Abstract
T cell immunoglobulin-domain and mucin-domain (TIM) proteins constitute a receptor family that was identified first on kidney and liver cells; recently it was also shown to be expressed on T cells. TIM-1 and -3 receptors denote different subsets of T cells and have distinct regulatory effects on T cell function. Ferritin is a spherical protein complex that is formed by 24 subunits of H- and L-ferritin. Ferritin stores iron atoms intracellularly, but it also circulates. H-ferritin, but not L-ferritin, shows saturable binding to subsets of human T and B cells, and its expression is increased in response to inflammation. We demonstrate that mouse TIM-2 is expressed on all splenic B cells, with increased levels on germinal center B cells. TIM-2 also is expressed in the liver, especially in bile duct epithelial cells, and in renal tubule cells. We further demonstrate that TIM-2 is a receptor for H-ferritin, but not for L-ferritin, and expression of TIM-2 permits the cellular uptake of H-ferritin into endosomes. This is the first identification of a receptor for ferritin and reveals a new role for TIM-2.
T cell immunoglobulin-domain and mucin-domain (TIM) proteins constitute a receptor family that was identified first on kidney and liver cells; recently it was also shown to be expressed on T cells (1–5). In humans, the TIM receptor family seems to include only three receptors, TIM-1, -3, and -4, whereas in the mouse it may include as many as eight (4). Human TIM-3 and -4 have apparent orthologs in mice, based on sequence homology, but human TIM-1 is almost equally homologous to mouse TIM-1 (41%) and mouse TIM-2 (36%), which are 66% homologous to each other.
In the mouse, the TIM gene family is linked to a locus (Tapr) that regulates airway hypersensitivity and the production of Th2 cytokines (6). In accord with a role for TIM receptors in immunity, TIM-1 is expressed preferentially by Th2 cells, and polymorphisms in human TIM-1 are associated with atopy, asthma, and rheumatoid arthritis (5–10). Mouse TIM-1 binds to TIM-4, which is expressed on antigen-presenting cells, and ligation of TIM-1 potentiates T cell activation (7, 11, 12). In contrast to TIM-1, TIM-3 is expressed preferentially on Th1 cells. Blockade or loss of this receptor in mice accelerates autoimmunity, which suggests that ligation of the receptor is immunosuppressive (13–15).
Although TIM receptors are of evident importance in immunity, their expression outside of the immune system indicates that these receptors may have broader functions. Thus, in primates and rodents, TIM-1 is expressed on renal tubular cells; in primates, an alternatively spliced form is expressed on liver cells, where it has been usurped as a receptor for hepatitis A (1–3, 16). The functions of TIM receptors on these nonhematopoietic cells are unknown.
Ferritin is a spherical protein complex that stores up to 4,000 iron atoms as an oxidized mineral core (17). It is a heteropolymer that is formed by 24 subunits of H- and L-ferritin; their ratios vary in different tissues and in response to iron, growth factors, inflammation, or malignancy (18). Ferritin primarily is expressed intracellularly, where it regulates iron mineralization and sequestration, and thereby buffers reactive oxygen species. This effect of H-ferritin is essential for the antiapoptotic effect of NF-κB, and the transcription of H-ferritin is up-regulated by NF-κB (19). However, ferritin also circulates, and earlier evidence suggests that H-ferritin acts as an immune regulator, through binding to subsets of lymphocytes and myeloid cells. Thus, ferritin inhibits T cell proliferation in response to mitogens, it impairs the maturation of B cells in vitro (20, 21), and it has immunosuppressive effects in vivo (22). Additionally, H-ferritin, but not L-ferritin, shows saturable binding to subsets of human T and B cells (23–26). Despite the evidence for H-ferritin receptors on the cell surface, none had been identified.
We demonstrate that TIM-2 is expressed at low levels on all splenic B cells and is expressed at higher levels on germinal center (GC) B cells. Outside the hematopoietic system, TIM-2 is expressed in liver, especially in bile duct epithelial cells, and in renal tubule cells. We further demonstrate that TIM-2 serves as a selective receptor for H-ferritin, but not for L-ferritin, and that binding of H-ferritin to TIM-2 leads to the endocytosis of extracellular H-ferritin. The expression of a surface receptor for H-ferritin is consistent with a role for H-ferritin in modulating cell function—beyond its role in storing iron—and the endocytic function of TIM-2 provides a new pathway for altering levels of H-ferritin independent of gene expression.
RESULTS
TIM-2 is expressed on all splenic B cells, with high levels on GC B cells
An expressed sequence tag for TIM-2 was isolated from the database based on its partial homology to the Ig domain of triggering receptor expressed on myeloid cells (TREM)-2 (27). By expression of this cDNA, we prepared a mAb against the extracellular domain of TIM-2. To define the levels of TIM-2 on lymphocyte cell subsets, mice were not immunized or were immunized with T-dependent antigens; staining with anti–TIM-2 was assessed on subsets that were defined by their surface phenotype. Studies from unimmunized or immunized mice revealed that although TIM-2 was expressed on follicular B cells, it was expressed at ∼2.5-fold higher levels on GC B cells (range 1.6–3.5) (Fig. 1 A). In contrast, TIM-2 was not detected on T cells (CD4+ or CD8+) (Fig. 1 A).
Figure 1.
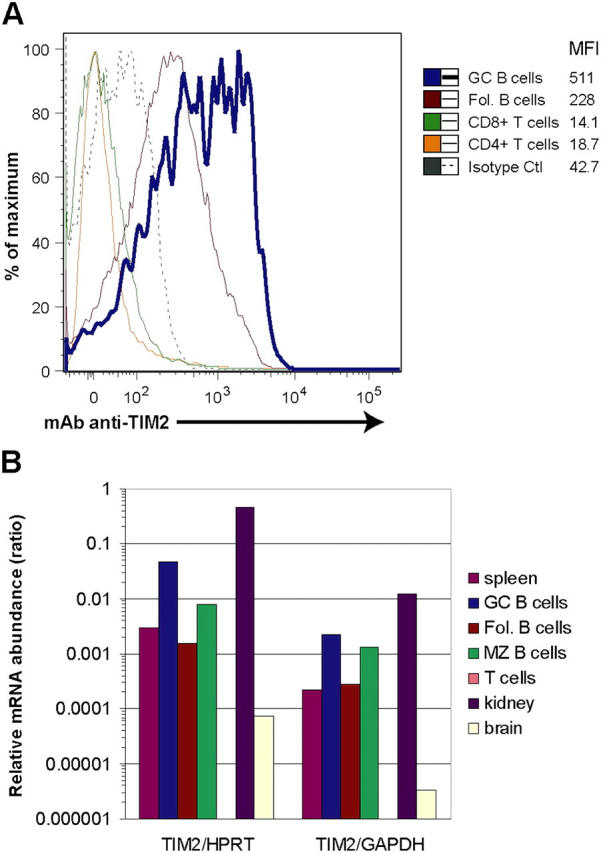
TIM-2 is expressed preferentially on GC B cells. (A) Flow cytometric analysis of TIM-2 expression on mouse spleen cells. Spleens were isolated 8 d after immunization with SRBCs. Each curve is normalized, so that the peaks are of equal height. Staining of GC cells is more than twice that of follicular B cells (Fol. B cells), although all B cells stain with anti–TIM-2, compared with an isotype control mAb (dotted line; the control shown is for GC B cells, but equivalent staining was seen with control staining of follicular B cells, not depicted). TIM-2 was not detected on CD4+ or CD8+ T cells. The numbers indicate geometric mean fluorescence intensity (MFI) for TIM-2 staining of each cell subset. (B) Quantitative RT-PCR analysis of TIM-2 expression. Total RNA was prepared from isolated subsets of spleen cells (sorted by FACS) or from isolated tissues, and was analyzed by quantitative RT-PCR. Expression of mRNA is shown as a ratio of TIM-2 mRNA/mRNA of two housekeeping genes, hypoxanthine-guanine phosphoribosyltransferase (HPRT) and GAPDH. Fol, follicular; MZ, marginal zone.
The expression of TIM-2 in splenic B cells, and its preferential expression in GC B cells was confirmed by quantitative RT-PCR, using SYBR Green. For these studies, splenic B cell subsets and T cells were isolated by fluorescence-activated cell sorting and used to prepare total RNA. Transcripts for TIM-2 were ∼10-fold more abundant in GC B cells than in follicular B cells, whereas levels of TIM-2 transcripts in the marginal zone B cells were between the two (Fig. 1 B). Little or no transcripts for TIM-2 were detected in splenic T cells.
