Abstract
Transcription factors of the nuclear factor (NF)-κB/Rel family translocate into the nucleus upon degradation of the IκBs. Postinduction repression of NF-κB activity depends on NF-κB–regulated resynthesis of IκBα, which dissociates NF-κB from DNA and exports it to the cytosol. We found that after activation, p65/RelA is degraded by the proteasome in the nucleus and in a DNA binding–dependent manner. If proteasome activity is blocked, NF-κB is not promptly removed from some target genes in spite of IκBα resynthesis and sustained transcription occurs. These results indicate that proteasomal degradation of p65/RelA does not merely regulate its stability and abundance, but also actively promotes transcriptional termination.
Keywords: NF-κB, Rel family, proteasome, transcriptional regulation
Introduction
Transcriptional induction of a large number of inflammatory genes, immune response genes, and genes promoting cell survival of both normal and cancer cells is regulated by the five transcription factors (TFs) of the nuclear factor (NF)-κB/Rel family, namely p65/RelA, cRel, RelB, p50, and p52 (1–3). Most homodimers and heterodimers generated by the NF-κB/Rel proteins are found in the cytoplasm of unstimulated cells in complexes with three major inhibitory proteins collectively indicated as IκB, namely IκBα, IκBβ, and IκBɛ (4). p105 and p100 (the proteins from which p50 and p52 are generated through limited proteasomal processing) also contribute to cytoplasmic retention of NF-κB/Rel dimers. IκBs contain an NH2-terminal regulatory region that is phosphorylated in response to stimulation and COOH-terminal ankyrin repeats that mediate association with NF-κB dimers. When extracellular signals transduced from several receptors activate the IκB kinase complex, it phosphorylates the IκBs at two amino-terminal serines, thus targeting them for polyubiquitination by the βTrCP proteins and subsequent proteasomal degradation (5). IκB degradation allows NF-κB to enter the nucleus and bind target genes. In vitro, the complex between NF-κB and the κB site is extremely stable, with a dissociation constant below 10−11 M (6) and a half-life of ∼45 min. This stable complex can be rapidly dissociated by the addition of IκBα, which reduces its half-life to 3 min (7). Several pieces of evidence indicate that IκBα is a master terminator of the NF-κB response. First, it is rapidly resynthesized in an NF-κB–dependent manner (8–10). Second, it can enter the nucleus as a free, NF-κB–unbound protein (11). Finally, it can export NF-κB from the nucleus (11–13). Therefore, according to the current model of NF-κB response termination, after nuclear translocation, NF-κB remains stably bound to target genes until resynthesized IκBα enters the nucleus, dissociates it from DNA, and shuttles it back to the cytoplasm, thus restoring the initial steady state. Analysis of IκBα−/− cells confirmed that resynthesis of IκBα provides a strong negative feedback and a fast down-regulation of the NF-κB response (14, 15), thus allowing the rapid termination of NF-κB activity after a transient TNF-α stimulation. IκBβ can fully compensate for IκBα deficiency when knocked in the IκBα locus and placed under control of the IκBα promoter, which indicates that the irreplaceable role of IκBα in NF-κB response termination simply reflects its unique temporal expression pattern (16). The main physiological role of IκBβ and IκBɛ, as deduced from computer modeling applied to the analysis of gene-deficient cells, is to prevent oscillations of the NF-κB response during long-lasting activations (15). Additional roles of IκBɛ in nucleo-cytoplasmic shuttling of NF-κB proteins cannot be ruled out, although IκBɛ is markedly less efficient than IκBα in this regard (17).
In addition to the global down-regulation of NF-κB activity due to IκBα resynthesis, gene-specific mechanisms of transcriptional shut-off exist that allow individual genes to be regulated in a selective fashion and independently of the behavior of the bulk of nuclear NF-κB. These mechanisms include the recruitment to target genes of NF-κB–induced transcriptional repressors, such as Twist 1/2 (18), and the replacement of an active NF-κB dimer with a dimer showing no transcriptional activity in the context of that specific gene (19).
The experiments described here were designed with the aim of identifying additional mechanisms of NF-κB response termination. We found that proteasome-dependent degradation of nuclear p65/RelA is a major mechanism of NF-κB response termination in the absence of IκBα. In cells containing IκBα, proteasomal degradation of p65/RelA provides an essential contribution to a prompt shut-off of the response, thus indicating that proteasome and IκBα synergistically act to efficiently and promptly terminate transcription of NF-κB–dependent genes.
