Abstract
The transcription factors encoded by the E2A and early B cell factor (EBF) genes are required for the proper development of B lymphocytes. However, the absence of B lineage cells in E2A- and EBF-deficient mice has made it difficult to determine the function or relationship between these proteins. We report the identification of a novel model system in which the role of E2A and EBF in the regulation of multiple B lineage traits can be studied. We found that the conversion of 70Z/3 pre-B lymphocytes to cells with a macrophage-like phenotype is associated with the loss of E2A and EBF. Moreover, we show that ectopic expression of the E2A protein E12 in this macrophage line results in the induction of many B lineage genes, including EBF, IL7Rα, λ5, and Rag-1, and the ability to induce κ light chain in response to mitogen. Activation of EBF may be one of the critical functions of E12 in regulating the B lineage phenotype since expression of EBF alone leads to the activation of a subset of E12-inducible traits. Our data demonstrate that, in the context of this macrophage line, E12 induces expression of EBF and together these transcription factors coordinately regulate numerous B lineage–associated genes.
Keywords: B cell development, transcription, lineage determination, gene expression
Blymphocytes, the cellular source of antibodies, develop from lineage unrestricted progenitors in the fetal liver and adult bone marrow (1). Commitment of multipotent progenitors to the B lineage is associated with their ability to proliferate in response to IL7, first in the presence of stromal cells and then alone (2, 3). In addition, committed B lineage cells can be identified by the expression of a number of B lineage–associated genes and rearrangement of the Ig heavy chain V, D, and J gene segments (4). The most immature, pro-B cells express the recombinase activating genes, Rag-1 and Rag-2, which function in the site-specific recombination of Ig gene segments (5–8). Once a functional Ig protein is synthesized it is expressed on the cell surface in association with the surrogate light chain proteins λ5 and VpreB and the signal transducing proteins Igα (mb-1) and Igβ (B29) which are also expressed in pro-B lymphocytes (9, 10). Expression of a functional Ig heavy chain and its associated proteins is required for the proper development of B cells since targeted inactivation of any of the genes whose products are required for Ig rearrangement or expression leads to an arrest before the development of pre-B lymphocytes (reviewed in reference 11). The majority of Igκ and Igλ light chain gene rearrangements occur in pre-B lymphocytes (12, 13).
The development of mature B lymphocytes from multipotent progenitors requires the coordinated action of a number of transcriptional regulators, including the products of the Scl/tal (14), Ikaros (15), Pu.1 (16, 17), E2A (18, 19), early B cell factor (EBF;1 reference 20), and B cell–specific activator protein (BSAP/Pax-5) genes (21). The SCL/tal, Ikaros, and Pu.1 genes function in the development of multipotent progenitors, whereas E2A, EBF, and Pax-5 are required specifically at the time of or after commitment to the B lineage. In the bone marrow of Pax-5–deficient mice, B cell development is arrested after the initiation of D-JH rearrangements but before V-DJH rearrangement (21). These B lineage progenitors express most pro-B cell–associated genes and respond to IL7 in the presence of stromal cells but fail to differentiate to more mature B cell stages (22). Mice that do not express the E2A or EBF gene show a block in B lineage progression at an earlier developmental stage, before the initiation of Ig gene rearrangement and expression of most B lineage–associated genes (18–20). The requirements for E2A and EBF in regulating B cell development are entirely unknown since deletion of most potential target genes results in an arrest at a later developmental stage than that observed in the absence of either factor (see reference 11).
The E2A gene encodes two basic helix-loop-helix (bHLH) transcription factors, E12 and E47, which arise through alternative splicing of exons encoding the bHLH domain (23, 24). E12 and E47 share >80% sequence identity and both proteins bind to the canonical E-box motif, CANNTG, found in the regulatory regions of many B lineage genes, including the Ig heavy and light chain enhancers (25, reviewed in 26). These proteins diverge primarily in the HLH motif, which mediates dimerization, and the basic region, which is required for binding to DNA (27, 28). Recent studies have demonstrated that both E12 and E47 are required for the proper development of B lineage cells indicating that these proteins are not functionally redundant (29). However, both proteins have the ability to commit lymphocytes to the B lineage.
E12 and E47 are class I bHLH proteins that are expressed in a wide range of cell types and can form heterodimers with cell-type specific, class II bHLH proteins. E2A proteins are known to play a central role in cell fate decisions as the dimerizing partners for the lineage-specific myogenic factors MyoD, Myf5, and myogenin, and the neurogenic/ pancreatic factor, NeuroD/Beta2 (30–33). However, during B lymphocyte development no lineage-specific bHLH partners for E2A have been identified and these proteins may function as homodimers (34, 35). Therefore, the specific B lineage defect in E2A-deficient mice may reflect the nonredundant functions of E2A homodimers in these cells.