The preferential expression of TIM-2 on GC B cells also was evident by immunohistochemistry (IHC). For these studies, we raised a rabbit antiserum against a peptide from the cytoplasmic domain of TIM-2. On fixed sections from spleen, this antiserum demonstrated clusters of TIM-2+ cells in the center of lymphoid follicles, although scattered cells were seen elsewhere, including the red pulp (Fig. 2 A, middle panel). No staining was seen with control antiserum (Fig. 2 A, left panel); staining by the anti–TIM-2 antiserum was blocked in the presence of the immunizing peptide, which demonstrated that binding was antigen-specific (Fig. 2 A, right panel). Immunofluorescent studies demonstrated that most, if not all, of the TIM-2+ cells that were clustered in the center of the follicles were B cells (Fig. 2 B). To identify the relationship of TIM-2+ cells to GCs more clearly, mice were immunized with sheep red cells 8 d before examination. By staining with peanut agglutinin, the clusters of TIM-2+ B cells were shown to be located within GCs (Fig. 2 C). The IHC and immunofluorescence studies did not reveal expression of TIM-2 on all B cells, as was detected by flow cytometry; we attribute this to the lower level of TIM-2 expression on non-GC B cells, which evidently is below the detection threshold of IHC. In accord with studies by flow cytometry and quantitative RT-PCR, T cell zones lacked TIM-2+ cells (Fig. 2 B, bottom row). As with IHC studies, staining with anti–TIM-2 was blocked in the presence of the immunizing peptide (unpublished data).
Figure 2.
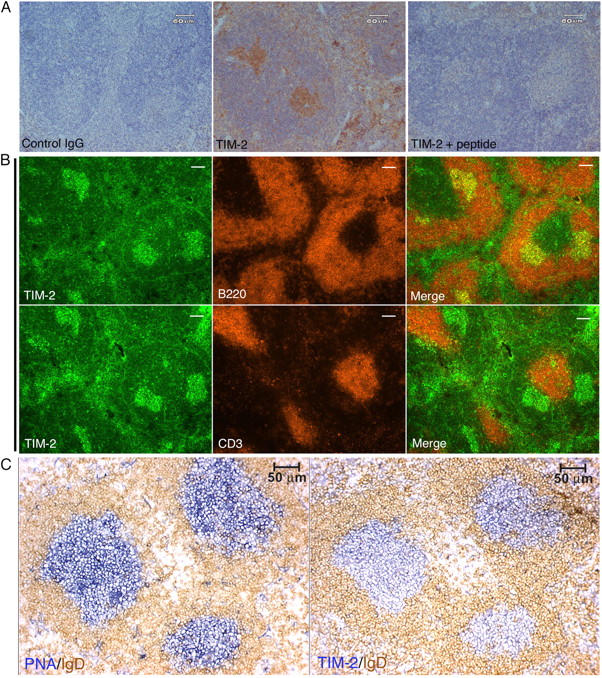
Immunohistochemistry and immunofluoresence for TIM-2 on spleen sections. (A) The anti–TIM-2 antiserum binds specifically to clusters of cells in splenic follicles. Paraffin-embedded sections from unimmunized mice were stained with an isotype-matched control rabbit antiserum (control IgG), anti–TIM-2 antiserum (TIM-2); or anti–TIM-2 antiserum blocked with an excess of the immunizing peptide (TIM-2 + peptide), followed by goat anti–rabbit IgG conjugated to horseradish peroxidase. The slides were counterstained with hematoxylin. (B) TIM-2–positive cells are predominantly B cells. Adjacent cryostat sections from unimmunized mice were co-stained for TIM-2 and B220 (top row) or TIM-2 and CD3 (bottom row). TIM-2 staining appears green (left columns), B220 and CD3 staining appear red (middle columns), and merged images are shown in the far right columns. Bars, 100 μm. (C) TIM-2–positive cells are localized to GCs. Adjacent cryostat sections from the spleen of a mouse that was immunized with SRBCs was stained for peanut agglutinin (PNA, blue, left panel) to show the GCs, and with IgD (brown, right panel) to show the surrounding follicular mantle zone. The section in the right panel was stained for TIM-2 (blue) and IgD (brown).
TIM-2 is expressed in localized areas of the liver and kidneys
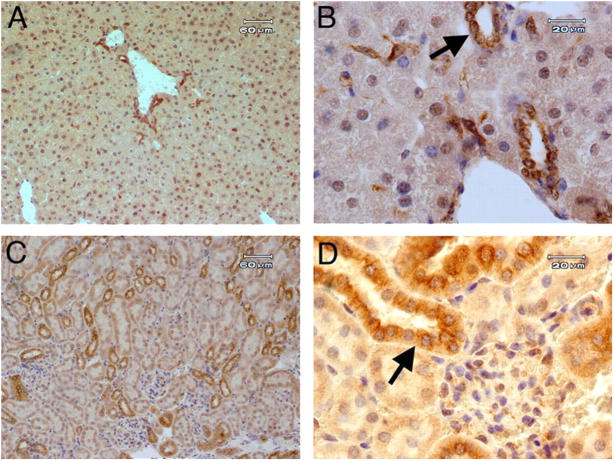
We used our rabbit antiserum to TIM-2 to demonstrate the localized expression of TIM-2 in sections of fixed tissue from liver and kidney. In the liver, the expression of TIM-2 was most prominent in bile duct epithelial cells (Fig. 3, A and B). To a lesser degree, staining of hepatocytes was seen, especially at junctures of hepatocytes. This suggested that TIM-2 may be expressed preferentially on bile canaliculi, which are formed as openings between hepatocytes. Variable staining of hepatocyte nuclei also was seen. All binding was blocked by the immunizing peptide (unpublished data). Thus, like human TIM-1, mouse TIM-2 is expressed in the liver, but it is particularly concentrated in bile duct epithelia and possibly bile canaliculi. In the kidney, expression of TIM-2 was concentrated in tubular epithelial cells with the morphology of distal tubular epithelial cells (Fig. 3, C and D). Staining was blocked by the immunizing peptide (unpublished data).
Figure 3.
Immunohistochemistry demonstrates expression of TIM-2 in kidney and liver. Paraffin-embedded sections from normal mice were stained with anti–TIM-2 antiserum and counterstained with hematoxylin. Representative sections are shown from liver (A and B) or kidney (C and D). A and C, bars, 60 μm; B and D, bars, 20 μm. Staining for TIM-2 was blocked by the immunizing peptide. In B, the arrow indicates bile duct epithelial cells, and in D, the arrow indicates distal tubular epithelial cells; both were stained with anti–TIM-2.
Identification of a ligand for TIM-2
A soluble ligand for TIM-2 is released by macrophage cell lines.
To identify ligands for mouse TIM-2, we created a chimeric TIM-2 receptor, in which the extracellular domain of TIM-2 (containing a FLAG epitope at the 5′ end) was linked to the cytoplasmic domain of CD3ζ, using the transmembrane region from CD8. This chimeric receptor then was expressed in the T cell line, BWZ.36 (BWZ), which contains a lacZ reporter gene under the control of three copies of an NFAT regulatory element (28). The CD3ζ cytoplasmic domain of the chimeric TIM-2 receptor contains three immunoreceptor tyrosine-based activation motifs. Activation of these immunoreceptor tyrosine-based activation motifs recruits the tyrosine kinases, ZAP-70 and Syk, and results in activation of the NFAT element and expression of lacZ. The expression of the chimeric TIM-2 receptor was confirmed by flow cytometry, using mAb's to FLAG or to TIM-2. Untransfected BWZ.36 cells did not express TIM-2, as demonstrated by flow cytometry or by PCR (unpublished data).
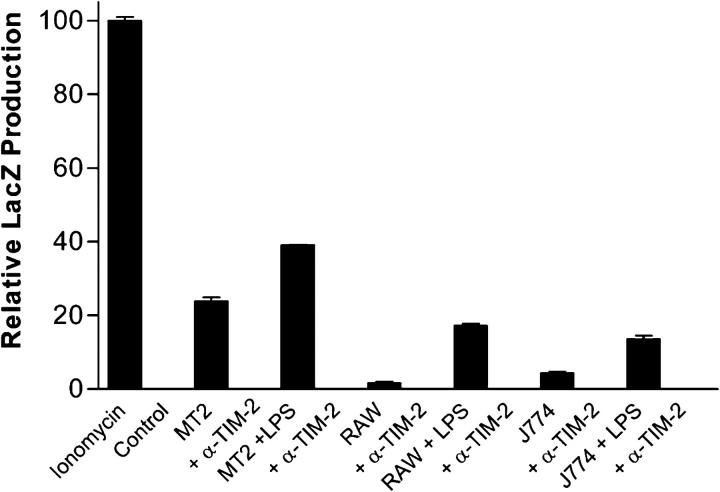
To screen cell lines for the expression of TIM-2 ligands, a variety of hematopoietic and nonhematopoietic cells lines was cocultured overnight with the BWZ.TIM-2/CD3ζ reporter cells, followed by assessment of lacZ activation. Three cell lines, all of macrophage lineage, stimulated the BWZ.TIM-2/CD3ζ cells: MT2, RAW264.7, and J774 (unpublished data). Furthermore, cell-free supernatants from these macrophage cell lines also activated lacZ production by BWZ.TIM-2/CD3ζ cells, and this activation was blocked by our mAb to TIM-2 (which does not activate TIM-2 unless it is cross-linked) (Fig. 4). The level of BWZ.TIM-2/CD3ζ activation by cell supernatant was increased when the macrophages were stimulated with LPS, and activation was blocked by anti–TIM-2. Thus, macrophage cell lines produce a soluble ligand for TIM-2.
Figure 4.