Materials and Methods
Antibodies and Reagents.
Anti-p65 (C20) was from Santa Cruz Biotechnology, Inc., anti-IκBα monoclonal was from Imgenex, anti-ubiquitin monoclonal antibody was from Zymed Laboratories, and the anti-Sug1 antiserum was from Affinity BioReagents, Inc. Anti-FLAG M2 was from Sigma-Aldrich. mTNF-α (R&D Systems) was used at a final concentration of 10 ng/ml.
Plasmids.
Human p65 was cloned in frame with an NH2-terminal FLAG epitope in a pCDNA3 (Invitrogen) derivative. The κB site binding-defective mutant of p65 was generated by mutagenesis using the QuikChange kit (Stratagene) and tagged at the NH2 terminus with either a FLAG or a green fluorescent protein (GFP) tag. The myc ubiquitin expression vector was from R. Kopito's lab (Stanford University, Stanford, CA).
Detection of Ubiquitin Conjugates.
10 mM N-ethylmaleimide (NEM) dissolved in ethanol was added to the culture medium 30 s before washing the cells in ice cold PBS containing 10 mM NEM. Cells were lysed in RIPA buffer containing 20 mM NEM.
Chromatin Immunoprecipitation (ChIP) Assays.
ChIP assays were performed as described previously (19). Sequences of promoter-specific primers and a detailed protocol are available upon request.
Results and Discussion
Proteasome-dependent Down-Regulation of the NF-κB Response in IκBα−/− Cells.
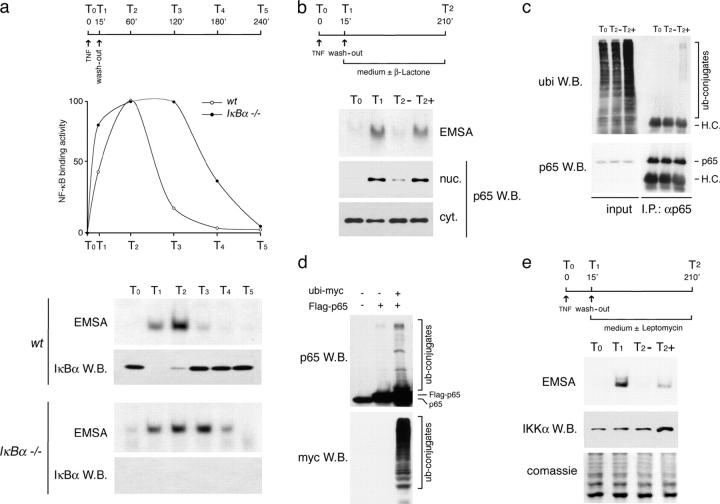
To identify IκBα-independent mechanisms of NF-κB response termination, we analyzed how the NF-κB response is terminated in the absence of IκBα. WT or IκBα−/− 3T3 cells were stimulated with a 15-min pulse of TNF-α, which induces the degradation of the IκBs, and a single wave of nuclear translocation of p50/p65, the most abundant NF-κB dimer in fibroblasts. Cells were then washed to remove TNF from the medium and terminate signaling. They were then placed in normal medium and returned to the incubator (Fig. 1 a). In agreement with published results (14, 15), in WT 3T3 cells NF-κB activity was strongly down-regulated concurrently with completion of IκBα resynthesis, whereas in IκBα−/− 3T3 cells the response was more sustained (Fig. 1 a). However, in spite of IκBα absence, in these cells nuclear NF-κB levels were also completely down-regulated in <4 h.
Figure 1.
