualitatively different Igs points toward different roles of the BCRs in their pathogenesis.
Results
IgVH and IgVL sequence analysis of MALT lymphomas
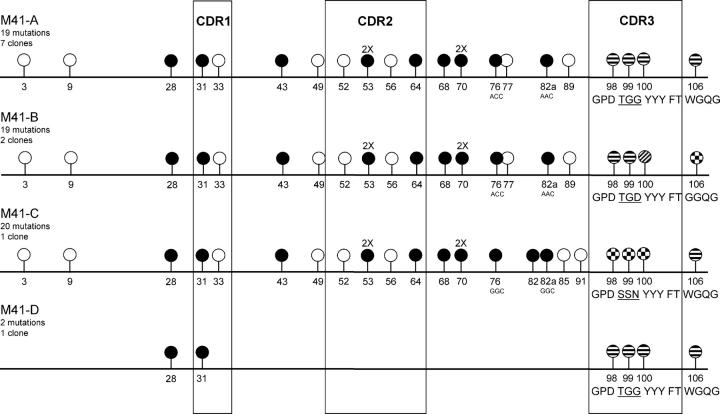
A panel of 24 MALT lymphomas was analyzed (Table I). All lymphomas were CD20+ and virtually all expressed the chemokine receptor CXCR3 and the mucosal homing integrin α4β7. The pulmonary lymphomas M19, M20, and M23 and the gastric lymphomas M56, M57, M58, M60, and M61 carried the t(11;18), involving the API2 and MALT1 genes (Table I). Except for M60, all IgV H and IgVκ genes analyzed were somatically mutated with means of 21 (range 0–68) and 18 mutations (range 1–58) per IgV gene, respectively (Tables II and III). Of M6, biopsies were available of two time points spanning a 9 mo interval. At relapse (M6′96), 15 somatic mutations were found in the expressed V3-7 IgV H gene while at presentation (M6′95) an additional replacement mutation in codon 13 had been present (Table II). Immunohistochemistry (Table I) and RT-PCR (Table II) indicated that the lung lymphoma M20 contained both IgM- and IgA-expressing tumor cells. The IgM- and the IgA-related IgV H sequences were identical and contained 5 mutations (Table II). In 17 of the 24 lymphomas (71%), the replacement versus silent (R/S) mutation ratios in the framework regions (FRs) of the IgV H genes, were significantly <1.5, implying that, in spite of the high mutation frequencies, selective forces had preserved the BCR in these lymphomas (unpublished data). Intraclonal variation (ICV) was determined for 16 IgV H genes and for 8 IgVκ genes (18). In 10 out of the 16 (63%) MALT lymphomas, significant ICV was found in IgV H (Table II). Except for M6, all lymphomas with ICV in IgV H genes also displayed ICV in the IgVκ genes, generally of a lower degree (Table III). In M15, lacking ICV in the IgV H gene, an exceptionally high degree of ICV was observed in its IgVκ gene (Tables II and III). Interestingly, M41 appeared a somatically diversified, V1-69/D3-22/JH4b-expressing lymphoma, harboring distinct subclones (M41-A, B, C, and D) with 19, 19, 20, and 2 mutations, respectively (Fig. 1).
Table I.
Clinical presentation, immunohistochemistry, and genetics of 24 MALT lymphomas
| Immunohistochemistry
|
PCR
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Sex | Age | Location | Clinical presentation | Ig isotype | Light chain | CXCR3 | α4β7 | t(11;18) |
| M4 | f | 60 | thyroid gland | Hashimoto's thyroiditis | NC | κ | − | − | − |
| M5 | f | 64 | parotid gland | Sjögren's syndrome | IgM | κ | + | + | − |
| M6'95 | f | 55 | stomach | gastritis | ND | κ | + | + | − |
| M6'96 | f | 56 | stomach | gastritis | IgM | κ | + | + | ND |
| M8 | m | 44 | parotid gland | unknown | IgM | κ | − | + | − |
| M9 | f | 58 | tonsil | unknown | IgM | κ | − | + | − |
| M11 | m | 38 | parotid gland | unknown | IgM | κ | + | + | − |
| M13 | m | 63 | ileum | unknown | NC | NC | + | + | − |
| M14 | f | 70 | parotid gland | Sjögren's syndrome | IgA | κ | + | + | − |
| M15 | f | 45 | lacrymal gland | Sjögren's syndrome | IgA | κ | + | + | − |
| M19 | f | 74 | lung | unknown | IgM | λ | + | + | + |
| M20 | m | 39 | lung | unknown | IgM/IgA | λ | + | + | + |
| M21 | f | 64 | parotid gland | Sjögren's syndrome | IgM | κ | + | + | − |
| M22a | m | 71 | groin lymph node | unknown | IgA | κ | + | + | − |
| M23 | m | 52 | lung | unknown | IgG | κ | + | + | + |
| M30 | f | 60 | stomach | gastritis | NC | κ | ND | ND | − |
| M41 | f | 55 | stomach | gastritis | ND | ND | ND | ND | − |
| M45 | m | 81 | stomach | gastritis | ND | ND | ND | ND | − |
| M46 | m | 65 | stomach | gastritis | ND | ND | ND | ND | − |
| M55 | m | 41 | lung | unknown | ND | ND | ND | ND | − |
| M56 | m | 78 | stomach | gastritis | IgM | κ | + | + | + |
| M57 | m | 81 | stomach | gastritis | IgM | κ | + | + | + |
| M58 | m | 74 | stomach | gastritis | IgM | NC | + | + | + |
| M60 | m | 71 | stomach | gastritis | IgM | λ | + | + | + |
| M61 | m | 38 | stomach | gastritis | IgM | λ | + | − | + |
Initially located in salivary gland.
NC, not clear.
Table II.
Immunoglobulin variable heavy chain genes of 24 MALT lymphomas
| Patient | Ig isotype (RT-PCR) |
VH family | Closest VH germline gene |
No. of mutations (%) | D gene | JH gene | Intraclonal variationa |
|---|---|---|---|---|---|---|---|
| M4 | γ | VH3 | V3-23 (DP47) | 68 (23) | NA | JH4b | 0.8 (5) |
| M5 | μ | VH3 | V3-7 (DP54) | 17 (5.8) | D3-3 | JH3b | 0.8 (5) |
| M6'95 | μ | VH3 | V3-7 (DP54) | 16 (5.4) | D3-22 | JH3b | ND |
| M6'96 | μ | VH3 | V3-7 (DP54) | 15 (5.1) | D3-22 | JH3b | 1.0 (5) |
| M8 | μ,δb | VH3 | V3-30 (DP49) | 26 (8.8) | D5-24 | JH5 | ND |
| M9 | μ,δb | VH1 | V1-69 (DP10) | 8 (2.7) | NA | JH4b | ND |
| M11 | μ | VH1 | V1-69 (DP10) | 11 (3.7) | D6-13 | JH4b | 3.0 (7) |
| M13 | μ | VH4 | V4-31 (DP65) | 14 (4.7) | D5-24 | JH4 | 1.8 (5) |
| M14 | α | VH3 | V3-23 (DP47) | 43 (15.0) | NA | JH6 | 2.3 (6) |
| M15 | α | VH1 | V1-18 (DP14) | 46 (16.0) | NA | JH4b | <0.4 (5) |
| M19 | μ,db | VH3 | V3-53 | 29 (10.0) | NA | JH4b | ND |
| M20 | μ,α | VH4 | V4-30.4 (DP78) | 5 (1.7) | NA | JH3b | ND |
| M21 | μ,δb | VH3 | V3-23 (DP47) | 23 (7.8) | D2-2 | JH4b | 0.7 (11) |
| M22 | α | VH1 | V1-69 (DP88) | 14 (4.8) | D4-17 | JH4b | <0.4 (15) |
| M23 | γ | VH1 | V1-69 (DP88) | 7 (2.4) | NA | JH4b | <0.4 (7) |
| M30 | γ | VH2 | V2-5 (VII-5) | 33 (11.1) | NA | JH1 | 0.7 (7) |
| M41 | ND | VH1 | V1-69 (DP10) | 19 (6.5) | D3-22 | JH4b | <0.4 (7)c |
| M45 | ND | VH1 | V1-3 (DP25) | 22 (7.5) | D3-10 | JH4b | ND |
| M46 | ND | VH3 | V3-30/30.5 (DP49) | 12 (4.1) | D5-12 | JH6b | ND |
| M55 | ND | VH3 | V3-7 (DP54) | 24 (8.1) | D3-22 | JH4b | ND |
| M56 | μ | VH1 | V1-69 (DP10) | 7 (2.4) | D1-14 | JH4b | <0.4 (11) |
| M57 | μ | VH1 | V1-18 (DP14) | 14 (4.8) | NA | JH6b | 1.9 (9) |
| M58 | μ | VH3 | V3-53 | 23 (7.9) | D3-22 | JH4b | ND |
| M60 | μ | VH1 | V1-69 (DP10) | 0 (0.0) | D2-15 | JH5b | <0.4 (9) |
| M61 | μ | VH1 | V1-18 (DP14) | 19d (6.5) | D2-15 | JH4b | 0.8 (4) |
The intraclonal variation is indicated as the mean number of nucleotide differences observed per clone, as compared with the consensus sequence. Numbers in parentheses indicate the number of clones that were sequenced.
IgD was not detected immunohistochemically.
M41 contained distinct subclones (Fig. 1).
M61 had a deletion of the three nucleotides of codon 29.
NA, the germline D gene could not definitely be assigned.
Table III.