Release of TIM-2 ligand by macrophage cell lines. MT2, RAW 264.7 (RAW), and J774 macrophage cell lines were cultured with or without LPS (100 ng/ml) for 24 h. Conditioned medium from these cells was used to stimulate the BWZ.TIM-2/CD3ζ reporter line for a period of 16 h. For blockade of ligand action, anti–TIM-2 (α-TIM-2) mAb was used at 50 μg/ml. All wells were supplemented with PMA (10 ng/ml). Data are plotted as lacZ production relative to that stimulated by ionomycin and normalized to PMA alone. Bars represent mean of triplicate samples, and error bars indicate standard error.
Previous studies indicated that TIM-2 binds to semaphorin4A (Sema4a), and mice that are genetically deficient in Sema4a have immune defects (29, 30). Therefore, we prepared a soluble Sema4A/FcIg fusion protein, using the same protein fragments used by Kumanogoh et al. This chimeric protein did not stimulate our reporter cell lines at concentrations up to 180 nM (unpublished data). Also, by flow cytometry, the same Sema4a/FcIg chimeric protein did not bind to the BWZ.TIM-2 cells or to CHO.TIM-2 cells, even at concentrations of 40 μg/ml of protein (∼330 nM; unpublished data). Thus, we have been unable to confirm the binding of Sema4A to TIM-2. Although this may require additional factors that are not represented in our detection systems, these results indicated that the TIM-2 ligand that was detected in macrophage supernatants was not Sema4A.
Molecular cloning of H-ferritin as a ligand for TIM-2.
To identify soluble ligands for TIM-2 that are produced by macrophages, we screened a cDNA expression library that was prepared from unstimulated MT2 macrophages. 144 pools of cDNA, each containing ∼180 different cDNA clones, were used to transfect 293T cells. After 5 d in culture, supernatants from these cells were screened for the activation of BWZ.TIM-2/CD3ζ cells. Two supernatants strongly stimulated the reporter cells, and the cDNAs from these pools were pursued to isolate single colonies. The cDNA that was isolated from both pools was identical and corresponded exactly to mouse H-ferritin (GenBank M60170).
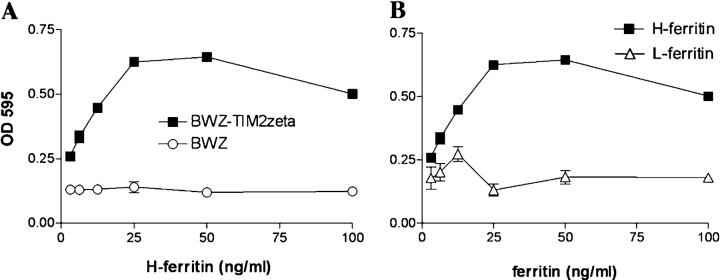
To verify that H-ferritin acts as a ligand for TIM-2, we produced and purified recombinant H- and L-ferritin in bacteria as described previously (31), except that the protein was passed over a polymyxin B column to remove endotoxin. Recombinant H-ferritin stimulated the BWZ.TIM-2/CD3ζ reporter cell lines, with maximal stimulation at ∼25 ng/ml (Fig. 5 A). H-ferritin did not stimulate untransfected BWZ cells (Fig. 5 A), or BWZ cells that were transfected with TIM-1/CD3ζ, TIM-3/CD3ζ, or TREM-2/CD3ζ, despite expression of these receptors at similar levels (unpublished data). Similar results were obtained with H-ferritin produced in eukaryotic (293T) cells (unpublished data). Furthermore, H-ferritin, but not L-ferritin, stimulated the BWZ.TIM-2/CD3ζ reporter cells (Fig. 5 B).
Figure 5.
H-ferritin but not L-ferritin ligates TIM-2. (A) H-ferritin activates BWZ.TIM-2/CD3ζ reporter cells (black squares) but not untransfected BWZ cells (open circles). The BWZ.TIM-2/CD3ζ reporter cells and the untransfected BWZ cells responded equally to PMA and ionomycin (unpublished data). (B) Recombinant H-ferritin (black squares) but not L-ferritin (open triangles) activates BWZ.TIM-2/CD3ζ reporter cells.
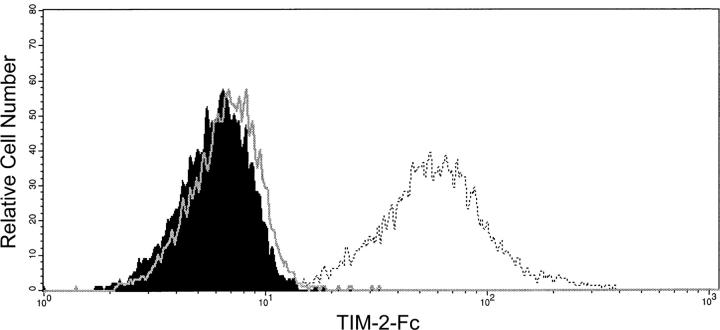
As an additional approach to documenting the interaction between TIM-2 and H-ferritin, we expressed H-ferritin on the surface of CHO cells by linking it to the CD8 transmembrane domain coupled to the CD3ζ cytoplasmic domain. We then prepared a soluble TIM-2–FcIg fusion protein, and examined binding by this protein to H-ferritin on the surface of CHO cells, as assessed by flow cytometry. The soluble TIM-2–FcIg fusion protein bound to CHO.H-ferritin cells, but not to untransfected CHO cells, and binding to the CHO.H-ferritin cells was blocked by antibody to TIM-2 (Fig. 6). In sum, by two approaches, TIM-2 binds H-ferritin. TIM-2 does not bind L-ferritin, and H-ferritin does not bind TIM-1 or -3.
Figure 6.
TIM-2–FcIg fusion protein binds to CHO cells expressing H-ferritin on the cell surface. CHO.H-ferritin cells were stained with TIM-2–Fc (open curve, dashed) or with secondary antibody only (filled curve). Addition of anti–TIM-2 monoclonal mAb (50 μg/ml; open curve) reduced binding to levels seen with secondary antibody alone.
TIM-2 is expressed on the cell surface and in endosomes.
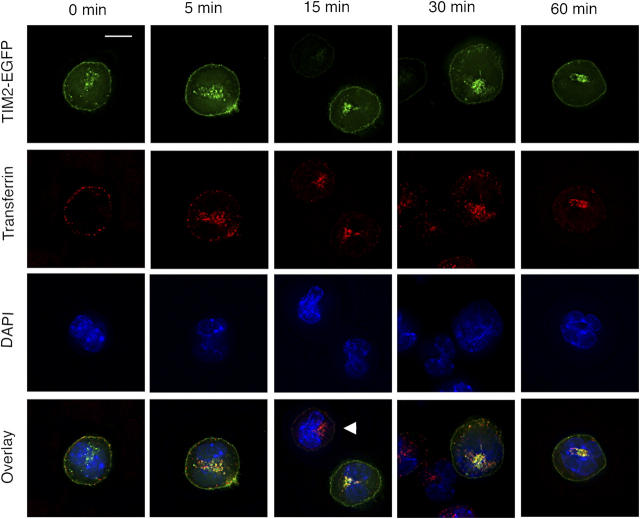
To examine the expression of TIM-2 further, we transfected BW5147 mouse T cells with the TIM-2 cDNA coupled to enhanced GFP (EGFP) at its cytoplasmic tail. After clonal selection, cells expressing TIM-2 were examined by deconvolution fluorescence microscopy. Transfected cells were mixed with untransfected cells on the same slide, as a control. In the absence of H-ferritin, TIM-2/EGFP+ cells demonstrated green fluorescence on the cell surface and in localized perinuclear regions (Fig. 7). To characterize the nature of the intracellular compartments in which TIM-2 is concentrated, we exposed TIM-2/EGFP+ cells to Alexa-568–conjugated transferrin, which serves as a marker for endosomes. As expected, transferrin was internalized by TIM-2/EGFP+ and TIM-2/EGFP− cells (Fig. 7). In the TIM-2/EGFP+ cells, transferrin and TIM-2 displayed punctate patterns that became progressively overlapping (Fig. 7). Notably, transferrin localized to the perinuclear compartment where TIM-2 is expressed prominently, which demonstrated that at least a substantial portion of this compartment represents membrane-derived endosomes.
Figure 7.
TIM-2 is expressed on the cell surface and in endosomes. BW5147 T cells (which lack TIM-2) were transfected with the gene for TIM-2, tagged at its COOH terminus with EGFP. Cells were exposed to transferrin, and coupled to the red dye Alexa-568, as a marker for endosomes. The figure shows the localization of TIM-2 and transferrin over the course of 1 h. As shown in the top row, TIM-2 localizes to the cell membrane and to localized intracellular compartments; this distribution is changed little by exposure to transferrin. As shown in the second row, transferrin is internalized largely within 15 min, identifying endosomes. In the third row, all cells are identified by staining of nuclei with DAPI. In the bottom row, fluorescence by TIM-2, transferrin, and DAPI are overlayed, demonstrating that the intracellular compartments expressing TIM-2 and transferrin are largely overlapping. Note that the panels for 15 and 30 min include, in addition to cells expressing TIM-2, cells that express little or no TIM-2 (arrowhead in 15-min panel). As expected, transferrin is endocytosed by TIM-2+ and TIM-2− cells. Bar, 10 μm.