E2A proteins also form heterodimers with class V HLH proteins, referred to as Id proteins, which lack a functional basic region and therefore inhibit the ability of E2A to bind DNA and activate transcription (36, 37). Four Id genes, Id1–4, have been identified in the mammalian genome. The Id gene products show overlapping and distinct distributions in the mouse embryo and adult tissues. Mice carrying an Id1 transgene expressed in B lineage cells, show an arrest in B cell development consistent with the requirement for E2A proteins (38). Within the hematopoietic system, Id proteins have been detected in both lymphoid and myeloid cells and shown to be important regulators of myeloid differentiation (39–43).
EBF is a novel transcription factor that is expressed in adipocytes, olfactory neurons, and all B lineage cells, with the exception of terminally differentiated plasma cells (44, 45). EBF contains a COOH-terminal domain that is related to the HLH domain and mediates EBF homodimer formation (46). However, this HLH-like domain does not appear to mediate strong dimerization with class I bHLH proteins (47). DNA binding by EBF homodimers is mediated by an NH2-terminal cysteine-rich region that contains a potential zinc coordination motif (46). This protein module can bind DNA as a monomer or homodimer on correctly spaced DNA half-sites.
Both E2A- and EBF-deficient mice are blocked at an early stage of B cell development (Fraction A). However, the functions of E2A and EBF in this developmental process have been difficult to deduce in mutant mice owing to the lack of B cell progenitors. To gain insight into how E2A and EBF control early B cell development, we have used a novel cell line model in which the ability of these factors to regulate B lineage–associated gene expression can be studied. We demonstrate that the expression of E12 in the 70Z/3 macrophage cell line results in the induction of a large number of B lineage–associated genes including EBF, λ5, IL7Rα, and Rag-1, and the ability to induce κ light chain in response to mitogens. The induction of most B lineage traits requires the NH2-terminal transactivation domains of E12 indicating that this phenotypic conversion is due to transcriptional activation of target genes by E12. In addition, we show that expression of EBF, in the absence of E12, leads to the activation of a subset of E12-inducible genes. Taken together, our data demonstrate that, in the context of this macrophage line, E12 induces the expression of EBF and that these transcription factors coordinately regulate numerous B lineage–associated genes.
Materials and Methods
Cell Lines and Culture Conditions.
All cell lines were maintained in Opti-MEM containing 5% FBS, 5.5 × 10−5 M β-mercaptoethanol, 100 μg/ml penicillin, and 100 μg/ml streptomycin. In some experiments the cells were cultured in the presence of 25 μg/ml LPS (GIBCO BRL, Gaithersburg, MD) for 36 h.
Protein Extracts and Electrophoretic Mobility Shift Assays.
Nuclear extracts were prepared as described (48). Protein concentrations were determined using Bio-Rad's protein assay reagent (Bio-Rad Laboratories, Hercules, CA). Double-stranded DNA probes were end labeled using T4 polynucleotide kinase and purified over a G25–Sepharose column. Electrophoretic mobility shift assays (EMSA) using the μE5, Pax-5, and Oct oligos were performed as described previously (49). The DNA–protein complexes were resolved by electrophoresis through a 5% polyacrylamide gel containing 45 mM Tris, 45 mM boric acid, and 1 mM EDTA, dried, and then exposed to autoradiographic film at −70°C. EMSA using the EBF probe were performed by incubating 10 μg nuclear extract in a binding buffer containing 10 mM Hepes, pH 7.9, 70 mM KCl, 70 mM ZnCl, 2.5 mM MgCl2, 1 mM dithiothreitol, 1 mM EDTA, 4% glycerol, 10 μg BSA and 0.2 μg poly(dIC-dIC) for 15 min at room temperature followed by 15 min at 4°C. The DNA–protein complexes were resolved by electrophoresis through a 5% polyacrylamide gel containing 25 mM Tris, 190 mM glycine, and 1 mM EDTA at 4°C as described (50). The sequences of the μE5 (51), EBF (50), Pax-5 (52), and Oct (53) oligonucleotides have been published previously.
Immunoprecipitations.
