Abstract
The mechanisms by which the hepatitis C virus (HCV) establishes persistence are not yet fully understood. Previous chimpanzee and now human studies suggest that mutations within MHC class I–restricted HCV epitopes might contribute to viral escape from cytotoxic T lymphocyte (CTL) responses. However, there are several outstanding questions regarding the role of escape mutations in viral persistence and their fate in the absence of immune selection pressure.
The hepatitis C virus is a small positive-stranded RNA virus within the Flaviviridae family that persists in ∼70% of infected individuals, considerably increasing their risk of developing cirrhosis and hepatocellular carcinoma. With an estimated 170 million infected individuals worldwide, HCV presents a major global health challenge (1). Studies of chimpanzees, the only nonhuman HCV host, have indicated that escape mutations in MHC class I–restricted epitopes may play a role in evasion of the antiviral CTL response (2, 3). In this issue, three reports demonstrate that CTL escape mutations also occur in human HCV infection (4–6) and reinforce the general relevance of this immune evasion mechanism to persistence of RNA viruses in humans.
Escape mutations in RNA viruses: precedents for HCV infection
The majority of RNA viruses produce RNA polymerases that lack proofreading activity, and thus encode viral genomes containing random base mutations. In the presence of immune selection pressure exerted by CTLs against wild-type virus, this genomic diversity could facilitate preferential expansion of mutant progeny encoding altered epitopes that evade recognition by effector T cells. The principle of “escape mutations” in MHC class I–restricted epitopes was first demonstrated in the model of murine lymphocytic choriomeningitis viral infection (7). However, this finding remained controversial, as escape was forced by the presence of relatively high affinity T cell receptor (TCR) transgenic T cells, and resultant mutations did not enable immune evasion from a broader epitope-specific response by nontransgenic CTLs. The relevance of this mechanism was ultimately established in studies of HIV and simian immunodeficiency virus (SIV) infections, where immune selection pressure by CD8+ T cells results in development of escape mutations (8, 9), particularly during the acute phase of infection (10, 11). The importance of escape mutations in immune evasion by these viruses has been confirmed in studies demonstrating that mutations in immunodominant epitopes are associated with progression to AIDS (12), with escape and subsequent disease progression after transfer of an autologous CTL clone (13), and with escape from partial control afforded by prior vaccination (14).
CTL escape in the chimpanzee model of HCV infection
Virus-specific CD8+ T cells are unquestionably important in the outcome of HCV infection because the onset of this response is temporally correlated with control of acute phase viremia, and antibody-mediated depletion of the CD8+ T cell subset prolongs viral replication in chimpanzees (1). HCV replication is directed by the error-prone RNA-dependent RNA polymerase encoded by the viral NS5b gene, which due to its propensity to introduce mutations into the viral genome might provide the same selective advantage enjoyed by HIV and SIV. This mechanism of immune escape may contribute to the remarkable ability of HCV to persist in infected individuals.
Initial investigations of HCV CTL mutational escape were performed in the chimpanzee. The major advantage of this model is that the sequence of the infecting inoculum is known, allowing for accurate analysis of viral evolution. The initial description of CTL escape in a single epitope was made relatively early after the discovery of HCV (2), but statistical proof that CD8+ T cells select for HCV escape mutations has only recently become available. In a study of three chimpanzees that developed persistent infection, mutations abrogating CTL function were described in multiple regions of the viral genome encoding known epitopes, and were largely confined to animals expressing the appropriate restricting MHC class I allele or a closely related subtype (3). The ratio of nonsynonymous base substitution (which changed the amino acid encoded) to synonymous base substitution (which left encoded amino acids unchanged) was higher in regions encoding these epitopes than in flanking sequences, consistent with Darwinian selection pressure (3). Thus, the limited data available from this sole animal model has supported the supposition that CTL escape mutations occur during the course of HCV infection.