Immunoglobulin variable light chain κ genes of the MALT lymphomas
| Patient | Vκ family |
Closest Vκ germline gene |
No. of mutations (%) |
Jκ gene |
Intraclonal variationa |
|---|---|---|---|---|---|
| M4 | Vκ1 | L9 (Ve+) | 58 (20.0) | Jκ4 | 0.5 (4) |
| M5 | Vκ3 | L2 (kv328, DPK21) | 9 (3.2) | Jκ1 | 1.0 (5) |
| M6'96 | Vκ3 | L2 (kv328, DPK21) | 5 (1.8) | Jκ1 | 0.4 (5) |
| M8 | Vκ1 | O12/O2 (DPK9) | 27 (9.5) | Jκ4 | ND |
| M9 | Vκ3 | A27 (kv325, DPK22) | 2 (0.69) | Jκ1 | ND |
| M11 | Vκ3 | A27 (kv325, DPK22) | 1 (0.35) | Jκ1 | 1.5 (6) |
| M14 | Vκ2 | A19/A3 (DPK15) | 51 (17.0) | Jκ4 | 1.2 (6) |
| M15 | Vκ4 | B3 (DPK24) | 17 (5.6) | Jκ4 | 9.3 (6) |
| M21 | Vκ3 | A27 (kv325, DPK22) | 13 (4.5) | Jκ1 | 0.5 (4) |
| M22 | Vκ3 | A27 (kv325, DPK22) | 6 (2.1) | Jκ2 | ND |
| M23 | Vκ1 | O12/O2 (DPK9) | 6 (2.1) | Jκ1 | 0.8 (8) |
The intraclonal variation is indicated as the mean number of nucleotide differences observed per clone, as compared to the consensus sequence. Numbers in parentheses indicate the number of clones that were sequenced.
Figure 1.
Schematic representation of the IgVH clones identified in M41. The lollipop-shaped symbols indicate nucleotide differences as compared with the V1-69 (DP10) germline IgV H gene. Except for the CDR3 region, replacement and silent mutations are indicated with closed and open circles, respectively, with codon numbering according to V-base indicated underneath. 2×, two mutations in the indicated codon. The mutations in codons 76 and 82a are different between M41-A/B and M41-C, respectively. In the CDR3 and in codon 106, interclonal differences are indicated by different filling patterns of the circles. In the CDR3, the deduced aa sequence is depicted in the one-letter code underneath. The CDR3 of M41-A/D, M41-B and M41-C displayed, respectively, 73, 82, and 91% homology to the CDR3 of RF-WOL. M41-D has only two mutations, shared with M41-A, B, and C.
IgVH-CDR3 amino acid sequences of MALT lymphomas and splenic MZBCL
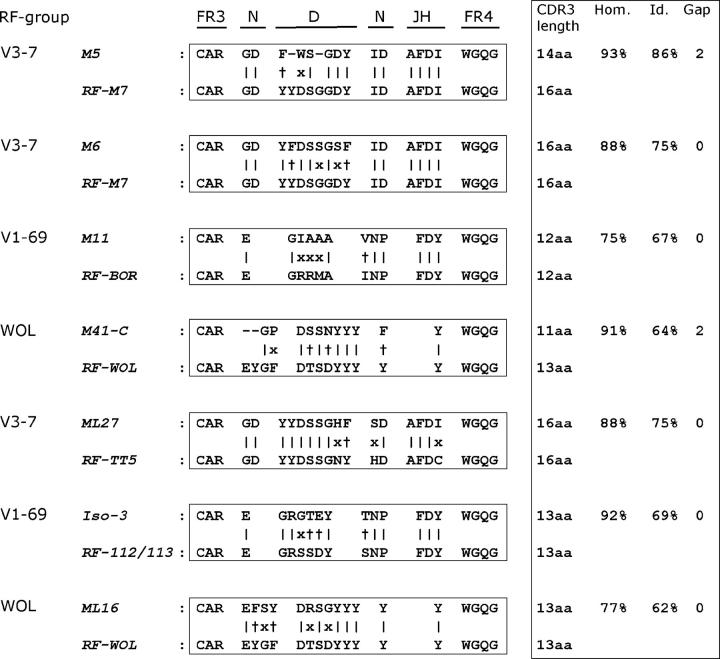
We compared the IgVH-CDR3 amino acid (aa) sequences of our panel of MALT lymphomas to IgVH-CDR3 aa sequences present on GenBank using the NCBI Protein-BLAST program with the option “search for short nearly exact matches” (BLASTP 2.2.6; reference 37). CDR3 regions consisting of at least 7 aa (all except M20) were analyzed (Table IV). A CDR3 sequence was considered to be homologous to previously reported CDR3 sequences on GenBank (a) if sharing at least 75% aa sequence homology and (b) a length difference between the CDR3 sequences not exceeding 3 aa (maximum gap of 3 aa). Applying these homology criteria, we found that the CDR3 of 4 MALT lymphomas (M23, M46, M56, and M60) displayed homology to different B cell clones analyzed previously in repertoire studies. Interestingly, 4 cases (M5, M6, M11, and M41) showed strong homology with CDR3 sequences of other MALT lymphomas, as well as with CDR3 sequences of rheumatoid factors (RF). Salivary gland lymphoma M5 and gastric lymphoma M6, both expressing a V3-7/JH3 rearrangement, were homologous to a plethora of IgVH-CDR3 aa sequences originating from salivary gland MALT lymphomas, gastric MALT lymphomas, HCV-associated lymphomas, B cell clones derived of benign myoepithelial sialadenitis, one DLBCL and two genuine V3-7–encoded RFs (Table S1, available at http://www.jem.org/cgi/content/full/jem.20050068/DC1). Salivary gland lymphoma M11, which expressed a V1-69/JH4 rearrangement, was homologous to five normal B cell clones, one salivary gland MALT lymphoma Isolate-4 (38), and three V1-69-encoded RFs (Table S1). Interestingly, Isolate-4 is one of the five cases described by Miklos et al. (38) having distinct CDR3 characteristics, indicating that M11 belongs to this group which is characterized by distinct aa motifs (ERG and NP) at the VH-DH- and the DH-JH-junctions, respectively. Gastric MALT lymphoma M41, which also expressed V1-69/JH4, was homologous to HCV-associated lymphomas of different histological subtypes, one splenic MZBCL, one salivary gland MALT lymphoma, one gastric MALT lymphoma and finally to one RF termed RF-WOL (reference 39 and Table S1).
Table IV.
Amino acid sequences of the variable heavy chain CDR3 of the 24 MALT lymphomas
| Patient | VH-D-JH rearrangement | CDR3 amino acid sequencea | CDR3 length |
|---|---|---|---|
| aa | |||
| M4 | V3-23 (DP47)/JH4b | CTK AHVPYFDGLSPSNV WGQG | 14 |
| M5 | V3-7 (DP54)/D3-3/JH3b | CAR GD FWSGDY ID AFDI WGQG | 14 |
| M6 | V3-7 (DP54)/D3-22/JH3b | CAR GD YFDSSGSF ID AFDI WGQG | 16 |
| M8 | V3-30 (DP49)/D5-24/JH5 | CAK DGSEFRLIY WFDS WGRG | 13 |
| M9 | V1-69 (DP10)/JH4b | CAR DWAHQGETRSNFLYY WGQG | 15 |
| M11 | V1-69 (DP10)/D6-13/JH4b | CAR E GIAAA VNP FDY WGQG | 12 |
| M13 | V4-31 (DP65)/D5-24/JH4 | CAG D RGGYN LL DC WGHG | 10 |
| M14 | V3-23 (DP47)/JH6 | CAK QMGLAGTQR FYGLDV WGKG | 15 |
| M15 | V1-18 (DP14)/JH4b | CAR ATLDLDGYM DF WGQG | 11 |
| M19 | V3-53/JH4b | CAT PISGTYHLY Y WGQG | 10 |
| M20 | V4-30.4 (DP78)/JH3b | CAR DQ AFDI WGQG | 6 |
| M21 | V3-23 (DP47)/D2-2/JH4b | CAK DLFFV GYCTTTGC NT FDY WGQG | 18 |
| M22 | V1-69 (DP88)/D4-17/JH4b | CAR GSN DYGDN VPVQPH Y WGQG | 15 |
| M23 | V1-69 (DP88)/JH4b | CAR VSGNSH FDY WGQG | 9 |
| M30 | V2-5 (VII-5)/JH1 | CAQ RGGFYDSSLGFYIAPFP H WGQG | 18 |
| M41-A | V1-69 (DP10)/D3-22/JH4b | CAR GP DTGGYYY F Y WGQG | 11 |
| M41-B | V1-69 (DP10)/D3-22/JH4b | CAR GP DTGDYYY F Y GGQG | 11 |
| M41-C | V1-69 (DP10)/D3-22/JH4b | CAR GP DSSNYYY F Y WGQG | 11 |
| M45 | V1-3 (DP25)/D3-10/JH4b | CAR GTKIRGIVKPFP DY WGQG | 14 |
| M46 | V3-30/30.5 (DP49)/D5-12/JH6b | CAK DSGYVNFYYT MDV WGQG | 13 |
| M55 | V3-7 (DP54)/D3-22/JH4b | CAK WDYENSAYFLH Y WGQE | 12 |
| M56 | V1-69 (DP10)/D1-14/JH4b | CAR DT GN H YFDY WGQG | 9 |
| M57 | V1-18 (DP14)/JH6b | CAT PPPRAGDGP YYYYGMDV WGQG | 17 |
| M58 | V3-53/D3-22/JH4b | CAR HSYDNNAY DF WGQG | 10 |
| M60 | V1-69 (DP10)/D2-15/JH5b | CAR DPVD CSGGSCY LS WFDP WGQG | 17 |
| M61 | V1-18 (DP14)/D2-15/JH4b | CAR D YCSGGICY GG DY WGQG | 13 |
The FR3 and FR4 are indicated in italics. The assignable nontemplated region-encoded amino acids are underlined.