TIM-2 is required for the cellular uptake of H-ferritin
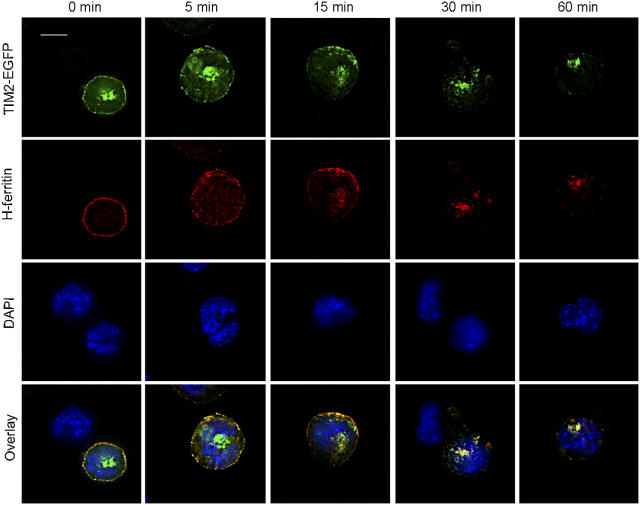
We next examined the consequences of exposing the TIM-2/EGFP+ cells to H-ferritin, tagged by conjugation to Alexa-568. H-ferritin bound only to TIM-2/EGFP+ cells, and confirmed that H-ferritin does not bind to the cells in the absence of TIM-2 (Fig. 8). With incubation at 37°, H-ferritin was internalized progressively by TIM-2/EGFP+ cells. Within 5 min, intracellular H-ferritin was evident in the form of punctate structures beneath the cell surface, where it colocalized with TIM-2; over the next 15–30 min, it colocalized increasingly with the perinuclear TIM-2/EGFP. Thus, TIM-2 transports H-ferritin to punctate structures within the cell, identified by transferrin as endosomes.
Figure 8.
H-ferritin is internalized by cells expressing TIM-2 but not by cells lacking TIM-2. The experiments used the same cell line as in Fig. 7, but here the cells were exposed to H-ferritin, identified by conjugation to Alexa-568. As shown in the top row, the distribution of TIM-2 on the cell surface becomes beaded within 5 min after incubation with H-ferritin; its expression on the cell surface declines thereafter. As shown in the second row, H-ferritin is internalized rapidly by cells expressing TIM-2. In the third row, all cells are identified by staining of nuclei with DAPI. In the bottom row, fluorescence by TIM-2, H-ferritin, and DAPI are overlayed. Note that the panels for 0 and 30 min include, in addition to cells expressing TIM-2, cells that express little or no TIM-2. Only cells expressing TIM-2 bind and internalize H-ferritin. The results are representative of four separate experiments. Bar, 10 μm.
DISCUSSION
We have demonstrated that TIM-2 is expressed in localized areas of the spleen, liver, and kidneys, and that it binds H-ferritin, but not L-ferritin. Further, after binding of H-ferritin to TIM-2 on the cell surface, H-ferritin is internalized into endosomes.
In the spleen, we found that TIM-2 is expressed on all B cells, with expression at higher levels on GC B cells. Thus, different members of the TIM family are expressed in different subsets of immune cells; TIM-2 is on B cells, whereas TIM-1 is inducible on Th2 cells, and TIM-3 is inducible on Th1 cells. TIM-4 initially was identified on splenic stromal cells, but recent studies indicated that it also is expressed on antigen-presenting cells, where it serves as a ligand for TIM-1 (7, 12). We have not detected TIM-2 on resting T cells, nor have we been able to induce its expression on T cells in vitro by treatment with mitogen (ConA) or by stimulation of T cells under conditions that induce Th1 or Th2 cells (unpublished data). However, the induction of TIM-1 or -3 on T cells requires repeated stimulation, and we may not have identified proper conditions to induce the expression of TIM-2 on T cells. Regardless, although TIM-2 is 66% homologous to TIM-1, these two TIM receptors differ in their expression on lymphocytes.
Our demonstration that TIM-2 binds to H-ferritin is the first identification of a cell surface receptor for ferritin, although the existence of ferritin receptors was postulated previously, based on saturable binding of ferritin to the surface of several cell types, including B and T lymphocytes. Thus, binding of H-ferritin to lymphocytes is increased in mitogen-stimulated cells, and purified H-ferritin forms surface patches on T cells, after which it is endocytosed into lysosomes (23, 24, 32). Studies using MOLT-4 human T cells indicated that they express a receptor for H-ferritin with an association constant of ∼7 × 10−7 L/mol, and that the receptor is increased on proliferating cells (25, 26). Biochemical evidence for an H-ferritin receptor also was found on liver cells, including activated liver lipocytes (33, 34). In addition, specific ferritin binding was demonstrated on erythroid precursor cells (35, 36), brain tissue (37, 38), and placental membranes (39).
In one report, a putative ferritin receptor was purified from human liver (40). Although the molecular weight of this receptor, 53 kD, is generally consistent with the predicted size of glycosylated TIM receptors, it was not identified further, and the specificity of this receptor for H-ferritin was not shown. The liver seems to express at least two binding sites for H-ferritin; one binds L- and H-ferritin with equal affinity, whereas the second, like TIM-2, binds only to H-ferritin (33, 41). In healthy individuals, circulating ferritin is predominantly L-ferritin. It was suggested that the first receptor, which binds H- and L-ferritin, may serve to regulate levels of serum ferritin, whereas the H-ferritin receptor may subserve independent cellular functions in response to a selective increase in H-ferritin.
Ferritin is increased in inflammation, and we showed previously that TNF-α and IL-1 activate transcription of the H-ferritin gene in an additive manner, providing at least one mechanism by which H-ferritin may be increased in inflammation (42, 43). The sources of circulating ferritin are not defined; however, our cloning of H-ferritin from a macrophage line is in accord with previous studies of H-ferritin production by macrophages, including studies that demonstrated that LPS stimulates H-ferritin production by J774 and RAW264.7 macrophage cells, both of which produced an LPS-inducible ligand for TIM-2 (44, 45). Recent studies have shown that transcription of H-ferritin is induced by NF-κB, and the consequent interaction of H-ferritin with iron serves to buffer the generation of reactive oxygen species, an activity that is required for the capacity of NF-κB to counter TNF-α–induced apoptosis (19). Our studies demonstrate that TIM-2 permits the endocytosis of H-ferritin, revealing that cellular levels of H-ferritin are not dependent solely on transcription. This finding opens a new pathway into understanding the role of ferritin in cell function.
The properties of TIM-2 that are required for endocytosis of H-ferritin by TIM-2 remain to be elucidated. The cytoplasmic domain of TIM-2 has three tyrosine residues. The membrane-proximal tyrosine is part of a YxxM motif, which has been associated with endocytic localization as well as with activation of PI3 kinase (46); however, the proximity of this motif to the inner leaflet of the cell membrane may render it nonfunctional. The other two tyrosines are not part of known signaling or targeting motifs, but one motif (E-[ED]-x-x-Y-x-x-E) is conserved through several TIM receptors, which suggests functional significance.
By IHC of the liver, mouse TIM-2 is expressed highly in bile ducts and, to a lesser extent, in hepatocytes. In accord with this, we find high levels of TIM-2 transcripts in the liver (unpublished data). The expression of TIM-2 in bile duct epithelial cells suggests that it may be involved in the transport of ferritin into or out of bile. Ferritin excretion into bile is believed to involve lysosomal exocytosis, but the mechanisms of excretion are not well defined (47, 48). From our studies, it is possible that this process involves TIM-2. Humans express high levels of TIM-1 in the liver; however, in the mouse, levels of transcripts for TIM-2 expression are 100–1,000 times higher than levels of transcripts for TIM-1 (unpublished data). Although the localization of TIM-1 in the human liver has not been described, these results suggest the possibility that, in the liver, mouse TIM-2 may be a functional ortholog of human TIM-1.
No human ortholog for mouse TIM-2 has been identified, although mouse TIM-2 is only slightly less homologous to human TIM-1 than is mouse TIM-1. Thus, human TIM-1 may share functions with mouse TIM-2 and -1, including the capacity to bind H-ferritin. Alternatively, the capacity of mouse TIM-2 to bind H-ferritin may have been usurped by a different human receptor. Whatever the nature of the human ferritin receptor, the expression of TIM-2 in mice roughly parallels that of known ferritin binding sites in humans. Thus, the expression of TIM-2 on mouse lymphocytes and hepatic cells corresponds with binding of H-ferritin to human lymphocytes and liver cells, except that we have not identified TIM-2 on resting T cells. However, TIM-2 is expressed on mouse EL-4 thymoma cells (unpublished data), and there may be conditions under which its expression is induced on fresh T cells. Studies are in progress to define the role of human TIMs in ferritin binding.