CTL escape mutation in human HCV infection
Until very recently, investigation of escape mutation in human HCV infections has been limited by practical considerations. As acute hepatitis C is often cryptic, patients usually do not present until late in infection. Study of viral evolution throughout the course of infection has hence proved extremely difficult. Furthermore, as the genomic sequence of the infecting inoculum is rarely known, interpretation of sequences obtained from circulating quasispecies in later phases of infection is problematic. Comparison to published prototypical HCV sequences partially circumvents these limitations, and this approach has provided preliminary indications that CTL escape mutations occur during human HCV infection (15).
Proof of the escape hypothesis would ideally require comparisons between the evolving viruses and the infectious inoculum, and not external reference sequences of published viruses. Much effort has therefore recently been directed to the surveillance of populations at high risk of HCV infection, including intravenous drug users, health care workers suffering needle stick injuries, and patients undergoing medical procedures. This allows prospective monitoring of HCV evolution, analysis that has to date been unavailable. In addition, cohorts of individuals infected by a single source of HCV of known sequence have also been studied. Such analyses allow definitive assessment of changes within the viral genome, which are critical to determining the role of immune selection pressure in viral evolution. Thus, in a recent publication in the Journal of Experimental Medicine, Timm et al. studied the evolution of an immunodominant HLA-B*08–restricted NS3 epitope during acute HCV infection in two HLA-B*08–positive patients, one infected by a needle stick injury, and the other by an undetermined route (16). CTL-mediated responses to this epitope were followed by the development of escape mutations in both individuals (16).
In this issue, three additional studies complement and extend this observation (4–6). These manuscripts in toto constitute a critical mass of evidence for CTL escape mutations in human HCV infection. Tester et al. followed two individuals acutely infected from a single source; in the recipient who did not spontaneously resolve infection, escape mutation in an immunodominant epitope was observed (6). Cox et al. analyzed the evolution of HCV by partial genome sequencing in eight acutely infected individuals, defining escape mutations in multiple CTL epitopes (4). In the third study, Ray et al. used the unique approach of comparing the sequences of viruses from 22 humans with chronic hepatitis C with the sequence of the single common virus that initiated these infections ∼20 year earlier (5). The expression of HLA-B*07, HLA-B*35, or HLA-B*37 alleles were found to be linked to the presence of mutations in epitopes presented by these alleles, indicating a likely role for CTL-mediated pressure in driving viral evolution (5).
Mechanisms of CTL escape
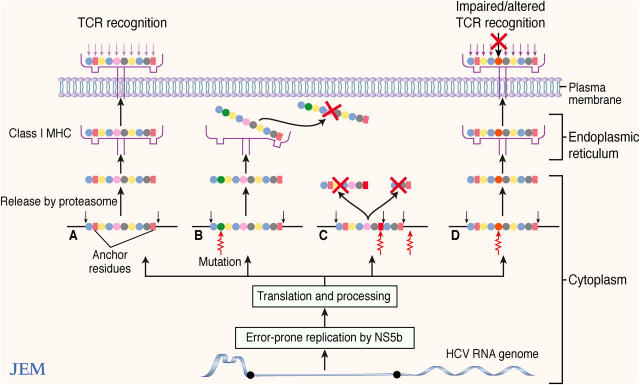
Thus, as with other highly mutable RNA viruses, escape mutations in MHC class I–restricted epitopes are a feature of HCV infection and can diminish CTL responses via several mechanisms. Most recently it has been demonstrated that amino acid substitutions within or adjacent to CTL epitopes can alter proteosomal processing causing epitope destruction before transport to the endoplasmic reticulum for MHC binding (16–18). A loss of epitope phenotype can also occur when amino acid anchor residues required for MHC binding are changed (3, 15). CTL recognition of epitopes may also be diminished despite apparent conservation of peptide MHC binding (15), perhaps due to reduced TCR recognition of the neo-epitope–MHC complex. Such mutated peptide–MHC complexes may alternatively antagonize responses to the wild-type epitope (15, 19, 20). These potential pathways of CTL escape are summarized in Fig. 1.
Figure 1.