We then, additionally, examined CDR3 of 35 gastric-, 26 salivary gland- and 15 pulmonary-MALT lymphomas as well as 32 splenic MZBCL from literature and/or GenBank (Table V). This revealed that overall 8 out of 45 (18%) gastric- and 13 out of 32 (41%) salivary gland-MALT lymphomas expressed IgVH-CDR3 with RF homology. Three major RF homology groups could be distinguished, i.e., nine encoded by V1-69/JH4 (V1-69-RF), eight by V3-7/JH3 (V3-7-RF) and three by V1-69/JH4 (RF-WOL) rearrangements, respectively (Table V). Intriguingly, in all cases homology areas included aa encoded by the nontemplated nucleotide (N) regions (Fig. 2). In addition, gastric MALT lymphoma ML15 (40) was homologous to another RF termed RF-C93 (41). Noteworthy, none of the 19 pulmonary MALT lymphomas expressed RF-homologous CDR3 whereas among the splenic MZBCLs, only one case expressed a CDR3 homologous to RF-WOL (Table V).
Table V.
Comparison of IgVH-CDR3 amino acid sequences of a panel of mature B-NHL with IgVH-CDR3 amino acid sequences from GenBank
| No. of cases with CDR3 homology
|
|||||||
|---|---|---|---|---|---|---|---|
| Type of lymphoma | Mean CDR3 length | N | Overalla | RFb | V1-69 RFc | V3-7 RFc | WOL-RFc |
| aa | (%) | (%) | |||||
| Gastric MALT | 13.6 | 45 | 16 (36) | 8 (18) | 1 | 4 | 2 |
| Salivary gland MALT | 14.5 | 32 | 15 (47) | 13 (41) | 8 | 4 | 1 |
| Pulmonary MALT | 12.7 | 19 | 2 (11) | 0 (0) | 0 | 0 | 0 |
| Other MALT | NI | 4 | 0 (0) | 0 (0) | 0 | 0 | 0 |
| Splenic MZBCL | 16.3 | 32 | 8 (25) | 1 (3) | 0 | 0 | 1 |
| MCL | 13.3 | 23 | 10 (44) | 0 (0) | 0 | 0 | 0 |
| B-CLL IgVH unmutated | 17.4 | 165 | 73 (44) | 2 (1) | 0 | 0 | 0 |
| B-CLL IgVH mutated | 13.7 | 143 | 24 (17) | 0 (0) | 0 | 0 | 0 |
| FL | 12.1 | 48 | 4 (8) | 0 (0) | 0 | 0 | 0 |
| DLBCL | 12.2 | 20 | 2 (10) | 1 (5) | 0 | 1 | 0 |
| BL | 12.6 | 48 | 5 (10) | 0 (0) | 0 | 0 | 0 |
| Myeloma | 13.0 | 31 | 5 (16) | 0 (0) | 0 | 0 | 0 |
The lymphomas used for the homology analyses are listed in the supplemental legend to this table (available at http://www.jem.org/cgi/content/full/jem.20050068/DC1).
Indicates the number of lymphomas that show at least 75% homology, according to the criteria described, to at least one IgVH-CDR3 sequence present in GenBank.
Indicates the number of lymphomas that show homology to known rheumatoid factor (RF) IgVH-CDR3 sequences.
Indicates the number of lymphomas with homology to canonical V1-69-, V3-7-encoded RFs and to WOL-RF. All the B-NHL that expressed RF homologous CDR3 regions are as follows: Gastric MALT lymphomas: ML13 homology to V1-69-RF; ML25, ML27, ML39a, and M6 homology to V3-7-RF; ML16 and M41 homology to WOL-RF; and ML15 homology to C93-RF. Salivary gland MALT lymphomas: Isolate 1–5, PO-1, BA-2.2, and M11 homology to V1-69-RF; G552, SH, Isolate 10, and M5 homology to V3-7-RF; and JA-1 homology to WOL-RF. Splenic MZBCL: Isolate 1 homology to WOL-RF. B-CLL unmutated: CLL011 homology to TB-3-D13-RF and ID-74 homology to SJ2-RF. DLBCL: EJ homology to V3-7-RF.
NI, not informative.
Figure 2.
IgVH-CDR3 amino acid sequences of selected cases of MALT lymphoma with homology to IgVH-CDR3 of rheumatoid factors. MALT lymphomas M5/M6, M11, and M41 share homology with V3-7-RF, V1-69-RF, and WOL-RF, respectively. In addition, RF CDR3 homology of three MALT lymphoma cases from literature (ML27, Iso-3, and ML16) with V3-7-RF, V1-69-RF, and WOL-RF, respectively, is depicted. The amino acid sequences are depicted by the single letter code. FR3 and FR4, framework region 3 and 4; N, amino acids encoded by the nontemplated nucleotides; D, gene segment; JH, gene segment; |, identical amino acids; +, similar amino acids; x, nonmatching amino acids; CDR3 length, length of the CDR3 region; Hom, percentage of homologous amino acids; Id, percentage of identical amino acids; Gap, length difference in amino acids of the compared IgVH-CDR3 regions.
Currently, 10 genuine V1-69/JH4-encoded RFs, isolated from rheumatoid arthritis patients or from healthy donors immunized with mismatched red blood cells (HID), have been described (42). V1-69/JH4 RFs typically contain a 12–14 aa CDR3 and are combined with an A27(kv325)-encoded IgVκ chain. Among the V1-69-RF-homologous MALT lymphomas, only of salivary gland lymphoma M11 the IgVκ is known, which indeed proved to be the canonical A27(kv325) IgVκ (Table III and Table S2, available at http://www.jem.org/cgi/content/full/jem.20050068/DC1). WOL-RF is also V1-69/JH4-encoded, but with a distinct 13 aa CDR3, again in combination with an A27(kv325)-encoded IgVκ chain. Five V3-7 RFs have been described, isolated from HIDs and an RA patient (42).V3-7-RFs are encoded by V3-7/D3-22/JH3 rearrangements, typically possessing a 16–17 aa CDR3 with the D3-22 in reading-frame 2, and in combination with an L2 (kv328)/Jκ1-encoded IgVκ chain. The V3-7/JH3-expressing salivary gland lymphomas M5 and SH (43) and gastric MALT lymphoma M6 indeed all coexpressed the canonical L2 (kv328)/Jκ1-encoded IgVκ chain (Table III and Table S2).
IgVH-CDR3 amino acid sequences of other mature B-non Hodgkin's lymphomas
For comparison, IgVH-CDR3 analyses were extended to a comprehensive panel of other mature B-NHLs available from literature and/or on GenBank. For most B-NHL entities, the average CDR3 aa length was comparable to that of mature naive B cells being 13,5 aa (44). Only splenic MZBCLs and the IgV H-unmutated B-CLLs expressed CDR3 of higher mean lengths (Table V). Of 48 FLs, 20 DLBCLs, 48 Burkitt lymphomas (BL) and 31 myelomas, only 16 cases (11%) displayed CDR3 aa homology with other CDR3 sequences present in GenBank. Thirteen of the 16 cases resembled those of normal B cells analyzed in repertoire studies (45–47). As mentioned above, one DLBCL EJ (48) showed homology to several gastric- and salivary gland-MALT lymphomas as well as to V3-7-RFs (Table V).
Analyses of 23 mantle cell lymphomas (MCL), deposited on GenBank, revealed that their CDR3 displayed a high frequency of homology (10 out of 23, 44%), mostly (7 cases) with CDR3 regions of unmutated normal B cells.
We analyzed a panel of 308 B-CLLs, 165 (54%) and 143 (46%) of the IgV-unmutated and the IgV-mutated subsets, respectively (Table V). Overall, the CDR3s of 97 out of the 308 B-CLLs (31%) displayed CDR3 homology, 59 of which (19%) with CDR3s of B-CLLs (inter B-CLL homology). Of the group of 97 B-CLLs with any homology, 75% belonged to the IgV-unmutated subset. This relative overrepresentation was even more outspoken among the group with inter-B-CLL homology in which 50 out of the 59 cases (85%) were unmutated. In fact, applying our criteria for homology, we distinguished eight CDR3-homology groups within 37 of these 59 B-CLLs, each of which with at least three representatives (Table VI). Very recently, B-CLL homology groups, except for our group 8, have been identified by other investigators as well based on distinct homology criteria (references 49–52 and supplemental legend to Table VI, available at http://www.jem.org/cgi/content/full/jem.20050068/DC1).
Table VI.
B-CLL IgVH-CDR3 amino acid sequence homology groups
| Group | Characteristic VH-D-JH | Mutation status |
CDR3 length |
No. of cases |
|---|---|---|---|---|
| aa | ||||
| 1 | (VH1, VH5, VH7)/D6-19 (frame 3)/JH4 | unmutated | 11–12 | 9 |
| 2 | (V1-69/V4-34)/D2-2 (frame 3)/JH6 | unmutated | 18–20 | 6 |
| 3 | V4-34/D5-5 (frame 1)/JH6 | mutated | 18 | 3 |
| 4 | V1-69/D3-16 (frame 3)/JH3 | unmutated | 19 | 3 |
| 5 | (V1-2/V1-3)/D1-26 (frame 3)/JH6 | unmutated | 15 | 3 |
| 6 | V4-39/D6-13 (frame 1)/JH5 | unmutated | 16-17 | 5 |
| 7 | V1-69/D3-10 (frame 3)/JH6 | unmutated | 18 | 5 |
| 8 | (V1-2/V1-46)/D3-22 (frame 2)/JH4 | unmutated | 17 | 3 |
The B-CLL cases of the eight IgVH-CDR3 homology groups and the resemblance of homology groups 1–7 to earlier described B-CLL homology groups are in the supplemental legend of this table.