107 cells were starved of methionine by incubating the cells for 1 h in 2 ml of minus methionine DME containing 10% dialyzed calf serum, 5.5 × 10−5 M β-mercaptoethanol, 100 μg/ml penicillin, 100 μg/ml streptomycin, and 10 mM glutamine in a humidified 37°C incubator containing 5% CO2. [35S]methionine (ICN) was added at a final concentration of 0.5 μCi/ml and the cells were incubated for 1 h with occasional agitation. Total cellular extracts were prepared in 500 μl of RIPA lysis buffer. The extracts were precleared for 1 h at 4°C with a 50-μl packed volume of protein A–Sepharose after which the supernatant was transferred to a new tube containing 5 μl of polyclonal rabbit anti-Id2 antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA) and rotated at 4°C for 1 h. A further 1 h incubation was carried out after the addition of 25 μl of protein A–Sepharose. The antigen-antibody-Sepharose conjugates were washed 4× with 1 ml of lysis buffer and dissociated by the addition of an equal volume of 2× SDS–polyacrylamide gel loading buffer and boiling for 10 minutes. The immunoprecipitated proteins were electrophoresed through a 12% SDS–polyacrylamide gel. The gels were fixed in 10% methanol/10% acetic acid for 10 min and then incubated for 30 min in 1 M sodium salicylate before being dried and exposed to autoradiographic film for 12 h.
Western Blot Analysis.
20 μg of nuclear extract was electrophoresed through an 8% SDS–polyacrylamide gel and transferred to Immobulon membranes. The membranes were stained with amido black for 2 min and subsequently washed with 10% methanol and 10% acetic acid in order to determine that equivalent amounts of nuclear protein had been transferred in each lane. The membranes were blocked for 1 h at room temperature in TBST (50 mM Tris, pH 7.5, 150 mM NaCl, and 0.5% Tween-20) containing 5% milk powder. The blots were then incubated for 2 h in TBST containing one of the following antibodies: 1:500 dilution of anti-E12 (Ab 382.6; PharMingen) or 1:250 dilution of anti-E47 (Ab 32.1; PharMingen). The blots were washed 3× for 15 min in TBST before addition of HRP-conjugated goat anti–mouse IgG at a dilution of 1:15,000 (GIBCO BRL) for 1 h. The blots were washed 3× for 15 min in TBST and developed using the ECL detection system (Amersham Pharmacia Biotech Inc., Piscataway, NJ).
Creation of Stable Cell Lines and Plasmid Construction.
3 × 107 cells were resuspended in 500 μl culture media containing 10 μg of the pHβAPneo, pHβAPneoE12, or pHβAPneoEBF plasmids and electroporated at 950 μF and 250 volts using a Gene Pulser (Bio-Rad Laboratories). The electroporated cells were cultured under limiting dilution conditions for 24 h before the addition of 1 mg/ml G418 to the culture media. After 2 to 3 wk, individual clones were harvested and expanded in 10- or 15-cm culture dishes. Both adherent and nonadherent cells were collected at each passage in order to avoid selection based on adherence. The pHβAPneo plasmid was obtained from Dr. S. Hedrick (University of California, San Diego, CA) and contains the neomycin resistance gene driven by the SV40 early promoter with cloning sites downstream of the human β-actin promoter (54). The pHβAPneoE12 expression construct was created by ligation of an E12, EcoRI-HindIII fragment, into the BamHI site of pHβAPneo after treatment of both digested fragments with Klenow and was generously donated by Dr. G. Bain. The pHbAPneoE12 bHLH construct was created by PCR amplification of a 395-bp fragment spanning nt 886–1279 (as numbered in reference 24) using primers bHLH for: 5′-GATCCCGGGCGGCTGACCACTCG-3′ and HLH rev: 5′-CCATCTAGAGCTGAAAGCACCATCTG-3′. This PCR fragment was digested with XbaI and cloned into pSS-Flag digested with XbaI and XmaI and designated as pSS-E12 bHLH. pSS-E12 bHLH was digested with XbaI, made blunt with Klenow, and subsequently digested with HindIII. This fragment was cloned into pHβAPneo digested with BamHI, made blunt with Klenow, and subsequently digested with HindIII resulting in pHβAPneoE12 bHLH. The pHβAPneoEBF construct was created by ligation of a SalI and HindIII fragment of the rat Olf/EBF cDNA (provided by Dr. R. Reed, Johns Hopkins University, Baltimore, MD) into pHβAPneo digested with SalI and HindIII.
FACS® Analysis.