Mechanisms of CTL escape after mutation in cognate epitopes. (A) Wild-type peptide is correctly processed, binds to MHC, and the resulting complex is recognized by the relevant TCR, triggering a CTL response. (B) Mutations in anchoring residues may lead to dissociation of the MHC–peptide complex. (C) Mutations in the epitope or in flanking regions can alter proteosomal processing, leading to destruction of the epitope. (D) Mutated epitopes may bind MHC; however, TCR recognition may be reduced or possibly altered, leading to antagonism against responses to the wild-type peptide.
It should be noted that CTL escape has not been observed in all studied epitopes (3, 5, 21). It is unclear why HCV-specific CTLs directed against these unaltered epitopes do not mediate viral clearance in persistently infected individuals, but recent studies have indicated that their antiviral functions may be impaired in chronic infection (1).
The factors constraining the emergence of escape mutations are incompletely understood. Even in chronically HCV-infected individuals, CD8+ T cell responses can sometimes be directed against a wide range of MHC class I–restricted epitopes (1). Indeed, one major criticism of the escape hypothesis has been the assumption that viral persistence would require almost simultaneous mutations within multiple epitopes targeted by CTLs. However, it is possible that the spontaneous loss of HCV-specific CD4+ T cells that is consistently observed in subjects who develop chronic HCV infection (1) could reduce the effectiveness of CD8+ T cells to a point where they select for viral escape variants, rather than containing infection. Direct support for this concept was obtained by antibody-mediated CD4+ T cell depletion of immune chimpanzees that had resolved prior HCV infections (22). Rechallenge of animals lacking CD4+ T cells with the same HCV strain resulted in viral persistence. Importantly, the frequency and breadth of the memory CD8+ T cell response was reduced in this setting, and persistent viruses developed escape mutations in multiple MHC class I–restricted epitopes (22). Consistent with the importance of CD4+ T cell responses in containing viral escape, in the study of Tester et al., mutational escape was associated with undetectable HCV-specific CD4+ T cell responses, whereas in the subject who resolved infection without mutational escape, broad sustained CD4+ T cell responses were demonstrated (6). In addition, a study of HCV-specific CD8+ T cell responses in HCV-infected chimpanzees indicated that the diversity of clonal TCR usage might also be a factor in the development of escape mutations (23). In a majority of cases, escape mutation was associated with reduced CTL clonal TCR diversity in comparison to epitopes in which escape mutations were not observed or those analyzed in chimpanzees that resolved infection (23). Similar analysis in SIV infection has also indicated a link between diverse clonotypic TCR repertoires and lack of mutation in the targeted epitope, whereas a more highly conserved clonotypic TCR repertoire was associated with viral escape (24).
Viral reversion and the meaning of consensus sequences
It is likely that the emergence of escape mutations is at least in part governed by the “fitness cost” imposed on HCV replication. However, although impairment of propagation may constrain the emergence of escape mutations in some structurally critical residues (25), evidence from studies in HIV and SIV indicates that compensatory viral mutations can occur, both within epitopes and in other regions, which allow for the generation of escape mutations in highly constrained sites (26–28). Consistent with mutational escape exacting fitness cost, recent studies of a limited number of HIV and SIV epitopes have indicated that reversion of epitopes to wild-type sequence can occur after transmission of mutated virus into a host that does not express the restricting MHC class I allele (29–31).
Although fitness cost is likely an important factor shaping HCV evolution, how individual amino acid substitutions alter replication is not known. In the acute infection cohort of Cox et al., sequences from the subjects' HCV genomes external to known CTL epitopes were compared with a corresponding HCV consensus sequence, and a tendency to mutate toward viral consensus was found in these regions (4). Similarly, in the study by Ray et al., mutation of the HCV genomes toward consensus was noted in regions outside known CTL epitopes. Additionally, in HLA-A*01- and B*08-negative individuals, absence of these alleles was associated with evolution toward consensus within epitopes restricted by these MHC molecules (5). These results have been interpreted as indicating that such mutations occur due to viral reversion to a more fit ancestral state (4, 5). Although the available data supports this model, conclusions are tempered by the absence of MHC haplotype information for the donor of the HCV inocula that would provide additional insight into the selective forces acting on the HCV genomes before and after transmission.