A significant fraction of B-CLLs derives from poly (auto) reactive B cells (26–30). In vitro RF reactivity has been proven for one representative of homology group 1 (POR) and one of group 2 (AIG) (30). Two B-CLLs, CLL-011 (53) and ID-74 (54) (CLL-011 belonging to homology group 1), showed CDR3 homology to two different, IgV H-unmutated, RFs termed TB-3-D13 (55) and RF-SJ2 (56), respectively (Table V). Poly-reactivity toward different auto-antigens, including IgG, has been demonstrated for homology group 4 member SMI (29). In addition, group 4 members share CDR3 homology with an anti-cardiolipine Ab (AF460965). It is noted that the B-CLLs that displayed in vitro RF reactivity (POR, AIG, and SMI) as well as the two B-CLLs (CLL-011 and ID-74) that displayed CDR3 homology to two unmutated RFs, all belong to the IgV-unmutated B-CLL subset. This clearly contrasts with the MALT lymphomas with V1-69-RF, V3-7-RF or WOL-RF homology as these RFs as well as the MALT lymphoma Igs are encoded by heavily mutated IgV H genes. Finally, none of the 308 B-CLLs showed CDR3 homology to V1-69-, V3-7-, or WOL-RFs.
RF activity of recombinant lymphoma-derived IgM antibodies
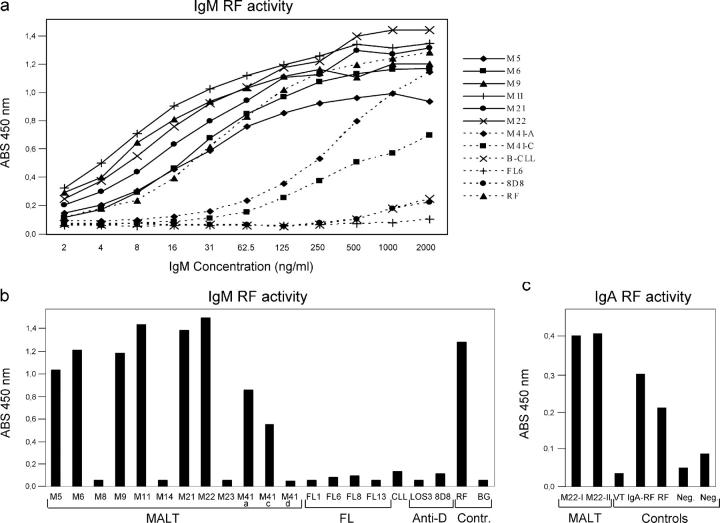
To prove that MALT lymphomas with RF-CDR3 homology possess IgG-binding activity, lymphoma-idiotype-derived Abs (LIDA) of IgM class were generated of M5, M6, M11, and M41-A, C, and D. Since, due to shortage of DNA, we could not resolve the IgVκ of M41 we combined each of the IgVH variants of the M41-A, C, and D subclones with the RF-canonical IgVκ chain of M22. Recombinant IgM LIDAs were also produced of 6 MALT lymphomas (M8, M9, M14, M21, M22, and M23), which are all devoid of RF-CDR3 homology. As additional controls, IgM LIDAs were generated of four follicular lymphomas (FL1, FL6′94, FL8′92, and FL13), one B-CLL, B-CLL26, and two anti-Rhesus(D) Ab producing B cell lines (8D8 and LOS3; reference 57). The IgVH and IgVκ sequences of these FLs and B-CLL have been previously reported by us (18, 58, 59). The lymphomas originally expressed IgM, with exception of M14, M22 (both IgA+), and M23 FL6′94, FL8′92 (both IgG+) (Tables I and II). RF-ELISA studies pointed out that the LIDA of M5, M6, and M11 were indeed strongly reactive with human IgG (Fig. 3). The LIDA of M9 and M22, both with a IgVH-CDR3 region not completely fulfilling our homology criteria but both with the RF-canonical V1-69/JH4-A27(kv325) combination of IgVH and IgVκ chains, also displayed strong RF activity (Tables II–IV and Fig. 3). Of note, also sera of patient M22, which contained high concentrations (22 mg/ml and 6.5 mg/ml) of lymphoma-related IgA paraprotein, displayed strong IgA-RF activity in ELISA (Fig. 3). This thus independently confirmed our finding with the M22-LIDA and underscored the validity of our approach of recombinant lymphoma Ab production in the eukaryotic expression system used. Moreover, the LIDA of M21, not even harboring an RF-canonical IgVH rearrangement but with an A27(kv325)-encoded IgVκ chain, also possessed strong RF activity. In contrast, none of the LIDAs of MALT lymphomas M8, M14, M23, the four FLs, the B-CLL nor the anti-Rhesus(D) Abs, all, except M23, lacking canonical IgVH RF rearrangements, bound to IgG (Fig. 3). Finally, LIDA M41-A/M22 (the dominant subclone with 19 mutations) and M41-C/M22 with 20 mutations bound IgG in ELISA whereas the LIDA of M41-D/M22 (a subclone with only two mutations) did not (Figs. 1–3).
Figure 3.
RF activity of lymphoma-idiotype-derived antibodies. (a) Titration of IgM LIDAs M5, M6, M9, M11, M21, M22, M41-A, M41-B, FL6'94, B-CLL26, 8D8, and RF control serum in the IgM-RF ELISA. The LIDAs M8, M14, M23, M41-D, FL1, FL8'92, FL13, and LOS3 did not react with human IgG and showed a similar binding curve as that of FL6'94. (b) Binding activity of IgM LIDAs, RF control serum, and anti-Rh(D) control IgM Abs (LOS3 and 8D8) in the IgM-RF ELISA. All samples were tested at a stratified concentration of 500 ng/ml IgM. The ABS 450 nm is plotted without subtraction of the background (BG) ABS 450 nm. (c) IgA-RF ELISA of two serum samples of M22 and as controls an IgA anti-Rh(D) Ab (VT-7G3), an IgA-RF-containing serum, the RF control serum that was also used in the IgM-RF ELISA, and two negative-control serum samples, respectively. All samples were tested at a stratified concentration of 20 μg/ml IgA. The net ABS 450 nm is plotted with subtraction of the background ABS 450 nm.
We next tested the binding capacities of the RF+ LIDAs of M5, M6, M9, and M11 with recombinant IgG1 and IgG3 preparations. The LIDA of M5, M6, and M9 reacted with IgG1 only. M11 reacted with both IgG1 and IgG3, and may thus be a pan-IgG reacting RF (unpublished data). In chronic gastritis, RF-expressing B cells may theoretically be stimulated by IgG coated on H. pylori or due to existence of cross-reacting epitopes between H. pylori and IgG-Fc. Upon comparison, one H. pylori (strain 26695, GenBank/EMBL/DDBJ accession no. AE000511) peptide, of the gene product “virulence-associated protein homologue VacB” (GenBank/EMBL/DDBJ accession no. AAD08293), was found to share 68% homology with aa 336–354 of the IgG1 Fc at the CH2-CH3 junction. However, none of M5, M6, M8, M9, M11, or M14 LIDA, with or without RF activity, reacted with a synthetic “336-354” peptide, nor did this peptide block the binding of the RF-LIDAs to IgG. In addition, no binding of any of these LIDAs to H. pylori–infected HM02 epithelial cells (strains 26695 and 1061) was observed. In addition, all LIDAs lacking RF-activity (M8, M14, M23, FL1, FL6′94, FL8′92, FL13, and B-CLL26) showed no antinuclear antibody (ANA) activity on Hep2 cells either (unpublished data).
To explore whether other BCR ligands are present within the tissue of MALT lymphomas or FLs, we also produced LIDAs of the IgG class of 3 (non-RF-CDR3-homologous) MALT lymphomas (M8, M14, and M15) and of 4 FLs (FL1, FL3′93, FL6′94, and FL63; reference 18). These LIDAs were FITC-labeled and tested immunohistochemically for reactivity on the corresponding lymphoma tissues. In none of these experiments however we detected reactivity with any tissue components (unpublished data).
Discussion
We systematically analyzed the immunoglobulin repertoires of a comprehensive panel of mature B-NHLs. Unbiased comparison of IgVH-CDR3–encoded aa sequences of individual B-NHLs with all IgVH-CDR3 presently available in GenBank revealed distinct patterns of the various B-NHL entities. This provided interesting clues concerning their potential ligands, which was functionally confirmed for the group of MALT lymphomas.
FLs, DLBCLs, BLs, and myelomas all exhibited a low degree of overall IgVH-CDR3 sequence homology (Table V). In none of these 147 B-NHLs, recurrent IgVH-CDR3 motifs were found. The majority of the homologous lymphomas expressed CDR3 that resembled those present in normal B cells. A few, however, shared homology with B-CLL and MALT lymphomas. One DLBCL displayed homology with gastric- and salivary gland-MALT lymphomas as well as with V3-7-RFs (Table V). Thus, in general FL, DLBCL, BL, and myelomas, all carrying significantly hypermutated IgV genes, seem to recognize unique epitopes, suggesting that they arise randomly out of the pool of B cells selected for nonself-antigens, most likely during the germinal center reaction. This is in accordance with previous observations that the germline IgV H gene usage of these B-NHLs is similar to that of normal peripheral B cells (18, 60). In contrast, B-CLL and MCL cases showed a high degree of overall CDR3 homology (31 and 44%, respectively) (Table V). Focusing on B-CLL that shared CDR3 homology with at least two other B-CLL (which held for 37 out of the 308 B-CLL analyzed), we distinguished eight CDR3-homology groups (Table VI). These homology groups in part overlap with B-CLL subgroups as reported by others (49–52). Inter-B-CLL homology was largely confined to the IgV H-unmutated subset, which shows a strong bias toward V1-69 usage (53): 62 out of 165 (38%) IgV H unmutated B-CLL expressed V1-69, most often (34/62, 55%) combined with JH6. In addition, the previously described poly-autoreactivity of a significant fraction of B-CLL was also clearly reflected in our study: Eighteen B-CLLs shared CDR3 homology with either of five B-CLL for which reactivity with auto-Ags such as IgG (RF), cardiolipin or myoglobulin has been reported (29, 30). Although the number of available MCL IgVH-CDR3 sequences was limited, we observed overall homology for almost half of the cases with CDR3 of unmutated IgV H genes of normal B cells. Still, MCLs are different from B-CLLs with respect to the IgV H repertoire bias, i.e., with preferential usage of V3-21 and V4-34 IgV H genes by MCLs (61, 62).