Termination of the NF-κB response in IκBα-deficient cells. (A) Kinetics of NF-κB down-regulation in WT and IκBα−/− 3T3 cells. Cells were stimulated with TNF-α as indicated and analyzed for NF-κB binding activity by electrophoretic mobility shift assay (EMSA) using a canonical κB site as a probe (5′-AGTTGAGGGGACTTTCCCAGGC-3′). Data were quantified with a phosphorimager. Kinetics of IκBα degradation and resynthesis are also shown. (B) Proteasome inhibition prevents NF-κB down-regulation in IκBα−/− cells. After TNF-α washout, cells were incubated with β lactone and assayed by EMSA. An anti-p65 Western blot (W.B.) on cytoplasmic and nuclear extracts is shown. (C) Endogenous p65 is ubiquitinated after NF-κB activation. Extracts from IκBα−/− cells were immunoprecipitated with an anti-p65 antibody and blotted with an anti-ubiquitin monoclonal antibody. H.C., heavy chains. (D) Reconstitution of p65 ubiquitination by cotransfection of FLAG-p65 and myc-ubiquitin expression vectors in HEK-293T cells. Blots obtained from whole cell extracts were probed with an anti-p65 polyclonal antibody and an anti-myc monoclonal antibody. (E) IκBα−/− cells were stimulated with TNF for 15 min, washed, and incubated with 10 ng/ml LMB for an additional 3.15 h. EMSAs were performed on nuclear lysates. As a positive control for LMB effects, accumulation of IκB kinase complex α in the nuclear fraction of LMB-treated cells is shown.
When fused to the Gal4 DNA binding domain, the activation domains of p65 and of other TFs confer instability to the chimeric protein (20). Instability depends on proteasomal degradation of the fusion protein, which directly correlates with the potency of the activation domain fused to Gal4. Moreover, the domain(s) required for transcriptional activity often overlaps with those triggering degradation (21). Therefore, we tested if p65 is degraded by the proteasome and if degradation may account for the decay of NF-κB activity in IκBα−/− cells. IκBα−/− 3T3 cells were stimulated with TNF for 15 min, washed, and further incubated with vehicle or clasto-lactacystin β lactone, a rapid and selective inhibitor of the chymotryptic and tryptic-like activities of the proteasome. In the presence of β lactone, nearly all nuclear NF-κB activity and p65 protein were preserved at >3 h after TNF washout (Fig. 1 b). Conversely, the cytoplasmic NF-κB fraction did not show any obvious change in abundance induced by proteasome inhibition.
To determine if p65 is a direct target of the proteasome, we examined whether endogenous p65 is polyubiquitinated. To this aim, p65 was immunoprecipitated from cells stimulated as described above. A ladder of high molecular weight forms of p65 was selectively recognized by a monoclonal antibody to ubiquitin in β lactone–treated cells (Fig. 1 c). To further demonstrate that p65 is polyubiquitinated, we cotransfected HEK-293T cells with expression vectors encoding p65 and myc-tagged ubiquitin. In these conditions, p65 exceeds the endogenous IκBs and is constitutively nuclear. High molecular weight forms of p65 were strongly augmented by cotransfection of the ubiquitin expression vector (Fig. 1 d), thus demonstrating that p65 undergoes polyubiquitination in vivo. Inhibition of Crm1-dependent nuclear export by leptomycin B (LMB) after a pulse of TNF only minimally interfered with complete down-regulation of NF-κB nuclear levels (Fig. 1 e), which indicates that although a minor fraction of NF-κB is probably exported in an IκBα-independent manner and degraded in the cytoplasm, most p65 molecules are in fact degraded in the nucleus.
These results indicate that if NF-κB does not rapidly reassociate with resynthesized IκBα, it is polyubiquitinated and degraded by the proteasome. Assuming that ∼120,000 p65 molecules enter nucleus after stimulation (22), that ∼3.5 h are required for their complete degradation, and that the kinetics of degradation is linear, it can be roughly estimated that ∼500–600 molecules of p65 are degraded every minute.
p65 Ubiquitination Requires Sequence-specific Binding to κB Sites.