Our studies also may bear on the role of H-ferritin in malignancy. H-ferritin is increased often in malignancy, and its expression correlates with poor prognosis. Thus, the expression of transcripts for H-ferritin by breast cancer cells is an adverse prognostic indicator (49). Similarly, in ovarian cancer, metastatic cells express higher transcripts for H-ferritin than do nonmetastatic cells (50). In melanoma, levels of circulating H-ferritin are elevated, and the levels of H-ferritin correlate with levels of CD4+CD25+CD69− regulatory T cells (51). Further, H-ferritin was shown to activate regulatory T cells through mechanisms that require dendritic cells (52). Studies in rats similarly showed an up-regulation of H-ferritin during the induction of hepatocellular carcinoma (53). Our studies identify a receptor in mice through which H-ferritin may selectively alter cell functions, including immune functions. It will be of interest to pursue the possibility that the production of H-ferritin by malignant cells alters the host response through the TIM-2 ferritin receptor.
As an additional note, while this paper was under review, Chakravarti, et al. demonstrated that transcripts for TIM-2 can be induced selectively in Th2 T cells (54). We did not detect elevated levels of TIM-2 transcripts in freshly isolated splenic T cells. In preliminary results we did, like Chakravarti, et al., find transcripts for TIM-2 in T cells after activation in vitro, but as noted in Results we have not defined conditions that will induce detectable levels of TIM-2 on the surface of T cells. We are testing the possibility that this may be regulated by H-ferritin.
MATERIALS AND METHODS
Mice.
All animal studies were approved by the Animal Studies Subcommittee of the Research & Development Committee, San Francisco Veterans Administration Medical Center, by the UCSF Animal Care Committee. C57BL/6 (B6), or congenic B6-Ly5.2 or B6-IgH a Thy1 a Gpi1 a mice were obtained from The Jackson Laboratory and the National Cancer Institute, and were maintained in a barrier facility. Where indicated, mice were immunized i.p. with 3.5 × 108 SRBCs (Colorado Serum Company), or with 50 μg 4-hydroxy-3-nitrophenyl–acetyl-chicken gamma globulin (NP30-CGG; Biosearch Technologies) precipitated in alum. Following euthanasia under approved protocols, tissues were obtained for flow cytometry and IHC at day 8 for SRBCs and day 14 for NP30-CGG, the peak of the GC response.
Cell lines.
The BWZ.36 (BWZ) mouse T cell lymphoma cell line (BW5147 cells transfected with the cDNA for lacZ under control of four copies of the NFAT promoter) was provided by N. Shastri (University of California Berkeley, Berkeley, CA) (28). The MT2 mouse macrophage cell line was provided by M. McKichan (University of California San Francisco, San Francisco, CA). All other cell lines are from the American Type Culture Collection. Cell lines other than MT2 were grown in RPMI 1640 supplemented with 10% heat-inactivated FBS, 25 μM 2-ME, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (cRPMI 1640). MT2 cells were grown in IMDM supplemented with 10% heat-inactivated FBS, 25 μM 2-ME, 2 mM l-glutamine, 100 U/ml penicillin. and 100 μg/ml streptomycin.
Production of a monoclonal antibody and of antisera to TIM-2.
For monoclonal antibody production, rats were immunized two times in the footpad with TIM-2–Fc fusion protein emulsified in Titermax adjuvant, followed by two immunizations with CHO cells expressing FLAG-tagged TIM-2. Lymphocytes from the popliteal lymph nodes were fused with YB-2/0 plasmacytoma cells, using standard techniques. Supernatants were screened for binding to BWZ.TIM-2 cells, but not BWZ cells. Subsequent studies demonstrated that the antibody did not bind to BWZ cells expressing TIM-1 or -3, as documented by expression of a FLAG epitope at the 5′ end (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20042433/DC1).
Rabbit antisera against the cytoplasmic domain of TIM-2 were produced by ProSci Inc. by immunization with a synthetic peptide (DQVYIIEDTPYPEEES), corresponding to the 16 terminal residues of the full-length TIM-2 protein. This sequence is not found in other members of the TIM family. Antisera were affinity purified with the immunizing peptide.
Flow cytometry and cell sorting.
Spleen cells were analyzed by flow cytometry or sorted by FACS as described (55). Cells were gated with wide forward scatter gates but narrow side scatter gates to include blast cells but to exclude granulocytes, and then were gated further on singlet cells using width and height parameters. Dead cells were excluded with propidium iodide. Follicular B cells and GC B cells were gated as depicted previously (55). For flow cytometry, follicular B cells were defined as CD19+, GL7−, Fas−, IgDhigh, CD4−, CD8−, and GC B cells were defined as CD19+, GL7high, Fas+, IgDlow, CD4−, CD8−. For cell sorting, follicular B cells were defined as CD19+, CD21med, CD23+; GC were defined as CD19+, GL7high, Fas+, IgDlow; and marginal zone B cells were defined as CD19+, CD21high, CD23−. The following monoclonal antibodies were used in these studies (from BD Biosciences unless otherwise indicated): rat anti-mouse CD4 (clone RM4-5, Alexa 405; Caltag), rat anti–mouse CD8 (clone 53–6.7, PerCP), rat anti–mouse CD19 (clone 1D3, PE-Cy7, APC, APC-Cy7), rat anti–mouse CD21 (clone 7G6, FITC), rat anti–mouse CD23 (clone B3B4, PE), hamster anti–mouse CD95/Fas (clone Jo2, PE-Cy7), rat anti–mouse T and B cell activation antigen (clone GL7, FITC), and rat anti–mouse IgD (clone 11–26, PE, Southern Biotechnology Associates Inc.). The monoclonal antibody to TIM-2 was biotinylated by using EZ-Link (Pierce Chemical Co.) according to the manufacturer's directions, and binding was detected with streptavidin APC (Invitrogen). Detection of FLAG-tagged TIM-2 used mAb M2 (biotinylated; Sigma-Aldrich). Detection of binding by the TIM-2/Fc chimeric protein was detected by using a PE-labeled goat anti–human IgG Fc-specific secondary antibody with minimal cross-reactivity to other species (Jackson ImmunoResearch Laboratories). Cells were analyzed on a FACSCalibur flow cytometer (Becton Dickinson), except that multicolor stains of fresh B cells were analyzed on a FACSAria flow cytometer (Becton Dickinson), using FlowJo software (Treestar). Fresh B cells were sorted on a Mo-Flo (DakoCytomation).
Quantitative PCR.
Total RNA and first-strand cDNA were prepared as described (55). For quantitative PCR, 4–8% of the cDNA was placed in a final volume of 50 μl containing SYBR green PCR Master Mix (Applied Biosystems, ABI) and primers (300 nM). Samples were analyzed on a 7300 real-time PCR system (ABI) with the following thermal-cycler conditions: 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s followed by 60°C for 1 min. Data were analyzed by the Comparative CT Method (ABI 7700), giving a relative ratio of target gene mRNA to housekeeping gene (hypoxanthine-guanine phosphoribosyltransferase, HPRT or GAPDH) mRNA. Primers were designed by using PrimerExpress version 2.0 and sequences were as follows: TIM-2: forward, CCAACACCAGCACACACAGAGACCT, reverse, TGGCTTCTGTGGAGGGATTACTTCA; HPRT: forward, AGGTTGCAAGCTTGCTGGT, reverse, TGAAGTACTCATTATAGTCAAGGGCA; GAPDH: forward, GGTCTACATGTTCCAGTATGACTCCAC, reverse, GGGTCTCGCTCCTGGAAGAT.
Immunohistochemistry.
Freshly harvested tissues were fixed by immersion in neutral 10% buffered formalin (Fisher Scientific) overnight, followed by embedding in paraffin. Tissue sections were cut at 10-μm thickness, mounted on precleaned glass slides, and baked for 1 h at 80°C. Slides were dewaxed with Citrisolv, equilibrated in distilled water, and subjected to antigen retrieval by microwaving at low power for 5 min in 10 mM citrate buffer pH 6.0 then cooling for 20 min. Endogenous peroxidase activity was blocked by incubating in 0.3% hydrogen peroxide for 5 min; nonspecific binding was blocked by incubating with 10% normal goat serum in 10 mM Tris-HCl, pH 7.5, 150 mM NaCl (TBS) for 1 h at room temperature (RT). Sections were incubated with affinity-purified anti–TIM-2 rabbit antiserum diluted 1:1,000 in TBS with 0.1% Tween 20 (TBST)/5% BSA for 1 h at RT, with or without immunizing peptide at 1 μg/ml; washed extensively in TBST; then incubated with goat anti–rabbit IgG conjugated to horseradish peroxidase (Caltag) diluted 1:1,000 in TBST/5% BSA for 30 min at RT. After washing in TBST, bound antibody was detected with 3,3′-diaminobenzidine (BioFX), counterstained with hematoxylin (Fisher Scientific), and mounted with Permount (Fisher Scientific). Slides were photographed under 10 and 40× objectives on an Olympus BX41 microscope equipped with a DP12 digital camera.
For experiments examining GCs, dual-color IHC on cryostat sections from mice that were immunized with SRBCs was performed as described (55). Slides were observed with a 10× objective on a Leica DMLB microscope equipped with an Optronics Engineering MDE1580 CCD camera.