MALT lymphomas were found to express a highly distinctive IgV H repertoire, confirming and extending earlier reports by the groups of Miklos et al. (38) and De Re et al. (63) on salivary gland MALT lymphomas and HCV-associated B cell lymphomas, respectively. Out of a total of 100 MALT lymphomas that we analyzed, 33 cases shared CDR3 aa homology with other, previously published, CDR3. Twenty-one of these 33 MALT lymphomas harbored, according to the criteria chosen, significant homology to RF-related CDR3 and, except for one case, could be classified into either of 3 canonical RF groups; V1-69-RFs, V3-7-RFs and WOL-RFs (Table V). In addition, 5 salivary gland MALT lymphomas were included, reported by Bahler et al. (43, 64) and Miklos et al. (38), which did not completely fulfill our stringent criteria for V1-69-RFs, but did express the typical V1-69/JH4 RF gene rearrangement. The RF-homology group solely involved gastric- and salivary gland-MALT lymphomas. The in vitro binding studies with the recombinant LIDA formally proved that MALT lymphomas with canonical RF IgVH- and IgVκ-chain rearrangements and RF-CDR3 homology indeed posses strong RF activity (Fig. 3). Moreover, MALT lymphomas M9, M21, and M22, which did not match our criteria for RF homology but of which M9 and M22 expressed the canonical RF V1-69/JH4 rearrangement, also exhibited strong IgG-binding capacity in vitro (Fig. 3). Thus, the actual proportion of MALT lymphomas with specificity for human IgG is likely to be higher than calculated on basis of our arbitrary homology criteria.
The degree of RF-CDR3 homology found in the 21 MALT lymphomas is strikingly high, taking into account that it concerns heavily mutated IgV H genes and that homology included N-region encoded aa residues (Fig. 2). This suggests a distinct pathogenesis and indicates that these lymphomas originate from precursors strongly selected for auto-IgG specificity. The latter notion is well supported by the finding that the LIDA of the major subclone of M41 (M41-A), with 19 somatic mutations, and M41-C, with 20 mutations, exhibited significant intrinsic IgG-binding activity of the expressed IgVH chains, whereas this could not be measured of a presumed ancestral subclone, M41-D, with 2 mutations (Figs. 1 and 3).
MALT lymphomas typically evolve in a background of chronic inflammation due to infection or autoimmunity. Evidence exists that the tumor B cells in gastric MALT lymphoma are not H. pylori–specific but largely depend on CD40 stimulation by H. pylori–specific T helper cells (23). It has recently been reported, in a murine model, that RF-expressing B cells can be selectively activated in a T cell independent manner by IgG-chromatin complexes through the synergistic engagement of the BCR and toll-like receptor 9 (TLR9; reference 65). TLR9 is expressed in the endoplasmic compartment and serves as pathogen sensor that binds unmethylated CpG DNA motifs which are more common in bacterial than in mammalian DNA. In the human system, CpG-DNA was shown to trigger T cell independent proliferation of memory B cells, but not of naive B cells, which correlated with the levels of TLR9 expression of memory and naive B cells, respectively (66, 67). Stimulation of TLR9 may thus, parallel to the CD40/CD40L pathway, operate in lymphoproliferations of MALT. In gastric MALT, RF B cells may receive synergistic signals of the RF-BCR by IgG–H. pylori complexes and of TLR9 by H. pylori DNA. Also in inflamed salivary gland tissue in Sjögren's syndrome, as well as in other autoimmune diseases, RF B cells may receive these signals of the BCR and TLR9 by complexes of IgG and DNA released during normal or pathological cell death. This scenario clearly lends support from the fact that virtually all Sjögren's syndrome patients produce antinuclear antibodies (ANA), including anti-SS-A and SS-B antibodies.
The most frequent genetic alteration found in MALT lymphoma is the t(11;18)(q21;q21) encoding an API2-MALT1 fusion product that constitutively activates the NF-κB pathway (68). The t(11;18) is present in ∼40 and ∼25% of pulmonary- and gastric-MALT lymphomas respectively whereas it is virtually absent in MALT lymphomas of the salivary gland (∼2%; references 69–71). We found that ∼40% of the salivary gland- and ∼20% of the gastric-MALT lymphomas express RF-like BCRs whereas we did not identify RF-like BCRs in any of the 19 pulmonary MALT lymphomas. In addition, none of the MALT lymphomas with a t(11;18) possessed RF-CDR3 homology (Table VII). Accordingly, the LIDA of the t(11;18)+ lung lymphoma M23 did not bind IgG in vitro (Fig. 3). This tentative inverse relation between RF-specificity and the t(11;18) suggests that MALT lymphomas containing t(11;18) do not depend for their expansion on BCR-, CD40-, or TLR9-mediated NF-κB activation (Table VII). The fact that t(11;18)+ gastric lymphomas are resistant to H. pylori eradication therapy is in support of this hypothesis (14, 15). By contrast, the t(11;18)− gastric and salivary gland MALT lymphomas with RF BCR may need chronic stimulation by IgG in Ag-Ab complexes in gastric- and salivary gland-MALT lymphomas e.g. as IgG–H. pylori and IgG-ANA complexes, respectively. Finally, the different Ig repertoire of t(11;18)+ MALT lymphomas, as compared with MALT lymphomas devoid of this translocation, indicates that this genetic alteration as such provides growth advantage and occurs before the selection process favoring RF-expressing clones.
Table VII.
Relation between the presence of t(11;18) and RF-CDR3 homology and/or RF activity of MALT lymphoma immunoglobulins
| t(11;18) and RF homology/ activity of 24 MALT lymphomas |
Frequencies of t(11;18) and RF homology among MALT lymphoma cohorts |
||||||
|---|---|---|---|---|---|---|---|
| t(11;18)a
|
|||||||
| n | + | − | n | t(11;18)b | n | RF-CDR3c | |
| (%) | (%) | ||||||
| Salivary gland | 6 | 0 (0) | 6 (4) | 114 | 2 (2) | 32 | 13 (41) |
| Gastric | 10 | 5 (0) | 5 (2) | 209 | 50 (24) | 45 | 8 (18) |
| Pulmonary | 4 | 3 (0) | 1 (0) | 113 | 47 (42) | 19 | 0 (0) |
| Other MALT | 4 | 0 (0) | 4 (1) | ND | NA | 4 | 0 (0) |
Numbers in parentheses indicate quantity of cases with RF-CDR3 homology and/or in vitro RF activity.
The data on t(11;18) and MALT lymphoma localization refer to combined data described previously (69–71).
Data adapted from Table V.
NA, not applicable.
Materials and Methods
Patient material and immunohistochemistry
Frozen or paraffin-embedded tissue of 23 low-grade and one large cell (M22) MALT lymphomas was obtained from the Westeinde Hospital, The Hague; the Free University Medical Center, Amsterdam; The Netherlands Cancer Institute, Amsterdam and the Academic Medical Center, Amsterdam, The Netherlands.
Tumor cell immunophenotypes were determined by immunohistochemical stainings on acetone-fixed cryostat sections and on formalin-fixed paraffin embedded sections using the highly sensitive Powervision+ detection system (ImmunoVision Technologies). Monoclonal antibodies used: IgM, κ- and λ-light chains (Becton Dickinson), IgG, IgA, CD20 (L26; DakoCytomation), CXCR3 (1C6; BD Biosciences), and α4β7 (Act-1).
M22 was a large cell lymphoma consisting of immunoblasts which had developed in a patient suffering from Sjögren's syndrome. The original diagnosis MALT lymphoma was not made in our hospital and unfortunately we were not able to recollect material from previous biopsies. This lymphoma most likely developed from a MALT-associated clone given the expression of IgA, CXCR3, the mucosa homing receptor α4β7 as well as the obvious plasmacytoid differentiation with the concurrent lymphoma-related paraproteinemia. These are characteristics highly compatible with extranodal marginal zone lymphomas but extraordinary for diffuse large B cell lymphomas.
This study was conducted in accordance with the ethical standards in our institutional medical ethical committee on human experimentation, as well as in agreement with the Helsinki Declaration of 1975, as revised in 1983.
DNA and RNA isolation; cDNA synthesis; and IgVH, IgVκ, and t(11;18) RT-PCR
DNA was isolated from paraffin sections by over night proteinase K digestion. RNA was isolated from frozen sections using the TRIzol reagent (Invitrogen) and cDNA was synthesized with Pd(N)6 random primers. The IgV H and IgVκ genes were amplified using IgVH and IgVκ family-specific leader primers combined with the appropriate reverse primer being either JH, Cμ, Cγ, Cα, Jκ, or Cκ. To determine the clonally expressed IgV H gene of the tumor B cells, the CDR3 region was also amplified, directly on cDNA and in nested PCRs on the IgVH family-specific PCR products, using a forward primer specific for the framework region 3 (FR3) in combination with one of the different nested downstream primers specific for JH, Cμ, Cγ, Cα or Cδ. The PCR programs and primers sequences were described previously (18, 58). Translocation t(11;18) was determined using the primers and the PCR program as described by Liu et al. (15).
Cloning and sequencing
IgV RT-PCR products of MALT lymphomas were either directly sequenced or cloned into pTOPO-TA-vectors and transformed into TOP10 bacteria (Invitrogen), to generate molecular IgV clones. Sequencing on both strands was performed by an ABI sequencer (Applied Biosystems) using the big dye-terminator cycle-sequencing kit. To identify the IgV germline gene used and the somatic mutations therein, the consensus sequence of each MALT lymphoma was compared with published germline sequences, using the Vbase database (72) and DNAplot on internet (http://www.mrc-cpe.cam.ac.uk). The IgV sequences of the MALT lymphomas were deposited on GenBank/EMBL/DDBJ (accession nos. AY281324, AY281325, AY281326, AY281327, AY281328, AY281329, AY281330, AY281331, AY281332, AY281333, AY281334, AY281335, AY281336, AY281337, AY281338, AY281339, AY281340, AY281341, AY281342, AY281343, AY466502, AY466503, AY561708 and AY927657, AY927658, AY927659, AY927660, AY927661, AY927662, AY927663, AY927664, AY927665, AY927666, AY927667, AY927668). The degree of intraclonal variation (ICV) of IgV H genes and IgVκ genes was calculated as the mean number of nucleotide differences of each molecular clone as compared with the consensus IgV H or IgVκ sequences (18). ICV was considered significant if exceeding 0.4 mutations/clone.