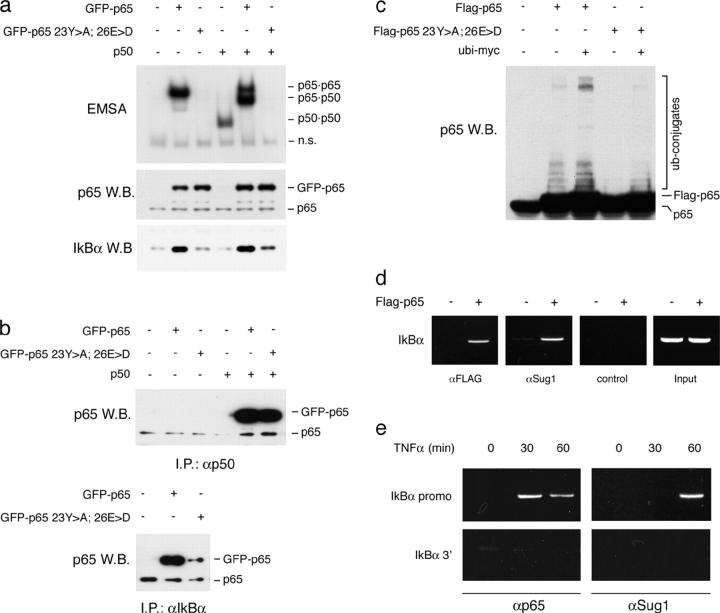
To investigate if sequence-specific DNA binding is required for p65 polyubiquitination, we generated a p65 mutant in which two residues essential for base-specific contacts, Tyr 23 and Glu 26 (corresponding to Tyr 36 and Glu 39 in mouse p65; reference 23), were mutated to Ala and Asp, respectively. The mutant p65 was virtually unable to bind canonical κB sites, both in homodimers and in heterodimers with p50 (Fig. 2 a), whereas it was able to bind p50 and IκBα (Fig. 2 b). The different amount of IκBα immunoprecipitated by the WT and the mutant p65 likely reflects its different abundance in cells overexpressing either of the two proteins. Indeed, although WT p65 increased expression of endogenous IκBα, the mutant was devoid of this activity (Fig. 2 a). The mutant p65 still retained a residual activity (∼15–20% of the WT protein) in luciferase assays (unpublished data). Polyubiquitination of the mutant p65 was much lower than that of the normal protein (Fig. 2 c), which indicates that polyubiquitination of p65 mainly occurs upon binding to specific target sites in the chromatin. However, from these experiments it is not possible to determine if p65 degradation is restricted to those molecules actively engaged in transcriptional activation.
Figure 2.
p65 ubiquitination requires sequence-specific binding to κB sites. (A) WT p65 and a p65 mutant bearing a double substitution in the Rel homology domain (23Y > A; 26E > D) were cloned with an NH2-terminal GFP tag and transfected in HEK-293 cells alone or with a p50 expression vector. Total lysates were made and assayed by EMSA using a canonical κB site as a probe. The GFP tag allows easy discrimination between p65 homodimers and p65/p50 heterodimers (n.s., nonspecific). An anti-p65 immunoblot shows the expression of endogenous p65 and transfected GFP-p65 in total lysates. Expression of IκBα is also shown. (B) Association of WT and mutant GFP-p65 with p50 and IκBα. 293T cells were transfected as indicated. Total cell extracts were immunoprecipitated with either an anti-p50 or an anti-IκBα antibody and then blotted with an anti-p65 antibody. (C) The κB site binding-deficient p65 mutant is not efficiently polyubiquitinated. HEK-293T were cotransfected with the indicated expression vectors. Whole cell extracts were assayed for the appearance of high molecular weight ubiquitinated forms of p65 by anti-p65 immunoblotting. (D) p65 induces recruitment of proteasome components to target genes. HEK-293 cells were transfected with empty vector or a FLAG-p65 expression vector. ChIP assays with an anti-FLAG antibody, an antibody against Sug1, or a control antibody were performed. Recruitment of FLAG-p65 and Sug1 to the endogenous IκBα gene promoter is shown. (E) Anti-p65 and anti-Sug1 ChIP assays on HEK-293 cells stimulated with TNF-α. Immunoprecipitated DNA was amplified with primers spanning the IκBα promoter or a region immediately downstream of the IκBα gene.
Recruitment of Proteasome Components to NF-κB Target Genes.
If polyubiquitination and proteasome-dependent degradation of p65 occur after recruitment to target genes, then the proteasome itself should be recruited to NF-κB–dependent promoters in a p65-dependent manner. To test this possibility, we transfected HEK-293 cells with a FLAG-p65 expression vector and performed a ChIP assay with an antibody recognizing the S8 component (Sug1) of the 19S proteasome complexes. p65 transfection induced a large increase in the association of Sug1 to the IκBα gene promoter, a typical NF-κB target (Fig. 2 d). Similarly, TNF-α stimulation of HEK-293 cells induced Sug1 recruitment to the IκBα promoter, but not to a region located immediately 3′ of the IκBα gene (Fig. 2 e), suggesting that NF-κB target genes may represent sites of proteasome-dependent degradation of p65.