The concept that consensus sequences represent genomes that are the most antigenic as well as replicatively fit is being challenged by studies of HIV evolution and CTL responses. Recent population-based studies have indicated that genetic polymorphisms within certain HIV CTL epitopes are significantly associated with expression of the restricting MHC class I alleles (32), indicating that common HLA alleles may leave a “footprint” on this virus, with loss of epitopes restricted by highly prevalent HLA class I alleles (33). Analysis of a limited set of HIV and SIV CTL escape mutations has indicated that for some mutations maintenance of a variant epitope sequence may be largely confined to subjects expressing the appropriate restricting MHC molecule (29–31). On the other hand, recent data indicates that at least some HIV CTL escape mutations persist within the viral genome in individuals who do not express the restricting MHC class I molecule (29, 34). Indeed, such escape mutations may even become prevalent enough to enter the consensus sequence (34), indicating that for these epitopes, marked fitness cost is not exacted by viral escape, and reversion to a more immunogenic ancestral state is not automatic upon passage to a host in which immune selection pressure is absent. Although similar evidence is not available for HCV infection, it is interesting to note that a mutation reported in an HLA-A*02–restricted epitope in which CTL escape was mediated by altered proteosomal processing was not observed at a statistically significant higher frequency among the HLA-A*02–positive population in comparison to the control HLA-A*02–negative population studied (17). It is tempting to speculate that this phenomenon might be due to low fitness cost associated with this particular mutation, thus allowing persistence of the variant sequence in the absence of immune selection pressure.
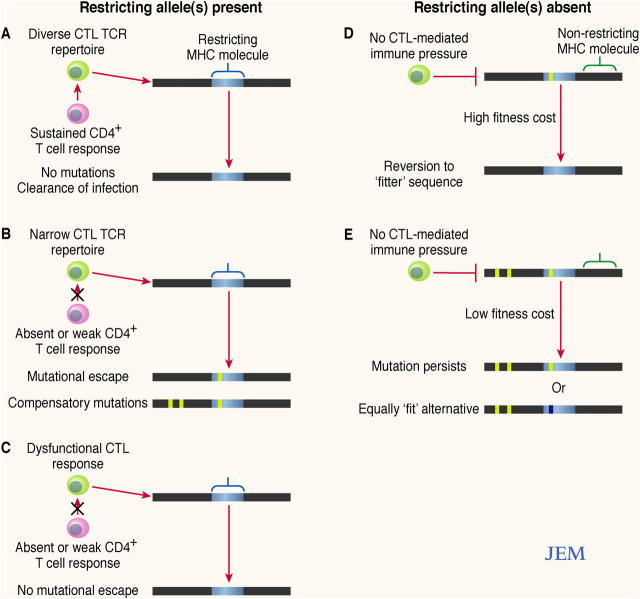
Thus, due to likely interplay between the opposing forces of immune selection pressure and viral fitness cost, a variety of outcomes are possible upon initial infection with HCV, or after subsequent transmission of the virus to a recipient in whom the initial MHC class I alleles are not expressed. A model indicating several potential scenarios of viral evolution is illustrated in Fig. 2.
Figure 2.
Evolution of HCV epitopes after infection. (A) Where a restricting allele is present, immune factors such as a sustained CD4+ T cell response and diverse clonal CTL TCR repertoire may constrain the development of escape mutations; this may be association with viral clearance. (B) Where the CD4+ T cell response fails and/or the clonal CTL TCR repertoire is narrow, escape mutations may emerge. Development of an escape mutation may require additional compensatory mutations. (C) If CTL responses are weak or absent, escape mutations may not develop. (D) In the absence of the restricting allele, where there is high fitness cost associated with the presence of an escape mutation, reversion to wild-type sequence is likely to occur because of minimal immune selection pressure. (E) When there is low associated fitness cost or well developed compensatory mutations in the absence of immune selection pressure, the mutated sequence may persist, or perhaps might mutate to an equally fit alternative.