Production of IgM antibodies derived of B-NHL
Recombinant IgMκ antibodies of the lymphomas (lymphoma-idiotype-derived Ab [LIDA]) of patients M5, M6, M8, M9, M11, M14, M21, M22, M23, M41, FL1, FL6′94, FL8′92, and B-CLL26 were produced using the pIgH(μ) and pIgL(κ) expression vectors as described previously (59). In brief, the IgVH and IgVκ sequences of each of these lymphomas, including one EBV B cell clone (8D8), which produces a human monoclonal antibody specific for the erythrocyte Rhesus(D) blood group antigen (57), were each cloned into the pIgH(μ) and pIgL(κ) vectors respectively. For production of recombinant antibody, 10 μg pIgH(μ) and 10 μg pIgL(κ) were linearized with PvuI and cotransfected into SP2/0 myeloma cells by electroporation. Subsequently, the transfected cells were selected in geneticin-containing medium. The IgVH of M41-A, C, and D were expressed with the IgVκ of M22. An Ig-secreting heterohybridoma of an IgMλ expressing FL FL13 (18), was produced by electrofusion with F3B6 (73) as described previously (74). LOS3 and VT-7G3 are an IgMκ and an IgAκ anti-Rhesus(D)-secreting EBV B cell clone, respectively (57). Supernatants were screened for IgMκ or IgMλ, using ELISAs as described previously (57). The pIgH(μ) and pIgL(κ) expression vectors were provided by J. van Es and T. Logtenberg (Utrecht Medical Center, Utrecht, The Netherlands).
LIDA reactivity in rheumatoid factor ELISA and on H. pylori–infected HM02 cells
LIDA reactivity with hIgG was determined using the IgM Rheumatoid Factor ELISA kit (Sanquin) according to the manufacturer's instructions. The plates were developed using TMB as substrate, as described previously (57). For IgA RF activity, a HRP-labeled rabbit anti-IgA Ab (DakoCytomation) was used. The “336-354” IgG1 Fc peptide was coated at 4 μg/ml, incubated with LIDA, followed by mAb anti-IgM-HRP (MH15/1-HRP; Sanquin), and developed as described previously (57). For blocking RF activity, LIDAs were tested in RF-ELISA in the presence of a 50–500 molar excess of the “336-354” IgG1 Fc peptide. To detect LIDA reactivity with H. pylori, ∼80% confluent cultures of HM02 cells were incubated with 2*106 CFU H. pylori (strains 1061 and 26695) and cultured for 1 wk. The cells were then fixed with methanol-aceton (1:1), immunocytochemically stained with LIDA, HRP labeled rabbit anti-IgM Ab (DakoCytomation) and developed with AEC as substrate.
Online supplemental material
Table S1 summarizes the IgVH-CDR3 amino acid sequence homology of MALT lymphomas M5, M6, M11, and M41 with other normal and malignant B cells as well as with RF-producing B cells. Table S2 gives an overview of the IgVκ-CDR3 amino acid sequences of the MALT lymphomas of Table III. The supplemental legend to Table V summarizes all lymphomas used in the IgVH-CDR3 homology analysis. The supplemental legend to Table VI depicts the B-CLL cases belonging to the eight IgVH-CDR3 homology groups and their resemblance to homology groups as described previously (49–52). Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20050068/DC1.
Acknowledgments
We thank M.E.C.M. Oud, E.J.M. Schilder-Tol, and Dr. M. Spaargaren for performing t(11;18) analyses; J.B.G. Mulder for technical advice and immunohistochemical stainings; Drs. T.A. Out and R.J. van de Stadt for providing sera; Dr. M.H. Delfau-Larue for sharing pathological information on pulmonary MALT lymphomas; Dr. D. Hamann for help with the RF ELISAs; Drs. K. van Amsterdam and A. van der Ende for providing H. pylori–infected HM02 cells; and C.C.H. Vellema, M.J.H. Berends, L.J.M.J. Delahaye, Dr. E.C.M. Ooms, and Dr. E.H. Jaspars for providing tissue material.
The authors have no conflicting financial interests.
Abbreviations used: Ag, antigen; B-CLL, B cell chronic lymphocytic leukemia; B-NHL, B non-Hodgkin's lymphoma; BCR, B cell antigen receptor; BL, Burkitt's lymphoma; D, diversity; DLBCL, diffuse large B cell lymphoma; FL, follicular lymphoma; FR, framework region; GC, germinal center; HCV, hepatitis C virus; HID, healthy immunized donor; ICV, intraclonal sequence variation; J, joining; LIDA lymphoma- idiotype-derived Ab; MALT, mucosa-associated lymphoid tissue; MCL, mantle cell lymphoma; MZBCL, marginal zone B cell lymphoma; RF, rheumatoid factor; V, variable.
References
- 1.Klein, U., T. Goossens, M. Fischer, H. Kanzler, A. Braeuninger, K. Rajewsky, and R. Küppers. 1998. Somatic hypermutation in normal and transformed human B cells. Immunol. Rev. 162:261–280. [DOI] [PubMed] [Google Scholar]
- 2.Hyjek, E., W.J. Smith, and P.G. Isaacson. 1988. Primary B-cell lymphoma of salivary glands and its relationship to myoepithelial sialadenitis. Hum. Pathol. 19:766–776. [DOI] [PubMed] [Google Scholar]
- 3.Hyjek, E., and P.G. Isaacson. 1988. Primary B cell lymphoma of the thyroid and its relationship to Hashimoto's thyroiditis. Hum. Pathol. 19:1315–1326. [DOI] [PubMed] [Google Scholar]
- 4.Wotherspoon, A.C., C. Ortiz-Hidalgo, M.R. Falzon, and P.G. Isaacson. 1991. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet. 338:1175–1176. [DOI] [PubMed] [Google Scholar]
- 5.Zucca, E., F. Bertoni, E. Roggero, G. Bosshard, G. Cazzaniga, E. Pedrinis, A. Biondi, and F. Cavalli. 1998. Molecular analysis of the progression from Helicobacter pylori-associated chronic gastritis to mucosa-associated lymphoid-tissue lymphoma of the stomach. N. Engl. J. Med. 338:804–810. [DOI] [PubMed] [Google Scholar]
- 6.Garbe, C., H. Stein, D. Dienemann, and C.E. Orfanos. 1991. Borrelia burgdorferi-associated cutaneous B cell lymphoma: clinical and immunohistologic characterization of four cases. J. Am. Acad. Dermatol. 24:584–590. [DOI] [PubMed] [Google Scholar]
- 7.Cerroni, L., S. Signoretti, G. Hofler, G. Annessi, B. Putz, E. Lackinger, D. Metze, A. Giannetti, and H. Kerl. 1997. Primary cutaneous marginal zone B-cell lymphoma: a recently described entity of low-grade malignant cutaneous B-cell lymphoma. Am. J. Surg. Pathol. 21:1307–1315. [DOI] [PubMed] [Google Scholar]
- 8.De Vita, S., C. Sacco, D. Sansonno, A. Gloghini, F. Dammacco, M. Crovatto, G. Santini, R. Dolcetti, M. Boiocchi, A. Carbone, and V. Zagonel. 1997. Characterization of overt B-cell lymphomas in patients with hepatitis C virus infection. Blood. 90:776–782. [PubMed] [Google Scholar]
- 9.Silvestri, F., C. Pipan, G. Barillari, F. Zaja, R. Fanin, L. Infanti, D. Russo, E. Falasca, G.A. Botta, and M. Baccarani. 1996. Prevalence of hepatitis C virus infection in patients with lymphoproliferative disorders. Blood. 87:4296–4301. [PubMed] [Google Scholar]
- 10.Ferreri, A.J., M. Guidoboni, M. Ponzoni, C. De Conciliis, S. Dell'Oro, K. Fleischhauer, L. Caggiari, A.A. Lettini, E. Dal Cin, R. Ieri, et al. 2004. Evidence for an association between Chlamydia psittaci and ocular adnexal lymphomas. J. Natl. Cancer Inst. 96:586–594. [DOI] [PubMed] [Google Scholar]
- 11.Lecuit, M., E. Abachin, A. Martin, C. Poyart, P. Pochart, F. Suarez, D. Bengoufa, J. Feuillard, A. Lavergne, J.I. Gordon, et al. 2004. Immunoproliferative small intestinal disease associated with Campylobacter jejuni. N. Engl. J. Med. 350:239–248. [DOI] [PubMed] [Google Scholar]
- 12.Drillenburg, P., R. van der Voort, G. Koopman, B. Dragosics, J.H.J.M. van Krieken, Ph.M. Kluin, J. Meenan, A.I. Lazarovits, T. Radaszkiewicz, and S.T. Pals. 1996. Preferential expression of the mucosal homing receptor integrin α4β7 in gastrointestinal non-Hodgkin's lymphomas. Am. J. Pathol. 150:919–927. [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, D., R.J. Benjamin, A. Shahsafaei, and D.M. Dorfman. 2000. The chemokine receptor CXCR3 is expressed in a subset of B-cell lymphomas and is a marker of B-cell chronic lymphocytic leukemia. Blood. 95:627–632. [PubMed] [Google Scholar]
- 14.Sugiyama, T., M. Asaka, T. Nakamura, S. Nakamura, S. Yonezumi, and M. Seto. 2001. API2-MALT1 chimeric transcript is a predictive marker for the responsiveness of H. pylori eradication treatment in low-grade gastric MALT lymphoma. Gastroenterology. 120:1884–1885. [DOI] [PubMed] [Google Scholar]