Removal of NF-κB from Target Genes in IκBα−/− Cells Is Proteasome Dependent.
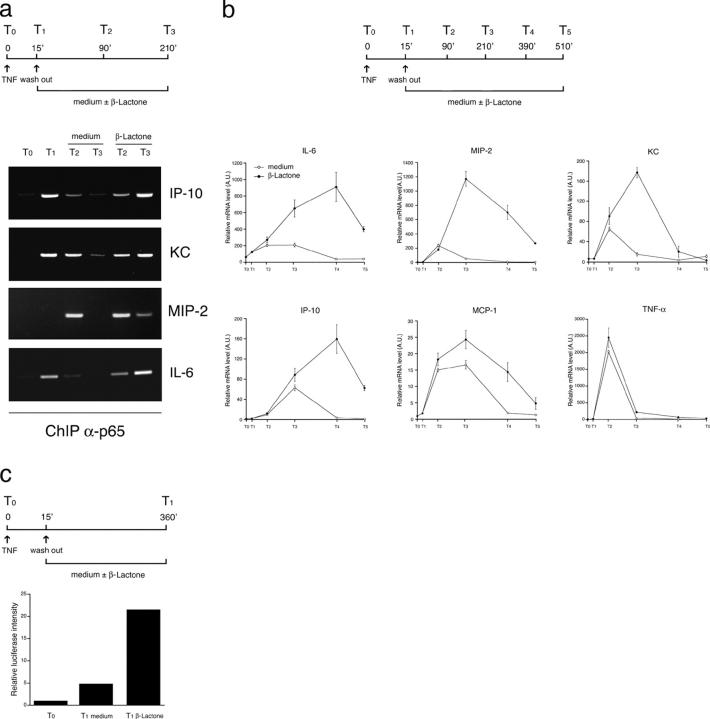
To determine if proteasomal degradation of p65 is required to remove it from target genes, we performed ChIP assays with an anti-p65 antibody. In IκBα−/− 3T3 cells, proteasome inhibition after a 15-min TNF-α treatment caused the persistence of p65 on the promoters of several validated p65 target genes (24), such as the chemokines KC, inducible protein (IP)-10, and macrophage inflammatory protein (MIP)-2, as well as IL-6 (Fig. 3 a). mRNA levels for these and several other NF-κB target genes tested, with the exception of TNF-α, were stronger and more persistently up-regulated by TNF-α in cells treated with β lactone than in cells where proteasome activity is intact (Fig. 3 b).
Figure 3.
Proteasome inhibition in IκBα-deficient cells determines persistent promoter occupancy and increased NF-κB–dependent transcriptional activity. (A) Anti-p65 ChIP in TNF-α–stimulated IκBα−/− cells. β lactone treatment after a pulse of TNF prolongs occupancy of all target genes tested and induces (B) increased and sustained transcription. (C) Proteasome inhibition increases κB site–directed transcription of a luciferase reporter in TNF-α–stimulated IκBα−/− cells.
To directly determine if proteasome activity is required to shut off NF-κB–dependent transcription in the absence of IκBα, we transfected IκBα−/− 3T3 cells with a luciferase reporter controlled by three κB sites. After transfection, cells were stimulated with TNF-α for 15 min, washed, and incubated for an additional 6 h in medium with or without β lactone. κB site–dependent transcription of the luciferase gene was up-regulated much stronger in cells treated with β lactone than in control cells (Fig. 3 c).
These observations indicate that in cells lacking IκBα, proteasomal degradation of p65 is required to remove it from target genes and terminate the response. Persistent promoter occupancy results in sustained NF-κB–dependent transcriptional activation.
p65 Degradation and NF-κB Response Shut Off in IκBα-containing Cells.