Immunofluorescence of frozen tissue sections.
Freshly harvested spleens were embedded rapidly in OCT compound (Miles Inc.), and immediately frozen in liquid nitrogen. Tissue sections were cut at 8-μm thickness, mounted on precleaned glass slides, and fixed for 10 min in ice-cold acetone. After rehydration in PBS, slides were treated with a biotin-blocking system (DakoCytomation), and nonspecific binding was blocked by incubating in TBS plus 10% normal goat serum for 1 h at RT. Sections were incubated with affinity-purified anti–TIM-2 rabbit antiserum diluted 1:250 in TBST/5% BSA as well as biotinylated antibodies against B220 (Caltag) or CD3 (BD Biosciences). After extensive washing in TBST, slides were incubated with goat anti–rabbit IgG conjugated to Alexa 488 and streptavidin conjugated to Alexa 546 (Invitrogen) at 1 μg/ml in TBST; mounted with Gelmount (Biomeda); and visualized on a Zeiss Axioskop 2 FS fluorescent microscope attached to a Hamamatsu Orca CCD camera. Digital images were acquired with a 10× objective, contrast and brightness were adjusted automatically using OpenLab3 defaults, and the unmodified TIFF files were exported.
Vector construction.
An expressed sequence tag corresponding to murine TIM-2 (AA575518) was identified in GenBank based on partial homology to the Ig domain of TREM-2, by using the tblastn algorithm. The construct was obtained from the American Type Culture Collecrtion and confirmed by sequencing. The full-length cDNA was expressed intact or with the addition of a FLAG epitope, using a vector provided by L. Lanier (University of California San Francisco, San Francisco, CA). In addition, the extracellular domain of TIM-2 was expressed as a chimeric molecule linked to the transmembrane domain of CD8 and the cytoplasm domain of CD3ζ. This was achieved by constructing a vector containing a CD8 leader sequence (amino acids 1–21, AAH25715) and FLAG epitope, followed by the extracellular domain of TIM-2 (amino acids 22–231), the CD8 transmembrane domain (amino acids 187–215, AAH25715), and the CD3ζ cytoplasmic domain (amino acids 216–327, AAF34793) (using a vector provided by A. Weiss, University of California San Francisco, San Francisco, California). A similar construct was used to express full-length mouse H-ferritin on the cell surface, substituting the H-ferritin cDNA sequence for the extracellular domain of TIM-2. These chimeric DNAs were shuttled into the pcDNA4 vector (Stratagene), and the integrity of the constructs was confirmed by sequencing. BWZ cells were transfected with either construct by electroporation, and were selected in cRPMI 1640 containing 0.75 mg/ml Zeocin (Invitrogen).
The extracellular domain of TIM-2 was expressed in soluble form as an Fc fusion protein. This construct was prepared in a pCDM8 vector into which amino acids 22–230 of TIM-2 were inserted between leader sequence for signaling lymphocytic activation molecule (amino acids 1–24, NP_038758.1) and the human IgG1 Fc domain (amino acids 243–473, CAA75030) (vector provided by L. Lanier). The chimeric cDNA was shuttled into the pcDNA4 vector (Stratagene), and the integrity of the construct was confirmed by sequencing. 293T cells in exponential growth were transfected with 3 μg plasmid DNA by using FuGENE (Roche); transfected cells were selected in cRPMI 1640 containing 0.75 mg/ml Zeocin (Invitrogen). Ig fusion protein was purified from conditioned medium using protein G affinity chromatography.
Preparation and characterization of H- and L-ferritin.
Mouse H- and L-ferritin were produced in Escherichia coli, and were purified by sucrose density gradient centrifugation as described (31). Endotoxin was removed by adsorption to immobilized polymyxin B (Detoxi-GelTM, Pierce Chemical Co.), according to the methods of the manufacturer. After adsorption, endotoxin levels were below 5 EU/ml, as assessed by the chromogenic end-point limulus amebocyte lysate assay (QCL-1000, Cambrex Bio Science). Additionally, mouse H-ferritin was produced in human 293T kidney cells by transfection with the cDNA, modified to use the signal sequence from CD8, followed by the sequence encoding a FLAG epitope (DYKDDDDK), and then the H-ferritin sequence. For the expression of H-ferritin on the cell surface, this construct was extended to include the CD8 transmembrane domain followed by the cytoplasmic domain of CD3ζ, and cells were transfected by electroporation. For fluorescence microscopy, H- and L-ferritin were labeled covalently with Alexa-568 by using a commercial kit (Invitrogen).
Reporter assay.
The BWZ line, derived by Sanderson and Shastri (28) from BW5147 T cells, contains a lacZ reporter construct regulated by four copies of an NFAT regulatory element. BWZ cells transfected with the TIM-2/CD3ζ chimeric receptor or (as a control) a TREM-2/CD3ζ chimeric receptor, were seeded in 96-well plates at 105 cells/well in cRPMI 1640 supplemented with 10 ng/ml PMA. To test for stimulation of these cells by macrophage (or other) cell lines, the stimulator cells were added at a ratio of 5:1 (i.e., 5 × 105 cells/well), in triplicate. Alternatively, 100 μl of “conditioned” medium from confluent cells was added to triplicate wells. For blocking experiments, anti–TIM-2 mAb was added to cells at 50 μg/ml at RT 15 min before ferritin. Plates were incubated for 16 h at 37°C in a 5% CO2 humidified atmosphere. Cells were washed once in PBS, and lacZ activity was determined by incubating the cells with 150 μM chlorophenolred-β-D-galactopyranoside in PBS supplemented with 100 mM 2-mercaptoethanol, 9 mM MgCl2, and 0.125% NP-40. After sufficient color development, absorbance was measured at 595 nm, and was corrected for background absorbance at 650 nm. Values were normalized by subtracting the absorbance of wells that were treated with PMA alone. Maximum stimulation was defined as the response to PMA plus 1 μM ionomycin. For stimulation of the reporter cell lines by purified mouse H- or L-ferritin, 5 × 105 cells/well were stimulated in RPMI 1640 supplemented with 1% FBS. For blocking experiments, anti–TIM-2 mAb or an isotype-matched control mAb (2C7, rat IgG2a anti-ovalbumin) was added to cells at 10 μg/ml at RT 15 min before ferritin. Cells were assayed as above.
Molecular cloning of TIM-2 ligands.
To screen for soluble TIM-2 ligands, a cDNA library that was prepared from MT2 macrophage cells was transformed into bacteria, and the bacterial titer was determined by growth on ampicillin plates. 144 pools of bacteria, each containing ∼180 CFUs, were grown overnight, and their DNA was isolated by using a Qiagen8 miniprep kit. Each miniprep was transfected into 293T to screen for soluble TIM-2 ligands as follows: 15,000 293T cells were plated in 96-well plates and allowed to adhere overnight. The next day, 0.5 μg DNA was mixed with 1.5 μl FuGENE in 100 μl OptiMEM medium and incubated for 30 min. Medium was aspirated from the 293T cells, and the OptiMEM/DNA mix was added. 5 h later, 100 μl RPMI 1640 supplemented with 20% FCS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin was added to each well. After 5 d, conditioned medium was collected and cell debris was removed by centrifugation. 100 μl of this conditioned medium was mixed with 105 BWZ.TIM-2-ζ reporter cells in the presence of 10 ng/ml PMA, and the plates were incubated for 16 h at 37°C in a 5% CO2 humidified atmosphere. Assay development was performed as detailed above. Positive minipreps were retransformed into bacteria, and the screening was repeated with pools containing 12 CFUs, and finally, with single colonies.
Expression of TIM-2 covalently linked to enhanced GFP.
To couple EGFP to the TIM-2 cytoplasmic (C−) terminus, the full-length TIM2 coding sequence, including the endogenous signal sequence, was amplified by PCR by using the primers 5′-CCGGGAATTCATGAATCAGATTCAAGTCTTC-3′ and 5′-TACCGTCGACTGGGACTCTTCTTCGGGGTAAGG-3′, and then cloned into the EcoRI and SalI sites of the pEGFP-N1 expression vector (BD Clontech). Clones were verified for integrity by sequencing and were transfected into BW5147 cells by using FuGENE6. Stable transfectants were selected with G418 (Cellgro) at 1 mg/ml in cRPMI 1640. Expression of the fusion construct was verified by flow cytometry, assessing expression of GFP and, on the cell surface, TIM-2.
Deconvolution microscopy.
BW5147 cells that were transfected with TIM2-EGFP were washed twice in PBS and incubated in uptake buffer (PBS supplemented with 10 mM Tris-HCl, 10 mM Hepes, 5 mM glucose, 1 mg/ml BSA, pH 7.4) for 30 min at 37°C. Cells were chilled on ice, and H-ferritin (1 μg/ml) or transferrin (50 μg/ml), each tagged with Alexa-568 (Invitrogen), was added. After 30 min on ice, cells were washed once with ice-cold PBS, and internalization was initiated by adding fresh uptake buffer prewarmed to 37°C. Aliquots were removed at defined time points and were added to excess ice-cold PBS with 0.02% sodium azide to stop internalization. Cells were pelleted, fixed in PBS/4% paraformaldehyde, and mounted on coverslips coated with 0.1% poly-l-lysine (Sigma-Aldrich). For some experiments, untransfected BW5147 cells were added as controls. Images were collected by an API DeltaVision DV3 Restoration microscope using a MicroMax 5 MHz cooled CCD camera (Roper Scientific); deconvolution was performed using API SoftWoRx software.