1 × 106 cells were stained as described previously (29). Cells were analyzed on a FACScan® (Becton Dickinson, Mountain View, CA) using the CellQuest software package. Live cells were gated on the basis of forward vs. side scatter. All antibodies were purchased from PharMingen (San Diego, CA) and include anti-Mac-1(FITC), anti-IgM(biotin), anti-κ(biotin), anti-Ly6A(PE), and the secondary reagent streptavidin (PE).
RNA Isolation and Northern Blot Analysis.
10 μg of total RNA was isolated by Trizol (GIBCO BRL) and separated through a 0.75% agarose gel buffered with 18 mM Na2HPO4, 2 mM NaH3PO4, and 6% formaldehyde after incubation for 5 min at 65°C in the same buffer containing 50% formamide. The gel was washed extensively with dH2O and the RNA was transferred to nytran membrane by capillary action using 10× SSC. The blot was hybridized with random primed, 32P-labeled gene-specific probes in 50% formamide, 5× SSC, 0.15% SDS, 1× Denhardt's, and 10% dextran sulfate containing 100 μg/ml heat-denatured herring sperm DNA at 42°C overnight. The blots were washed 2× for 30 min at 42°C in 2× SSC/0.2% SDS and 2× 15 min at 60°C in 0.2× SSC/0.2% SDS before exposure to autoradiographic film at −70°C for 12–24 h.
Reverse Transcriptase Polymerase Chain Reaction Assays.
2 μg of total RNA was treated with 10 U DNAse (Boehringer Mannheim Corp., Indianapolis, IN) at 37°C for 10 min in reverse transcriptase (RT) buffer (GIBCO BRL) followed by heat inactivation at 70°C for 10 min. DNA-free RNA was reverse transcribed and amplified as described previously (29). PCR reactions to detect Rag-1 and Ikaros were amplified for 25 cycles. PCR reactions to detect actin were amplified for 13 cycles of 1 min at 94°C, 1 min at 59°C, and 1 min at 72°C. The sequence of the Rag-1, actin, and Ikaros PCR primers have been published previously (18, 55).
Results
The Transition of 70Z/3 Pre-B Lymphocytes to the Macrophage Phenotype Is Associated with a Loss of the E2A and EBF Transcription Factors.
A number of B lineage cell lines and a subpopulation of chronic lymphocytic and acute leukemias have been shown to lose some of their B lineage characteristics and acquire characteristics of macrophages (56– 63). The 70Z/3 pre-B line is unique among these lineage switching cells in that it undergoes spontaneous alterations in vitro which result in the emergence of macrophage-like cells (56). Previous studies have suggested that this switch occurs through the reproducible loss of sets of B lineage genes (64). We reasoned that the loss of B lineage characteristics may be the result of a loss of key regulators of B lineage genes. Therefore, this cell line would provide novel insights as to which transcriptional regulators are involved in the maintenance of the B lineage program.
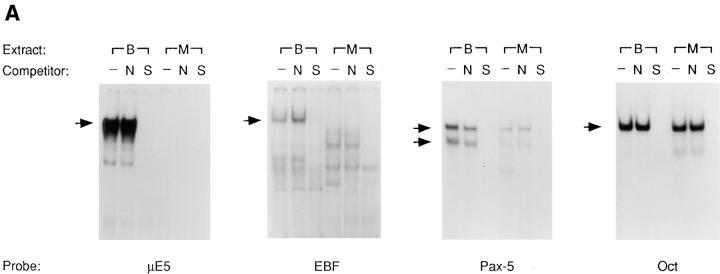
The E2A, EBF, and Pax-5 transcription factors have been shown to be required for progression through the earliest stages of B cell development. To determine whether these proteins were expressed in the 70Z/3 pre-B or macrophage cell lines we examined the ability of nuclear extracts to complex with specific DNA binding sites by EMSA. Nuclear extract from the pre-B line contained proteins that bound to the E2A-binding site (E-box) in the μE5 oligo (Fig. 1 A). This complex was competed by an excess of unlabeled μE5 and was supershifted with antibodies specific for the E2A proteins (data not shown). By contrast, the 70Z/3 macrophage line did not show specific binding to the μE5 probe, indicating an absence of functional E2A (Fig. 1 A). The pre-B cell extract also contained proteins that bound specifically to the EBF- and the Pax-5–binding sites (Fig. 1 A). The macrophage nuclear extract lacked the EBF complex although a faster mobility complex was detected. The identity of the protein(s) in this faster mobility complex is unknown. However, it is unlikely to contain EBF since EBF mRNA is not detected in the macrophage line (see Fig. 3 A). The macrophage nuclear extract did contain Pax-5 binding activity, although the amount of binding was approximately fivefold less than that detected in the pre-B cell extract (Fig. 1 A). Equivalent amounts of Octamer binding activity were detected in extracts from both lines (Fig. 1 A). Therefore, the progression of 70Z/3 pre-B lymphocytes to the macrophage phenotype is associated with a loss of E2A and EBF DNA binding activity and a reduction in Pax-5.