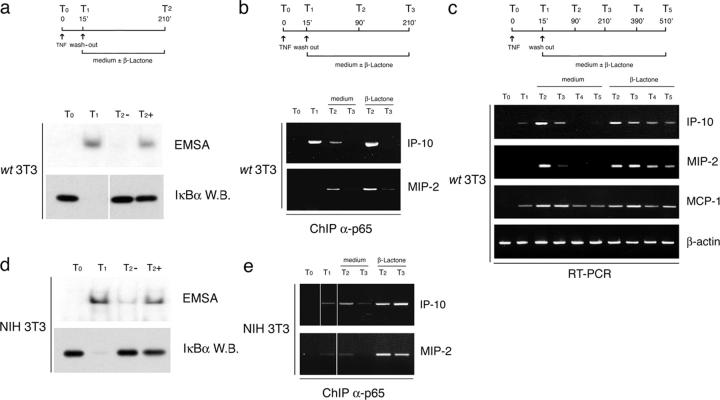
Next, we tested if proteasome activity also collaborates to NF-κB response termination in cells expressing IκBα. In WT 3T3 cells, proteasome inhibition after a 15-min pulse of TNF-α prevented complete down-regulation of nuclear NF-κB activity (Fig. 4 a). This result indicates that termination of the NF-κB response in normal cells reflects the combination of two separate activities: resynthesis of IκBα and degradation of NF-κB. In WT 3T3 cells, β lactone rescued ∼25% of maximal NF-κB activity (measured between 30 and 60 min after TNF-α).
Figure 4.
Effects of proteasome inhibition on NF-κB response termination in IκBα-containing cells. (A) Proteasome inhibition impairs down-regulation of nuclear NF-κB activity (assayed by EMSA) in WT 3T3 cells. IκBα degradation and resynthesis is also shown. (B) Anti-p65 ChIP assay and mRNA analysis (C) in TNF-stimulated WT 3T3 cells treated with β lactone or vehicle as indicated. The effects of β lactone treatment on IP-10 and MIP-2 occupancy by p65 and on their transcriptional activity are shown. (D) Effects of proteasome inhibition on NF-κB response down-regulation and p65 occupancy of target genes (E) in NIH3T3 cells.
To directly monitor if proteasome inhibition affects p65 occupancy of target gene promoters in normal cells, we performed ChIP assays in WT 3T3 cells stimulated with a pulse of TNF-α and then treated with β lactone.
Proteasome inhibition after NF-κB activation impaired or delayed the removal of p65 from the target genes MIP-2 and IP-10 (Fig. 4 b). The most obvious differences between control and β lactone–treated cells in p65 removal from IP-10 were observed at the earlier time point (Fig. 4, T2), whereas at MIP-2 the effect was more sustained. Analysis of the corresponding mRNAs showed that although MCP-1 was similarly induced and down-regulated in control cells and cells treated with β lactone, MIP-2 and IP-10 were induced in a more sustained fashion when proteasome activity was blocked (Fig. 4 c). The lack of any detectable effect on the accumulation of MCP-1 mRNA (a p65 target itself) indicates that the effects of proteasome inhibition may in part be gene specific.
Overall, proteasome inhibition exerted more dramatic effects in IκBα-deficient cells than in normal cells (Fig. 3 a), indicating a synergistic activity of proteasome and resynthesized IκBα in response termination. Indeed, in the absence of either IκBα or proteasome activity, NF-κB response down-regulation was slower than normal.
The observation that proteasome inhibition impairs or delays p65 removal from target genes even in the presence of a normal IκBα resynthesis is intriguing and may indirectly provide additional mechanistic insights into how NF-κB is dissociated from chromatin. Several mechanisms may plausibly explain why resynthesized IκBα is unable to promptly remove NF-κB from some target genes if proteasome is blocked. First, posttranslational modifications of p65, such as acetylation (25) or prolyl isomerization (26), have been shown to inhibit p65 interaction with IκBα. If a promoter is loaded with p65 molecules bearing these modifications, it will likely depend on proteasomal degradation for p65 removal unless the posttranslational modification is erased. Second, when bound to some promoters in the context of large complexes of TFs, NF-κB might be not accessible to IκBα. In this case, proteasome may either disassemble and destroy the whole enhanceosome (including p65) or indirectly facilitate IκBα activity by removing peripherally located TFs, thus exposing DNA-bound p65.
In different cell types, in response to different stimuli, and at the level of different promoters, the relative contribution of the two pathways to response termination may vary. Indeed, the behavior of NIH-3T3 and HeLa cells differed from that of WT 3T3 cells. In NIH-3T3 cells, β lactone rescued ∼50% of the maximal NF-κB binding activity (Fig. 4 d) and exerted a stronger and more sustained effect on p65 occupancy of MIP-2 and IP-10 promoters than that observed in WT 3T3 cells (Fig. 4 e). In HeLa cells, β lactone partially impaired NF-κB down-regulation but only transiently, as response was anyway completely terminated at late time points (unpublished data).