Online supplemental material.
Fig. S1 shows the binding of anti–TIM-2 mAb to TIM-2, but not to TIM-1 or -3. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20042433/DC1.
Acknowledgments
We thank S. Hayashi, J. Brown, and Y. Xu for expert technical assistance, and E. Theil for helpful discussions.
This work was supported by the Veterans Administration and by grants R01 CA87922-01A1 (to W.E. Seaman), DK42412 (to S.V. Torti and F.M. Torti), and GM038093 (to F.M. Brodsky) from the National Institutes of Health. T.T. Chen was supported by an Advanced Career Development Award from the Veterans Administration. C.D.C. Allen was supported by a Howard Hughes Medical Institute predoctoral fellowship. M.C. Nakamura was supported by a Research Scholar Grant RSG-01-167-01-LIB from the American Cancer Society.
The authors have no conflicting financial interests.
Abbreviations used: GC, germinal center; IHC, immunohistochemistry; RT, room temperature; TIM, T cell immunoglobulin domain and mucin domain; TREM, triggering receptor expressed on myeloid cells.
T.T. Chen and L. Li contributed equally to this work.
M.R. Daws' present address is Department of Anatomy, University of Oslo, Oslo, Norway NO317.
References
- 1.Feigelstock, D., P. Thompson, P. Mattoo, Y. Zhang, and G.G. Kaplan. 1998. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J. Virol. 72:6621–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ichimura, T., J.V. Bonventre, V. Bailly, H. Wei, C.A. Hession, R.L. Cate, and M. Sanicola. 1998. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J. Biol. Chem. 273:4135–4142. [DOI] [PubMed] [Google Scholar]
- 3.Ichimura, T., C.C. Hung, S.A. Yang, J.L. Stevens, and J.V. Bonventre. 2004. Kidney injury molecule-1: a tissue and urinary biomarker for nephrotoxicant-induced renal injury. Am. J. Physiol. Renal Physiol. 286:F552–563. [DOI] [PubMed] [Google Scholar]
- 4.Kuchroo, V.K., D.T. Umetsu, R.H. DeKruyff, and G.J. Freeman. 2003. The TIM gene family: emerging roles in immunity and disease. Nat. Rev. Immunol. 3:454–462. [DOI] [PubMed] [Google Scholar]
- 5.Khademi, M., Z. Illes, A.W. Gielen, M. Marta, N. Takazawa, C. Baecher-Allan, L. Brundin, J. Hannerz, C. Martin, R.A. Harris, et al. 2004. T Cell Ig- and mucin-domain-containing molecule-3 (TIM-3) and TIM-1 molecules are differentially expressed on human Th1 and Th2 cells and in cerebrospinal fluid-derived mononuclear cells in multiple sclerosis. J. Immunol. 172:7169–7176. [DOI] [PubMed] [Google Scholar]
- 6.McIntire, J.J., S.E. Umetsu, O. Akbari, M. Potter, V.K. Kuchroo, G.S. Barsh, G.J. Freeman, D.T. Umetsu, and R.H. DeKruyff. 2001. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat. Immunol. 2:1109–1116. [DOI] [PubMed] [Google Scholar]
- 7.Meyers, J.H., S. Chakravarti, D. Schlesinger, Z. Illes, H. Waldner, S.E. Umetsu, J. Kenny, X.X. Zheng, D.T. Umetsu, R.H. Dekruyff, et al. 2005. TIM-4 is the ligand for TIM-1, and the TIM-1-TIM-4 interaction regulates T cell proliferation. Nat. Immunol. 6:455–464. [DOI] [PubMed] [Google Scholar]
- 8.McIntire, J.J., S.E. Umetsu, C. Macaubas, E.G. Hoyte, C. Cinnioglu, L.L. Cavalli-Sforza, G.S. Barsh, J.F. Hallmayer, P.A. Underhill, N.J. Risch, et al. 2003. Immunology: hepatitis A virus link to atopic disease. Nature. 425:576. [DOI] [PubMed] [Google Scholar]
- 9.Chae, S.C., J.H. Song, Y.C. Lee, J.W. Kim, and H.T. Chung. 2003. The association of the exon 4 variations of Tim-1 gene with allergic diseases in a Korean population. Biochem. Biophys. Res. Commun. 312:346–350. [DOI] [PubMed] [Google Scholar]
- 10.Chae, S.C., J.H. Song, S.C. Shim, K.S. Yoon, and H.T. Chung. 2004. The exon 4 variations of Tim-1 gene are associated with rheumatoid arthritis in a Korean population. Biochem. Biophys. Res. Commun. 315:971–975. [DOI] [PubMed] [Google Scholar]
- 11.Umetsu, S.E., W.L. Lee, J.J. McIntire, L. Downey, B. Sanjanwala, O. Akbari, G.J. Berry, H. Nagumo, G.J. Freeman, D.T. Umetsu, and R.H. Dekruyff. 2005. TIM-1 induces T cell activation and inhibits the development of peripheral tolerance. Nat. Immunol. 6:447–454. [DOI] [PubMed] [Google Scholar]
- 12.Shakhov, A.N., S. Rybtsov, A.V. Tumanov, S. Shulenin, M. Dean, D.V. Kuprash, and S.A. Nedospasov. 2004. SMUCKLER/TIM4 is a distinct member of TIM family expressed by stromal cells of secondary lymphoid tissues and associated with lymphotoxin signaling. Eur. J. Immunol. 34:494–503. [DOI] [PubMed] [Google Scholar]
- 13.Monney, L., C.A. Sabatos, J.L. Gaglia, A. Ryu, H. Waldner, T. Chernova, S. Manning, E.A. Greenfield, A.J. Coyle, R.A. Sobel, et al. 2002. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature. 415:536–541. [DOI] [PubMed] [Google Scholar]
- 14.Sabatos, C.A., S. Chakravarti, E. Cha, A. Schubart, A. Sanchez-Fueyo, X.X. Zheng, A.J. Coyle, T.B. Strom, G.J. Freeman, and V.K. Kuchroo. 2003. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nat. Immunol. 4:1102–1110. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Fueyo, A., J. Tian, D. Picarella, C. Domenig, X.X. Zheng, C.A. Sabatos, N. Manlongat, O. Bender, T. Kamradt, V.K. Kuchroo, et al. 2003. Tim-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nat. Immunol. 4:1093–1101. [DOI] [PubMed] [Google Scholar]
- 16.Bailly, V., Z. Zhang, W. Meier, R. Cate, M. Sanicola, and J.V. Bonventre. 2002. Shedding of kidney injury molecule-1, a putative adhesion protein involved in renal regeneration. J. Biol. Chem. 277:39739–39748. [DOI] [PubMed] [Google Scholar]
- 17.Theil, E.C. 2003. Ferritin: at the crossroads of iron and oxygen metabolism. J. Nutr. 133:1549S–1553S. [DOI] [PubMed] [Google Scholar]
- 18.Torti, F.M., and S.V. Torti. 2002. Regulation of ferritin genes and protein. Blood. 99:3505–3516. [DOI] [PubMed] [Google Scholar]
- 19.Pham, C.G., C. Bubici, F. Zazzeroni, S. Papa, J. Jones, K. Alvarez, S. Jayawardena, E. De Smaele, R. Cong, C. Beaumont, et al. 2004. Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell. 119:529–542. [DOI] [PubMed] [Google Scholar]
- 20.Matzner, Y., C. Hershko, A. Polliack, A.M. Konijn, and G. Izak. 1979. Suppressive effect of ferritin on in vitro lymphocyte function. Br. J. Haematol. 42:345–353. [DOI] [PubMed] [Google Scholar]
- 21.Morikawa, K., F. Oseko, and S. Morikawa. 1995. A role for ferritin in hematopoiesis and the immune system. Leuk. Lymphoma. 18:429–433. [DOI] [PubMed] [Google Scholar]
- 22.Harada, T., M. Baba, I. Torii, and S. Morikawa. 1987. Ferritin selectively suppresses delayed-type hypersensitivity responses at induction or effector phase. Cell. Immunol. 109:75–88. [DOI] [PubMed] [Google Scholar]
- 23.Renau-Piqueras, J., F. Miragall, and J. Cervera. 1985. Endocytosis of cationized ferritin in human peripheral blood by resting T-lymphocytes. Cell Tissue Res. 240:743–746. [DOI] [PubMed] [Google Scholar]
- 24.Fargion, S., A.L. Fracanzani, B. Brando, P. Arosio, S. Levi, and G. Fiorelli. 1991. Specific binding sites for H-ferritin on human lymphocytes: modulation during cellular proliferation and potential implication in cell growth control. Blood. 78:1056–1061. [PubMed] [Google Scholar]