Figure 1.
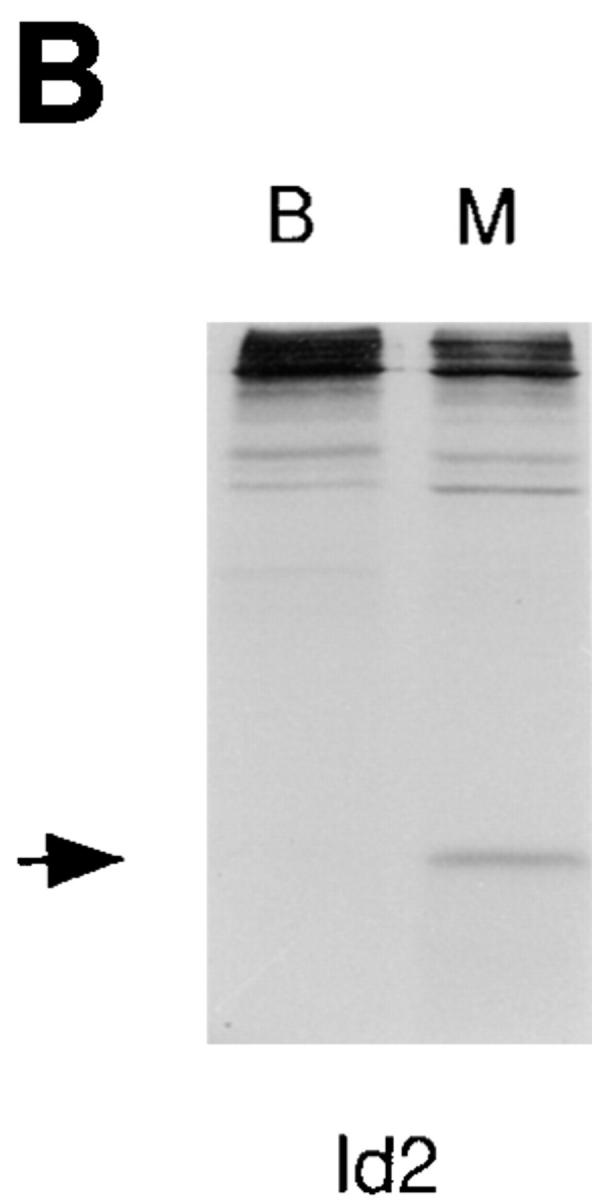
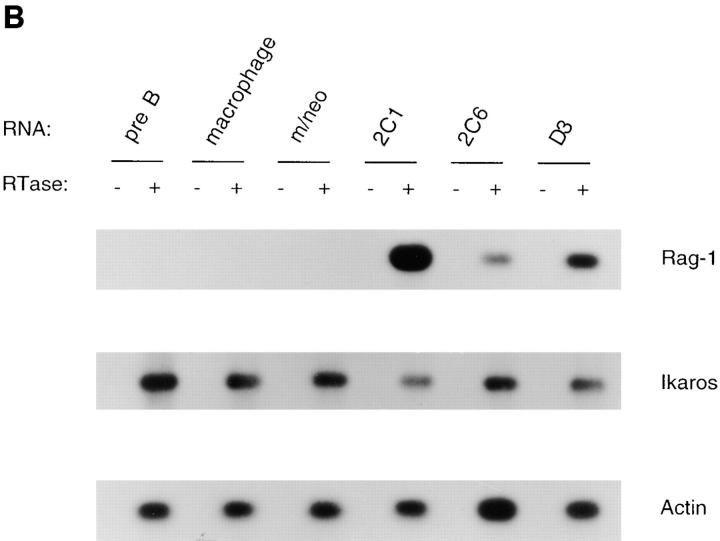
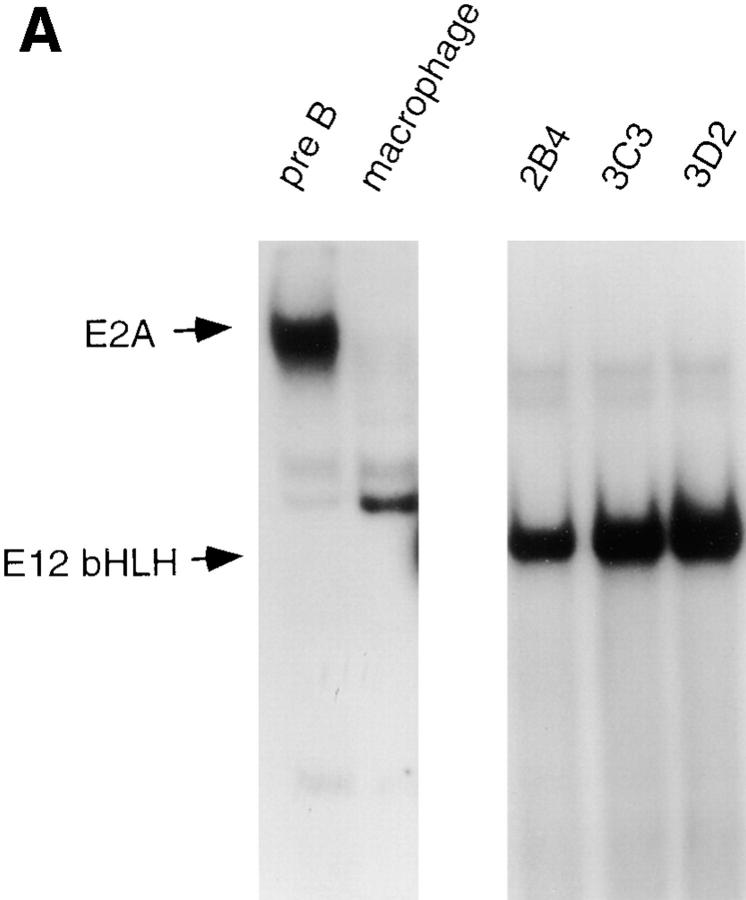
A reduction in E2A, EBF and Pax-5 DNA binding activity is associated with the transition of 70Z/3 pre-B cells to the macrophage phenotype. (A) Electrophoretic mobility shift analysis of nuclear extracts prepared from the 70Z/3 pre-B (B) and macrophage (M) cell lines. 10 μg of nuclear extract was incubated with a 32P-labeled oligonucleotide probe containing an E2A- (μE5), EBF-, Pax-5–, or Oct-binding site. The extracts were incubated in the absence (−) or presence of