Conclusions.
An adequate control of NF-κB response termination is of paramount importance to prevent a sustained production of inflammatory mediators as well as an extended transcription of the many other genes controlled by the NF-κB system. The absolute requirement for a stringent control of NF-κB activity is clearly indicated by the multiple phenotypic abnormalities and the neonatal lethality observed in IκBα-deficient mice (14).
Here we show that p65 polyubiquitination and proteasomal degradation is a dominant mechanism of posttranscriptional repression in the absence of IκBα. More importantly, this mechanism acts in synergism with resynthesized IκBα to guarantee a timely termination of the response in normal cells. Because proteasome inhibition selectively affects the nuclear fraction of p65, it is clear that degradation of p65 is at least in great part linked to its activation. More specifically, p65 polyubiquitination requires binding to κB sites as indicated by the inefficient ubiquitination of a p65 mutant that is devoid of high affinity κB site binding activity.
Remarkably, the proteasome-dependent pathway of NF-κB response termination is conserved from Drosophila to mammals. Loss of function mutations in different components of the Drosophila SCF-E3 ubiquitin ligases cause increased levels of both full-length and processed Relish (a Drosophila NF-κB homologue) and constitutive induction of the target gene diptericin (27).
The interplay between ubiquitin, proteasome, and transcriptional regulation is extremely complex. A model compatible with many observations is that promoter-bound TFs recruit ubiquitin ligases, which ubiquitinate both TFs and RNApolI (28). Ubiquitination of TFs enhances their activity and promotes proteasome recruitment, which destroys the TFs and at the same time may exert nonproteolytic activities that stimulate transcription (29). As a consequence of this mechanism, TFs recruited to target promoters would trigger only a single round of transcriptional initiation and would be subsequently destroyed and eventually reloaded. In this context, the high number of p65-containing dimers (>100,000) that enter the nucleus after activation may serve as a reservoir of NF-κB molecules available for the repetition of the cycle of recruitment, transcriptional activation, and degradation. The evaluation of the possible role of p65 ubiquitination in transcriptional activation will require the identification of the p65 ubiquitin ligase(s) acting at the promoter level. SOCS-1 has been recently reported to polyubiquitinate p65 and promote its degradation (26), but because SOCS-1 is mainly cytoplasmic it is unlikely to act in a transcription-coupled ubiquitination/degradation pathway.
In addition to its role in response termination, degradation of chromatin-bound NF-κB molecules may also promote an exchange of NF-κB dimers at target genes (19), thus indirectly impacting on their transcriptional activity.
Acknowledgments
We thank A. Hoffmann and G. Ghosh for hints and suggestions.
This work was supported by the Swiss National Science Foundation, the Swiss Federation Against Cancer, and the Fondazione Ticinese per la Ricerca sul Cancro.
References
- 1.Siebenlist, U., G. Franzoso, and K. Brown. 1994. Structure, regulation and function of NF-kappa B. Annu. Rev. Cell Biol. 10:405–455. [DOI] [PubMed] [Google Scholar]
- 2.Verma, I.M., J.K. Stevenson, E.M. Schwarz, D. Van Antwerp, and S. Miyamoto. 1995. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev. 9:2723–2735. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin, A.S., Jr. 1996. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649–683. [DOI] [PubMed] [Google Scholar]
- 4.Whiteside, S.T., and A. Israel. 1997. I kappa B proteins: structure, function and regulation. Semin. Cancer Biol. 8:75–82. [DOI] [PubMed] [Google Scholar]
- 5.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18:621–663. [DOI] [PubMed] [Google Scholar]
- 6.Baeuerle, P.A., and D. Baltimore. 1989. A 65-kappaD subunit of active NF-kappaB is required for inhibition of NF-kappaB by I kappaB. Genes Dev. 3:1689–1698. [DOI] [PubMed] [Google Scholar]
- 7.Zabel, U., and P.A. Baeuerle. 1990. Purified human I kappa B can rapidly dissociate the complex of the NF-kappa B transcription factor with its cognate DNA. Cell. 61:255–265. [DOI] [PubMed] [Google Scholar]