- 15.Liu, H., H. Ye, A. Ruskone-Fourmestraux, D. de Jong, S. Pileri, C. Thiede, A. Lavergne, H. Boot, G. Caletti, T. Wundisch, et al. 2002. T(11;18) is a marker for all stage gastric MALT lymphomas that will not respond to H. pylori eradication. Gastroenterology. 122:1286–1294. [DOI] [PubMed] [Google Scholar]
- 16.Hermine, O., F. Lefrere, J.P. Bronowicki, X. Mariette, K. Jondeau, V. Eclache-Saudreau, B. Delmas, F. Valensi, P. Cacoub, C. Brechot, et al. 2002. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N. Engl. J. Med. 347:89–94. [DOI] [PubMed] [Google Scholar]
- 17.Casato, M., C. Mecucci, V. Agnello, M. Fiorilli, G.B. Knight, C. Matteucci, L. Gao, and J. Kay. 2002. Regression of lymphoproliferative disorder after treatment for hepatitis C virus infection in a patient with partial trisomy 3, Bcl-2 overexpression, and type II cryoglobulinemia. Blood. 99:2259–2261. [DOI] [PubMed] [Google Scholar]
- 18.Aarts, W.M., R.J. Bende, E.J. Steenbergen, P.M. Kluin, E.C.M. Ooms, S.T. Pals, and C.J.M. van Noesel. 2000. Variable heavy chain gene analysis of follicular lymphomas: correlation between heavy chain isotype expression and somatic mutation load. Blood. 95:2922–2929. [PubMed] [Google Scholar]
- 19.Qin, Y., A. Greiner, M.J.F. Trunk, B. Schmausser, M.M. Ott, and H.K. Muller-Hermelink. 1995. Somatic hypermutation in low-grade mucosa-associated lymphoid tissue-type B-cell lymphoma. Blood. 86:3528–3534. [PubMed] [Google Scholar]
- 20.Du, M.-Q., C.-F. Xu, T.C. Diss, H.-Z. Peng, A.C. Wotherspoon, P.G. Isaacson, and L.-X. Pan. 1996. Intestinal dissemination of gastric mucosa-associated lymphoid tissue lymphoma. Blood. 88:4445–4451. [PubMed] [Google Scholar]
- 21.Du, M., T.C. Diss, C. Xu, H. Peng, P.G. Isaacson, and L. Pan. 1996. Ongoing mutation in MALT lymphoma immunoglobulin gene suggests that antigen stimulation plays a role in the clonal expansion. Leukemia. 10:1190–1197. [PubMed] [Google Scholar]
- 22.Hallas, C., A. Greiner, K. Peters, and H.K. Müller-Hermelink. 1998. Immunoglobulin VH genes of high-grade mucosa-associated lymphoid tissue lymphomas show a high load of somatic mutations and evidence of antigen-dependent affinity maturation. Lab. Invest. 78:277–287. [PubMed] [Google Scholar]
- 23.Hussell, T., P.G. Isaacson, J.E. Crabtree, and J. Spencer. 1996. Helicobacter pylori-specific tumour-infiltrating T cells provide contact dependent help for the growth of malignant B cells in low-grade gastric lymphoma of mucosa-associated lymphoid tissue. J. Pathol. 178:122–127. [DOI] [PubMed] [Google Scholar]
- 24.Hussell, T., P.G. Isaacson, J.E. Crabtree, A. Dogan, and J. Spencer. 1993. Immunoglobulin specificity of low grade B cell gastrointestinal lymphoma of mucosa-associated lymphoid tissue (MALT) type. Am. J. Pathol. 142:285–292. [PMC free article] [PubMed] [Google Scholar]
- 25.Greiner, A., C. Knörr, Y. Qin, A. Schultz, A. Marx, R.A. Kroczek, and H.K. Müller-Hermelink. 1998. CD40 ligand and autoantigen are involved in the pathogenesis of low-grade B-cell lymphomas of mucosa-associated lymphoid tissue. Dev. Immunol. 6:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sthoeger, Z.M., M. Wakai, D.B. Tse, V.P. Vinciguerra, S.L. Allen, D.R. Budman, S.M. Lichtman, P. Schulman, L.R. Weiselberg, and N. Chiorazzi. 1989. Production of autoantibodies by CD5-expressing B lymphocytes from patients with chronic lymphocytic leukemia. J. Exp. Med. 169:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borche, L., A. Lim, J.-L. Binet, and G. Dighiero. 1990. Evidence that chronic lymphocytic leukemia B lymphocytes are frequently committed to production of natural autoantibodies. Blood. 76:562–569. [PubMed] [Google Scholar]
- 28.Broker, B.M., A. Klajman, P. Youinou, J. Jouquan, C.P. Worman, J. Murphy, L. Mackenzie, R. Quartey-Papafio, M. Blaschek, and P. Collins. 1988. Chronic lymphocytic leukemic (CLL) cells secrete multispecific autoantibodies. J. Autoimmun. 1:469–481. [DOI] [PubMed] [Google Scholar]
- 29.Martin, T., S.F. Duffy, D.A. Carson, and T.J. Kipps. 1992. Evidence for somatic selection of natural autoantibodies. J. Exp. Med. 175:983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritsch, O., C. Magnac, G. Dumas, C. Egile, and G. Dighiero. 1993. V gene usage by seven hybrids derived from CD5+ B-cell chronic lymphocytic leukemia and displaying autoantibody activity. Blood. 82:3103–3112. [PubMed] [Google Scholar]
- 31.Bogdan, C.A., A.A. Alexander, M.K. Gorny, R. Matute, N. Marjanovic, S. Zolla-Pazner, P.D. Walden, H.M. Furneaux, G.S. Sidhu, and D.R. Jacobson. 2003. Chronic lymphocytic leukemia with prostate infiltration mediated by specific clonal membrane-bound IgM. Cancer Res. 63:2067–2071. [PubMed] [Google Scholar]
- 32.Musante, L., M. Ulivi, G. Cutrona, N. Chiorazzi, S. Roncella, G. Candiano, and M. Ferrarini. 1999. Identification of HSP-60 as the specific antigen of IgM produced by BRG-lymphoma cells. Electrophoresis. 20:1092–1097. [DOI] [PubMed] [Google Scholar]
- 33.Cunto-Amesty, G., G. Przybylski, M. Honczarenko, J.G. Monroe, and L.E. Silberstein. 2000. Evidence that immunoglobulin specificities of AIDS-related lymphoma are not directed to HIV-related antigens. Blood. 95:1393–1399. [PubMed] [Google Scholar]
- 34.Nobuoka, A., S. Sakamaki, K. Kogawa, K. Fujikawa, M. Takahashi, Y. Hirayama, N. Takayanagi, H. Ikeda, S. Sekiguchi, and Y. Niitsu. 1999. A case of malignant lymphoma producing autoantibody against platelet glycoprotein Ib. Int. J. Hematol. 70:200–206. [PubMed] [Google Scholar]
- 35.Mann, D.L., P. DeSantis, G. Mark, A. Pfeifer, M. Newman, N. Gibbs, M. Popovic, M.G. Sarngadharan, R.C. Gallo, J. Clark, and W. Blattner. 1987. HTLV-I-associated B-cell CLL: indirect role for retrovirus in leukemogenesis. Science. 236:1103–1106. [DOI] [PubMed] [Google Scholar]
- 36.Quinn, E.R., H.C. Chunghuang, K.G. Hadlock, S.K.H. Foung, M. Flint, and S. Levy. 2001. The B-cell receptor of a hepatitis C virus (HCV)-associated non-Hodgkin lymphoma binds the viral E2 envelope protein, implicating HCV in lymphomagenesis. Blood. 98:3745–3749. [DOI] [PubMed] [Google Scholar]
- 37.Altschul, S.F., T.L. Madden, A.A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D.J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miklos, J.A., S.H. Swerdlow, and D.W. Bahler. 2000. Salivary gland mucosa-associated lymphoid tissue lymphoma immunoglobulin VH genes show frequent use of V1-69 with distinctive CDR3 features. Blood. 95:3878–3884. [PubMed] [Google Scholar]
- 39.Andrews, D.W., and J.D. Capra. 1981. Complete amino acid sequence of variable domains from two monoclonal human anti-gamma globulins of the Wa cross-idiotypic group: suggestion that the J segments are involved in the structural correlate of the idiotype. Proc. Natl. Acad. Sci. USA. 78:3799–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertoni, F., G. Cazzaniga, G. Bosshard, E. Roggero, R. Barbazza, M. De Boni, C. Capella, E. Pedrinis, F. Cavalli, A. Biondi, and E. Zucca. 1997. Immunoglobulin heavy chain diversity genes rearrangement pattern indicates that MALT-type gastric lymphoma B cells have undergone an antigen selection process. Br. J. Haematol. 97:830–836. [DOI] [PubMed] [Google Scholar]
- 41.Ermel, R.W., T.P. Kenny, A. Wong, A. Solomon, P.P. Chen, and D.L. Robbins. 1994. Preferential utilization of a novel V lambda 3 gene in monoclonal rheumatoid factors derived from the synovial cells of rheumatoid arthritis patients. Arthritis Rheum. 37:860–868. [DOI] [PubMed] [Google Scholar]
- 42.Borretzen, M., I. Randen, J.B. Natvig, and K.M. Thompson. 1995. Structural restriction in the heavy chain CDR3 of human rheumatoid factors. J. Immunol. 155:3630–3637. [PubMed] [Google Scholar]
- 43.Bahler, D.W., J.A. Miklos, and S.H. Swerdlow. 1997. Ongoing Ig gene hypermutation in salivary gland mucosa-associated lymphoid tissue-type lymphomas. Blood. 89:3335–3344. [PubMed] [Google Scholar]
- 44.Wardemann, H., S. Yurasov, A. Schaefer, J.W. Young, E. Meffre, and M.C. Nussenzweig. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. [DOI] [PubMed] [Google Scholar]
- 45.Wang, X., and B.D. Stollar. 1999. Immunoglobulin VH gene expression in human aging. Clin. Immunol. 93:132–142. [DOI] [PubMed] [Google Scholar]