- 25.Moss, D., L.W. Powell, P. Arosio, and J.W. Halliday. 1992. Effect of cell proliferation on H-ferritin receptor expression in human T lymphoid (MOLT-4) cells. J. Lab. Clin. Med. 120:239–243. [PubMed] [Google Scholar]
- 26.Moss, D., L.W. Powell, P. Arosio, and J.W. Halliday. 1992. Characterization of the ferritin receptors of human T lymphoid (MOLT-4) cells. J. Lab. Clin. Med. 119:273–279. [PubMed] [Google Scholar]
- 27.Daws, M.R., L.L. Lanier, W.E. Seaman, and J.C. Ryan. 2001. Cloning and characterization of a novel mouse myeloid DAP12-associated receptor family. Eur. J. Immunol. 31:783–791. [DOI] [PubMed] [Google Scholar]
- 28.Sanderson, S., and N. Shastri. 1994. LacZ inducible, antigen/MHC-specific T cell hybrids. Int. Immunol. 6:369–376. [DOI] [PubMed] [Google Scholar]
- 29.Kumanogoh, A., S. Marukawa, K. Suzuki, N. Takegahara, C. Watanabe, E. Ch'ng, I. Ishida, H. Fujimura, S. Sakoda, K. Yoshida, and H. Kikutani. 2002. Class IV semaphorin Sema4A enhances T-cell activation and interacts with Tim-2. Nature. 419:629–633. [DOI] [PubMed] [Google Scholar]
- 30.Kumanogoh, A., T. Shikina, K. Suzuki, S. Uematsu, K. Yukawa, S. Kashiwamura, H. Tsutsui, M. Yamamoto, H. Takamatsu, E.P. Ko-Mitamura, et al. 2005. Nonredundant roles of Sema4A in the immune system: defective T cell priming and Th1/Th2 regulation in Sema4A-deficient mice. Immunity. 22:305–316. [DOI] [PubMed] [Google Scholar]
- 31.Rucker, P., F.M. Torti, and S.V. Torti. 1997. Recombinant ferritin: modulation of subunit stoichiometry in bacterial expression systems. Protein Eng. 10:967–973. [DOI] [PubMed] [Google Scholar]
- 32.Moss, D., A.R. Hibbs, D. Stenzel, L.W. Powell, and J.W. Halliday. 1994. The endocytic pathway for H-ferritin established in live MOLT-4 cells by laser scanning confocal microscopy. Br. J. Haematol. 88:746–753. [DOI] [PubMed] [Google Scholar]
- 33.Moss, D., S. Fargion, A.L. Fracanzani, S. Levi, M.D. Cappellini, P. Arosio, L.W. Powell, and J.W. Halliday. 1992. Functional roles of the ferritin receptors of human liver, hepatoma, lymphoid and erythroid cells. J. Inorg. Biochem. 47:219–227. [DOI] [PubMed] [Google Scholar]
- 34.Ramm, G.A., R.S. Britton, R. O'Neill, and B.R. Bacon. 1994. Identification and characterization of a receptor for tissue ferritin on activated rat lipocytes. J. Clin. Invest. 94:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gelvan, D., E. Fibach, E.G. Meyron-Holtz, and A.M. Konijn. 1996. Ferritin uptake by human erythroid precursors is a regulated iron uptake pathway. Blood. 88:3200–3207. [PubMed] [Google Scholar]
- 36.Meyron-Holtz, E.G., B. Vaisman, Z.I. Cabantchik, E. Fibach, T.A. Rouault, C. Hershko, and A.M. Konijn. 1999. Regulation of intracellular iron metabolism in human erythroid precursors by internalized extracellular ferritin. Blood. 94:3205–3211. [PubMed] [Google Scholar]
- 37.Hulet, S.W., S. Powers, and J.R. Connor. 1999. Distribution of transferrin and ferritin binding in normal and multiple sclerotic human brains. J. Neurol. Sci. 165:48–55. [DOI] [PubMed] [Google Scholar]
- 38.Hulet, S.W., S. Menzies, and J.R. Connor. 2002. Ferritin binding in the developing mouse brain follows a pattern similar to myelination and is unaffected by the jimpy mutation. Dev. Neurosci. 24:208–213. [DOI] [PubMed] [Google Scholar]
- 39.Liao, Q.K., P.A. Kong, J. Gao, F.Y. Li, and Z.M. Qian. 2001. Expression of ferritin receptor in placental microvilli membrane in pregnant women with different iron status at mid-term gestation. Eur. J. Clin. Nutr. 55:651–656. [DOI] [PubMed] [Google Scholar]
- 40.Adams, P.C., L.W. Powell, and J.W. Halliday. 1988. Isolation of a human hepatic ferritin receptor. Hepatology. 8:719–721. [DOI] [PubMed] [Google Scholar]
- 41.Anderson, G.J., W.P. Faulk, P. Arosio, D. Moss, L.W. Powell, and J.W. Halliday. 1989. Identification of H- and L-ferritin subunit binding sites on human T and B lymphoid cells. Br. J. Haematol. 73:260–264. [DOI] [PubMed] [Google Scholar]
- 42.Miller, L.L., S.C. Miller, S.V. Torti, Y. Tsuji, and F.M. Torti. 1991. Iron-independent induction of ferritin H chain by tumor necrosis factor. Proc. Natl. Acad. Sci. USA. 88:4946–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei, Y., S.C. Miller, Y. Tsuji, S.V. Torti, and F.M. Torti. 1990. Interleukin 1 induces ferritin heavy chain in human muscle cells. Biochem. Biophys. Res. Commun. 169:289–296. [DOI] [PubMed] [Google Scholar]
- 44.Recalcati, S., D. Taramelli, D. Conte, and G. Cairo. 1998. Nitric oxide-mediated induction of ferritin synthesis in J774 macrophages by inflammatory cytokines: role of selective iron regulatory protein-2 downregulation. Blood. 91:1059–1066. [PubMed] [Google Scholar]
- 45.Kim, S., and P. Ponka. 2000. Effects of interferon-gamma and lipopolysaccharide on macrophage iron metabolism are mediated by nitric oxide-induced degradation of iron regulatory protein 2. J. Biol. Chem. 275:6220–6226. [DOI] [PubMed] [Google Scholar]
- 46.Wu, H., D.A. Windmiller, L. Wang, and J.M. Backer. 2003. YXXM motifs in the PDGFbeta receptor serve dual roles as PI 3-kinase binding motifs and tyrosine-based endocytic sorting signals. J Biol Chem. 278:40425–40428 [erratum published in 278:45040]. [DOI] [PubMed]
- 47.Zuyderhoudt, F.M., P. Vos, G.G. Jorning, and J. Van Gool. 1985. Ferritin in liver, plasma and bile of the iron-loaded rat. Biochim. Biophys. Acta. 838:381–386. [DOI] [PubMed] [Google Scholar]
- 48.Cleton, M.I., J.W. Sindram, L.H. Rademakers, F.M. Zuyderhoudt, W.C. De Bruijn, and J.J. Marx. 1986. Ultrastructural evidence for the presence of ferritin-iron in the biliary system of patients with iron overload. Hepatology. 6:30–35. [DOI] [PubMed] [Google Scholar]
- 49.Yang, D.C., F. Wang, R.L. Elliott, and J.F. Head. 2001. Expression of transferrin receptor and ferritin H-chain mRNA are associated with clinical and histopathological prognostic indicators in breast cancer. Anticancer Res. 21:541–549. [PubMed] [Google Scholar]
- 50.Tripathi, P.K., and S.K. Chatterjee. 1996. Elevated expression of ferritin H-chain mRNA in metastatic ovarian tumor. Cancer Invest. 14:518–526. [DOI] [PubMed] [Google Scholar]
- 51.Gray, C.P., P. Arosio, and P. Hersey. 2003. Association of increased levels of heavy-chain ferritin with increased CD4+ CD25+ regulatory T-cell levels in patients with melanoma. Clin. Cancer Res. 9:2551–2559. [PubMed] [Google Scholar]
- 52.Gray, C.P., P. Arosio, and P. Hersey. 2002. Heavy chain ferritin activates regulatory T cells by induction of changes in dendritic cells. Blood. 99:3326–3334. [DOI] [PubMed] [Google Scholar]
- 53.Wu, C.G., M. Groenink, A. Bosma, P.H. Reitsma, S.J. van Deventer, and R.A. Chamuleau. 1997. Rat ferritin-H: cDNA cloning, differential expression and localization during hepatocarcinogenesis. Carcinogenesis. 18:47–52. [DOI] [PubMed] [Google Scholar]
- 54.Chakravarti, S., C.A. Sabatos, S. Xiao, Z. Illes, E.K. Cha, R.A. Sobel, X.X. Zheng, T.B. Strom, and V.K. Kuchroo. 2005. Tim-2 regulates T helper type 2 responses and autoimmunity. J. Exp. Med. 202:437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen, C.D., K.M. Ansel, C. Low, R. Lesley, H. Tamamura, N. Fujii, and J.G. Cyster. 2004. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 5:943–952. [DOI] [PubMed] [Google Scholar]