Are CTL escape mutations in HCV important for viral persistence?
Although it is clear that CTL escape mutations occur in HCV genomes, the relevance of this mechanism to viral persistence is an open question. Mutations usually occur within the first 3–4 months of infection (3, 4), consistent with the delayed generation of CD8+ T cell responses that is an unusual feature of acute HCV infections (1). Such observations are compatible with release from early immune selection pressure as viral escape is established, and perhaps suggest a role for CTL escape mutations in the genesis of chronic infection. However, as observed in SIV infection (9), the rate at which escape mutations occur during HCV infection can be highly variable, occurring between 10 weeks to 2 years postinfection in chimpanzees (3). It is notable that once established, escape mutations in HCV tend to remain fixed within the circulating quasispecies of an individual (3, 15), perhaps driven by persisting CD8+ T cell responses, which are often concentrated within the liver (1). The cause of this variability in the timing of viral escape remains to be determined and may well be governed largely by the stochastic nature of viral evolution and a requirement for the development of compensatory mutations for viruses containing certain escape mutations to remain viable. However, differences in the kinetics of the immune response might also be involved in the variable dynamics of viral evolution. Indeed, future detailed prospective study of factors such as the temporal relationship between the emergence of escape mutations and breadth and persistence of the HCV-specific CD4+ T cell response, already demonstrated as important in disease resolution and containment of mutational escape (1, 22), might not only further elucidate a role for these cells in mutational escape, but also the role of mutational escape in viral persistence.
Concluding remarks
Although recent data confirms the generation of escape mutations in HCV, it is clear that this field is in its infancy. As is becoming increasingly apparent from the immunodeficiency virus literature, studies comparing circulating viral genomes to consensus sequences are extremely difficult to interpret. Further detailed prospective studies of acute and evolving HCV infection are required to delineate the role of escape mutations in HCV persistence, as well as to define both the immunological and virological factors governing viral evolution. In particular, careful dissection of these factors will be critical to the design of any effective CTL-based vaccine.
Acknowledgments
Our research was supported by Public Health Service grants RO1 A147367 and U19 AI48231 to C.M. Walker. D.G. Bowen was supported by a CJ Martin Fellowship from the National Health and Medical Research Council of Australia and an AstraZeneca Fellowship in Medical Research from the Royal Australasian College of Physicians.
References
- 1.Shoukry, N.H., A.G. Cawthon, and C.M. Walker. 2004. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu. Rev. Microbiol. 58:391–424. [DOI] [PubMed] [Google Scholar]
- 2.Weiner, A., A.L. Erickson, J. Kansopon, K. Crawford, E. Muchmore, A.L. Hughes, M. Houghton, and C.M. Walker. 1995. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc. Natl. Acad. Sci. USA. 92:2755–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erickson, A.L., Y. Kimura, S. Igarashi, J. Eichelberger, M. Houghton, J. Sidney, D. McKinney, A. Sette, A.L. Hughes, and C.M. Walker. 2001. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity. 15:883–895. [DOI] [PubMed] [Google Scholar]
- 4.Cox, A.L., T. Mosbruger, Q. Mao, Z. Liu, X.-H. Wang, H.-C. Yang, J. Sidney, A. Sette, D. Pardoll, D.L. Thomas, and S.C. Ray. Cellular immune selection with hepatitis C virus persistence in humans. J. Exp. Med. 201:1741–1752. [DOI] [PMC free article] [PubMed]