- 8.Scott, M.L., T. Fujita, H.C. Liou, G.P. Nolan, and D. Baltimore. 1993. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 7:1266–1276. [DOI] [PubMed] [Google Scholar]
- 9.Ito, C.Y., A.G. Kazantsev, and A.S. Baldwin, Jr. 1994. Three NF-kappa B sites in the I kappa B-alpha promoter are required for induction of gene expression by TNF alpha. Nucleic Acids Res. 22:3787–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiao, P.J., S. Miyamoto, and I.M. Verma. 1994. Autoregulation of I kappa B alpha activity. Proc. Natl. Acad. Sci. USA. 91:28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arenzana-Seisdedos, F., J. Thompson, M.S. Rodriguez, F. Bachelerie, D. Thomas, and R.T. Hay. 1995. Inducible nuclear expression of newly synthesized I kappa B alpha negatively regulates DNA-binding and transcriptional activities of NF-kappa B. Mol. Cell. Biol. 15:2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arenzana-Seisdedos, F., P. Turpin, M. Rodriguez, D. Thomas, R.T. Hay, J.L. Virelizier, and C. Dargemont. 1997. Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J. Cell Sci. 110:369–378. [DOI] [PubMed] [Google Scholar]
- 13.Tam, W.F., L.H. Lee, L. Davis, and R. Sen. 2000. Cytoplasmic sequestration of rel proteins by IkappaBalpha requires CRM1-dependent nuclear export. Mol. Cell. Biol. 20:2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beg, A.A., W.C. Sha, R.T. Bronson, and D. Baltimore. 1995. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 9:2736–2746. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann, A., A. Levchenko, M.L. Scott, and D. Baltimore. 2002. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 298:1241–1245. [DOI] [PubMed] [Google Scholar]
- 16.Cheng, J.D., R.P. Ryseck, R.M. Attar, D. Dambach, and R. Bravo. 1998. Functional redundancy of the nuclear factor κB inhibitors I κBα and I κBβ. J. Exp. Med. 188:1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, S.H., and M. Hannink. 2002. Characterization of the nuclear import and export functions of Ikappa B(epsilon). J. Biol. Chem. 277:23358–23366. [DOI] [PubMed] [Google Scholar]
- 18.Sosic, D., J.A. Richardson, K. Yu, D.M. Ornitz, and E.N. Olson. 2003. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 112:169–180. [DOI] [PubMed] [Google Scholar]
- 19.Saccani, S., S. Pantano, and G. Natoli. 2003. Modulation of NF-kappaB activity by exchange of dimers. Mol. Cell. 11:1563–1574. [DOI] [PubMed] [Google Scholar]
- 20.Molinari, E., M. Gilman, and S. Natesan. 1999. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 18:6439–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salghetti, S.E., M. Muratani, H. Wijnen, B. Futcher, and W.P. Tansey. 2000. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. USA. 97:3118–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hottiger, M.O., L.K. Felzien, and G.J. Nabel. 1998. Modulation of cytokine-induced HIV gene expression by competitive binding of transcription factors to the coactivator p300. EMBO J. 17:3124–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, F.E., D.B. Huang, Y.Q. Chen, and G. Ghosh. 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 391:410–413. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, A., T.H. Leung, and D. Baltimore. 2003. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 22:5530–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, L., W. Fischle, E. Verdin, and W.C. Greene. 2001. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 293:1653–1657. [DOI] [PubMed] [Google Scholar]
- 26.Ryo, A., F. Suizu, Y. Yoshida, K. Perrem, Y.C. Liou, G. Wulf, R. Rottapel, S. Yamaoka, and K.P. Lu. 2003. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell. 12:1413–1426. [DOI] [PubMed] [Google Scholar]
- 27.Khush, R.S., W.D. Cornwell, J.N. Uram, and B. Lemaitre. 2002. A ubiquitin-proteasome pathway represses the Drosophila immune deficiency signaling cascade. Curr. Biol. 12:1728–1737. [DOI] [PubMed] [Google Scholar]
- 28.Salghetti, S.E., A.A. Caudy, J.G. Chenoweth, and W.P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science. 293:1651–1653. [DOI] [PubMed] [Google Scholar]
- 29.Muratani, M., and W.P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4:192–201. [DOI] [PubMed] [Google Scholar]