- 46.Bauer, K., M. Zemlin, M. Hummel, S. Pfeiffer, J. Karstaedt, G. Steinhauser, X. Xiao, H. Versmold, and C. Berek. 2002. Diversification of Ig heavy chain genes in human preterm neonates prematurely exposed to environmental antigens. J. Immunol. 169:1349–1356. [DOI] [PubMed] [Google Scholar]
- 47.Geiger, K.D., U. Klein, A. Brauninger, S. Berger, K. Leder, K. Rajewsky, M.L. Hansmann, and R. Kuppers. 2000. CD5-positive B cells in healthy elderly humans are a polyclonal B cell population. Eur. J. Immunol. 30:2918–2923. [DOI] [PubMed] [Google Scholar]
- 48.Ottensmeier, C.H., A.R. Thompsett, D. Zhu, B.S. Wilkins, J.W. Sweetenham, and F.K. Stevenson. 1998. Analysis of VH genes in follicular and diffuse lymphoma shows ongoing somatic mutation and multiple isotype transcripts in early disease with changes during disease progression. Blood. 91:4292–4299. [PubMed] [Google Scholar]
- 49.Tobin, G., U. Thunberg, K. Karlsson, F. Murray, A. Laurell, K. Willander, G. Enblad, M. Merup, J. Vilpo, G. Juliusson, et al. 2004. Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood. 104:2879–2885. [DOI] [PubMed] [Google Scholar]
- 50.Widhopf, G.F., L.Z. Rassenti, T.L. Toy, J.G. Gribben, W.G. Wierda, and T.J. Kipps. 2004. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood. 104:2499–2504. [DOI] [PubMed] [Google Scholar]
- 51.Ghiotto, F., F. Fais, A. Valetto, E. Albesiano, S. Hashimoto, M. Dono, H. Ikematsu, S.L. Allen, J. Kolitz, K.R. Rai, et al. 2004. Remarkably similar antigen receptors among a subset of patients with chronic lymphocytic leukemia. J. Clin. Invest. 113:1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Messmer, B.T., E. Albesiano, D.G. Efremov, F. Ghiotto, S.L. Allen, J. Kolitz, R. Foa, R.N. Damle, F. Fais, D. Messmer, et al. 2004. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J. Exp. Med. 200:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fais, F., F. Ghiotto, S. Hashimoto, B. Sellars, A. Valetto, S.L. Allen, P. Schulman, V.P. Vinciguerra, K. Rai, L.Z. Rassenti, et al. 1998. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Invest. 102:1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamblin, T.J., Z. Davis, A. Gardiner, D.G. Oscier, and F.K. Stevenson. 1999. Unmutated Ig VH genes are associated with a more aggressive from of chronic lymphocytic leukemia. Blood. 94:1848–1854. [PubMed] [Google Scholar]
- 55.Djavad, N., S. Bas, X. Shi, J. Schwager, M. Jeannet, T. Vischer, and E. Roosnek. 1996. Comparison of rheumatoid factors of rheumatoid arthritis patients, of individuals with mycobacterial infections and of normal controls: evidence for maturation in the absence of an autoimmune response. Eur. J. Immunol. 26:2480–2486. [DOI] [PubMed] [Google Scholar]
- 56.Pascual, V., I. Randen, K. Thompson, M. Sioud, O. Forre, J. Natvig, and J.D. Capra. 1990. The complete nucleotide sequences of the heavy chain variable regions of six monospecific rheumatoid factors derived from Epstein-Barr virus-transformed B cells isolated from the synovial tissue of patients with rheumatoid arthritis. Further evidence that some autoantibodies are unmutated copies of germ line genes. J. Clin. Invest. 86:1320–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bende, R.J., G.J. Jochems, T.H. Frame, M.R. Klein, R.V.W. van Eijk, R.A.W. van Lier, and W.P. Zeijlemaker. 1992. Effects of IL-4, IL-5 and IL-6 on growth and immunoglobulin production of Epstein-Barr virus-infected human B cells. Cell. Immunol. 143:310–323. [DOI] [PubMed] [Google Scholar]
- 58.Aarts, W.M., R.J. Bende, S.T. Pals, and C.J.M. van Noesel. 1999. Analysis of variable heavy and light chain genes in follicular lymphomas of different heavy chain isotype. Curr. Top. Microbiol. Immunol. 246:217–224. [DOI] [PubMed] [Google Scholar]
- 59.Bende, R.J., W.M. Aarts, S.T. Pals, and C.J. van Noesel. 2002. Immunoglobulin diversification in B cell malignancies: internal splicing of heavy chain variable region as a by-product of somatic hypermutation. Leukemia. 16:636–644. [DOI] [PubMed] [Google Scholar]
- 60.Lossos, I.S., C.Y. Okada, R. Tibshirani, R. Warnke, J.M. Vose, T.C. Greiner, and R. Levy. 2000. Molecular analysis of immunoglobulin genes in diffuse large B-cell lymphomas. Blood. 95:1797–1803. [PubMed] [Google Scholar]
- 61.Walsh, S.H., M. Thorselius, A. Johnson, O. Soderberg, M. Jerkeman, E. Bjorck, I. Eriksson, U. Thunberg, O. Landgren, M. Ehinger, et al. 2003. Mutated VH genes and preferential VH3-21 use define new subsets of mantle cell lymphoma. Blood. 101:4047–4054. [DOI] [PubMed] [Google Scholar]
- 62.Kienle, D., A. Krober, T. Katzenberger, G. Ott, E. Leupolt, T.F. Barth, P. Moller, A. Benner, A. Habermann, H.K. Muller-Hermelink, et al. 2003. VH mutation status and VDJ rearrangement structure in mantle cell lymphoma: correlation with genomic aberrations, clinical characteristics, and outcome. Blood. 102:3003–3009. [DOI] [PubMed] [Google Scholar]
- 63.De Re, V., S. De Vita, A. Marzotto, M. Rupolo, A. Gloghini, B. Pivetta, D. Gasparotto, A. Carbone, and M. Boiocchi. 2000. Sequence analysis of the immunoglobulin antigen receptor of hepatitis C virus-associated non-Hodgkin lymphomas suggests that the malignant cells are derived from the rheumatoid factor-producing cells that occur mainly in type II cryoglobulinemia. Blood. 96:3578–3584. [PubMed] [Google Scholar]
- 64.Bahler, D.W., and S.H. Swerdlow. 1998. Clonal salivary gland infiltrates associated with myoepithelial sialoadenitis (Sjögren's syndrome) begin as nonmalignant antigen-selected expansions. Blood. 91:1864–1872. [PubMed] [Google Scholar]
- 65.Leadbetter, E.A., I.R. Rifkin, A.M. Hohlbaum, B.C. Beaudette, M.J. Shlomchik, and A. Marshak-Rothstein. 2002. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 416:603–607. [DOI] [PubMed] [Google Scholar]
- 66.Bernasconi, N.L., E. Traggiai, and A. Lanzavecchia. 2002. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 298:2199–2202. [DOI] [PubMed] [Google Scholar]
- 67.Bernasconi, N.L., N. Onai, and A. Lanzavecchia. 2003. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 101:4500–4504. [DOI] [PubMed] [Google Scholar]
- 68.Lucas, P.C., M. Yonezumi, N. Inohara, L.M. McAllister-Lucas, M.E. Abazeed, F.F. Chen, S. Yamaoka, M. Seto, and G. Nunez. 2001. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J. Biol. Chem. 276:19012–19019. [DOI] [PubMed] [Google Scholar]
- 69.Ye, H., H. Liu, A. Attygalle, A.C. Wotherspoon, A.G. Nicholson, F. Charlotte, V. Leblond, P. Speight, J. Goodlad, A. Lavergne-Slove, et al. 2003. Variable frequencies of t(11;18)(q21;q21) in MALT lymphomas of different sites: significant association with CagA strains of H. pylori in gastric MALT lymphoma. Blood. 102:1012–1018. [DOI] [PubMed] [Google Scholar]
- 70.Okabe, M., H. Inagaki, K. Ohshima, T. Yoshino, C. Li, T. Eimoto, R. Ueda, and S. Nakamura. 2003. API2-MALT1 fusion defines a distinctive clinicopathologic subtype in pulmonary extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue. Am. J. Pathol. 162:1113–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Streubel, B., I. Simonitsch-Klupp, L. Mullauer, A. Lamprecht, D. Huber, R. Siebert, M. Stolte, F. Trautinger, J. Lukas, A. Puspok, et al. 2004. Variable frequencies of MALT lymphoma-associated genetic aberrations in MALT lymphomas of different sites. Leukemia. 18:1722–1726. [DOI] [PubMed] [Google Scholar]
- 72.Cook, G.P., and I.M. Tomlinson. 1995. The human immunoglobulin VH repertoire. Immunol. Today. 16:237–242. [DOI] [PubMed] [Google Scholar]
- 73.Foung, S.K., S. Perkins, A. Raubitschek, J. Larrick, G. Lizak, D. Fishwild, E.G. Engleman, and F.C. Grumet. 1984. Rescue of human monoclonal antibody production from an EBV-transformed B cell line by fusion to a human-mouse hybridoma. J. Immunol. Methods. 70:83–90. [DOI] [PubMed] [Google Scholar]
- 74.van Duijn, G., J.P. Langedijk, M. de Boer, and J.M. Tager. 1989. High yields of specific hybridomas obtained by electrofusion of murine lymphocytes immunized in vivo or in vitro. Exp. Cell Res. 183:463–472. [DOI] [PubMed] [Google Scholar]