- 5.Ray, S.C., L. Fanning, X.-H. Wang, D.M. Netski, E. Kenny-Walsh, and D.L. Thomas. Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J. Exp. Med. 201:1753–1759. [DOI] [PMC free article] [PubMed]
- 6.Tester, I., S. Smyk-Pearson, P. Wang, A. Wertheimer, E. Yao, D.M. Lewinsohn, J.E. Tavis, and H.R. Rosen. Immune evasion versus recovery after acute hepatitis C virus infection from a shared source. J. Exp. Med. 201:1725–1731. [DOI] [PMC free article] [PubMed]
- 7.Pircher, H., D. Moskophidis, U. Rohrer, K. Burki, H. Hengartner, and R.M. Zinkernagel. 1990. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature. 346:629–633. [DOI] [PubMed] [Google Scholar]
- 8.Phillips, R.E., S. Rowland-Jones, D.F. Nixon, F.M. Gotch, J.P. Edwards, A.O. Ogunlesi, J.G. Elvin, J.A. Rothbard, C.R. Bangham, C.R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 354:453–459. [DOI] [PubMed] [Google Scholar]
- 9.Evans, D.T., D.H. O'Connor, P. Jing, J.L. Dzuris, J. Sidney, J. da Silva, T.M. Allen, H. Horton, J.E. Venham, R.A. Rudersdorf, et al. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270–1276. [DOI] [PubMed] [Google Scholar]
- 10.Borrow, P., H. Lewicki, X. Wei, M.S. Horwitz, N. Peffer, H. Meyers, J.A. Nelson, J.E. Gairin, B.H. Hahn, M.B. Oldstone, and G.M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205–211. [DOI] [PubMed] [Google Scholar]
- 11.Allen, T.M., D.H. O'Connor, P. Jing, J.L. Dzuris, B.R. Mothe, T.U. Vogel, E. Dunphy, M.E. Liebl, C. Emerson, N. Wilson, et al. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 407:386–390. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P.J., R.E. Phillips, R.A. Colbert, S. McAdam, G. Ogg, M.A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217. [DOI] [PubMed] [Google Scholar]
- 13.Koenig, S., A.J. Conley, Y.A. Brewah, G.M. Jones, S. Leath, L.J. Boots, V. Davey, G. Pantaleo, J.F. Demarest, C. Carter, et al. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330–336. [DOI] [PubMed] [Google Scholar]
- 14.Barouch, D.H., J. Kunstman, M.J. Kuroda, J.E. Schmitz, S. Santra, F.W. Peyerl, G.R. Krivulka, K. Beaudry, M.A. Lifton, D.A. Gorgone, et al. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature. 415:335–339. [DOI] [PubMed] [Google Scholar]
- 15.Chang, K.M., B. Rehermann, J.G. McHutchison, C. Pasquinelli, S. Southwood, A. Sette, and F.V. Chisari. 1997. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J. Clin. Invest. 100:2376–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timm, J., G.M. Lauer, D.G. Kavanagh, I. Sheridan, A.Y. Kim, M. Lucas, T. Pillay, K. Ouchi, L.L. Reyor, J.S. Zur Wiesch, et al. 2004. CD8 epitope escape and reversion in acute HCV infection. J. Exp. Med. 200:1593–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seifert, U., H. Liermann, V. Racanelli, A. Halenius, M. Wiese, H. Wedemeyer, T. Ruppert, K. Rispeter, P. Henklein, A. Sijts, et al. 2004. Hepatitis C virus mutation affects proteasomal epitope processing. J. Clin. Invest. 114:250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura, Y., T. Gushima, S. Rawale, P. Kaumaya, and C.M. Walker. 2005. Escape mutations alter proteasome processing of major histocompatibility complex class I-restricted epitopes in persistent hepatitis C virus infection. J. Virol. 79:4870–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko, T., T. Moriyama, K. Udaka, K. Hiroishi, H. Kita, H. Okamoto, H. Yagita, K. Okumura, and M. Imawari. 1997. Impaired induction of cytotoxic T lymphocytes by antagonism of a weak agonist borne by a variant hepatitis C virus epitope. Eur. J. Immunol. 27:1782–1787. [DOI] [PubMed] [Google Scholar]
- 20.Tsai, S.L., Y.M. Chen, M.H. Chen, C.Y. Huang, I.S. Sheen, C.T. Yeh, J.H. Huang, G.C. Kuo, and Y.F. Liaw. 1998. Hepatitis C virus variants circumventing cytotoxic T lymphocyte activity as a mechanism of chronicity. Gastroenterology. 115:954–965. [DOI] [PubMed] [Google Scholar]
- 21.Urbani, S., B. Amadei, P. Fisicaro, M. Pilli, G. Missale, A. Bertoletti, and C. Ferrari. 2005. Heterologous T cell immunity in severe hepatitis C virus infection. J. Exp. Med. 201:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grakoui, A., N.H. Shoukry, D.J. Woollard, J.H. Han, H.L. Hanson, J. Ghrayeb, K.K. Murthy, C.M. Rice, and C.M. Walker. 2003. HCV persistence and immune evasion in the absence of memory T cell help. Science. 302:659–662. [DOI] [PubMed] [Google Scholar]
- 23.Meyer-Olson, D., N.H. Shoukry, K.W. Brady, H. Kim, D.P. Olson, K. Hartman, A.K. Shintani, C.M. Walker, and S.A. Kalams. 2004. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J. Exp. Med. 200:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price, D.A., S.M. West, M.R. Betts, L.E. Ruff, J.M. Brenchley, D.R. Ambrozak, Y. Edghill-Smith, M.J. Kuroda, D. Bogdan, K. Kunstman, et al. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 21:793–803. [DOI] [PubMed] [Google Scholar]
- 25.Peyerl, F.W., H.S. Bazick, M.H. Newberg, D.H. Barouch, J. Sodroski, and N.L. Letvin. 2004. Fitness costs limit viral escape from cytotoxic T lymphocytes at a structurally constrained epitope. J. Virol. 78:13901–13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelleher, A.D., C. Long, E.C. Holmes, R.L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, et al. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27–restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peyerl, F.W., D.H. Barouch, W.W. Yeh, H.S. Bazick, J. Kunstman, K.J. Kunstman, S.M. Wolinsky, and N.L. Letvin. 2003. Simian-human immunodeficiency virus escape from cytotoxic T-lymphocyte recognition at a structurally constrained epitope. J. Virol. 77:12572–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedrich, T.C., C.A. Frye, L.J. Yant, D.H. O'Connor, N.A. Kriewaldt, M. Benson, L. Vojnov, E.J. Dodds, C. Cullen, R. Rudersdorf, et al. 2004. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J. Virol. 78:2581–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie, A.J., K.J. Pfafferott, P. Chetty, R. Draenert, M.M. Addo, M. Feeney, Y. Tang, E.C. Holmes, T. Allen, J.G. Prado, et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289. [DOI] [PubMed] [Google Scholar]
- 30.Friedrich, T.C., E.J. Dodds, L.J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D.T. Evans, R.C. Desrosiers, B.R. Mothe, J. Sidney, et al. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10:275–281. [DOI] [PubMed] [Google Scholar]
- 31.Barouch, D.H., J. Powers, D.M. Truitt, M.G. Kishko, J.C. Arthur, F.W. Peyerl, M.J. Kuroda, D.A. Gorgone, M.A. Lifton, C.I. Lord, et al. 2005. Dynamic immune responses maintain cytotoxic T lymphocyte epitope mutations in transmitted simian immunodeficiency virus variants. Nat. Immunol. 6:247–252. [DOI] [PubMed] [Google Scholar]
- 32.Moore, C.B., M. John, I.R. James, F.T. Christiansen, C.S. Witt, and S.A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 296:1439–1443. [DOI] [PubMed] [Google Scholar]
- 33.Altfeld, M., T.M. Allen, E.T. Kalife, N. Frahm, M.M. Addo, B.R. Mothe, A. Rathod, L.L. Reyor, J. Harlow, X.G. Yu, et al. 2005. The majority of currently circulating human immunodeficiency virus type 1 clade B viruses fail to prime cytotoxic T-lymphocyte responses against an otherwise immunodominant HLA-A2-restricted epitope: implications for vaccine design. J. Virol. 79:5000–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leslie, A., D. Kavanagh, I. Honeyborne, K. Pfafferott, C. Edwards, T. Pillay, L. Hilton, C. Thobakgale, D. Ramduth, R. Draenert, et al. 2005. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J. Exp. Med. 201:891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]