a 100-fold excess of specific (S) or nonspecific (N) competitive oligo. Arrows indicate the specific DNA-binding complexes corresponding to E2A, EBF, Pax-5, or Oct-1. Two complexes are expected with Pax-5 oligo that contains both a Pax-5– and ets-binding site (52). (B) Immunoprecipitation of Id2 from 35S-methionine–labeled 70Z/3 pre-B (B) and macrophage (M) cell lines. (C) Western blot analysis of 70Z/3 pre-B (B) and macrophage (M) nuclear extracts. E2A proteins were detected with an anti-E12 (mAb 382.1) or anti-E47 (mAb 32.1) monoclonal antibody.
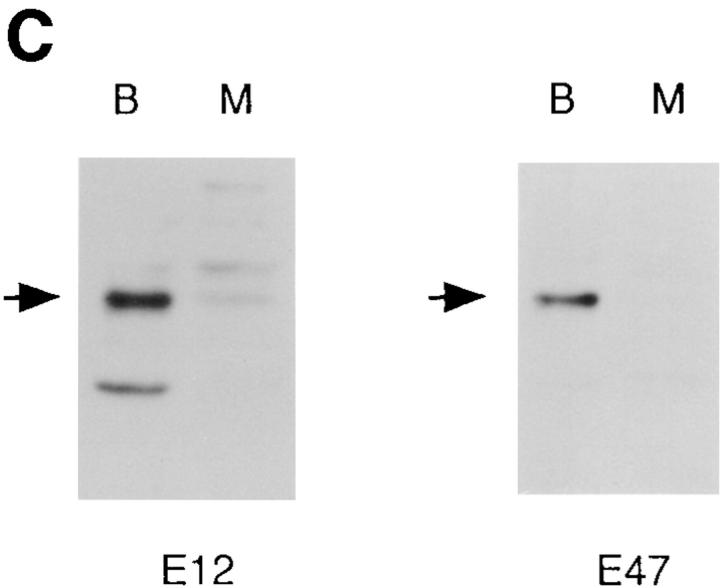
Figure 3.
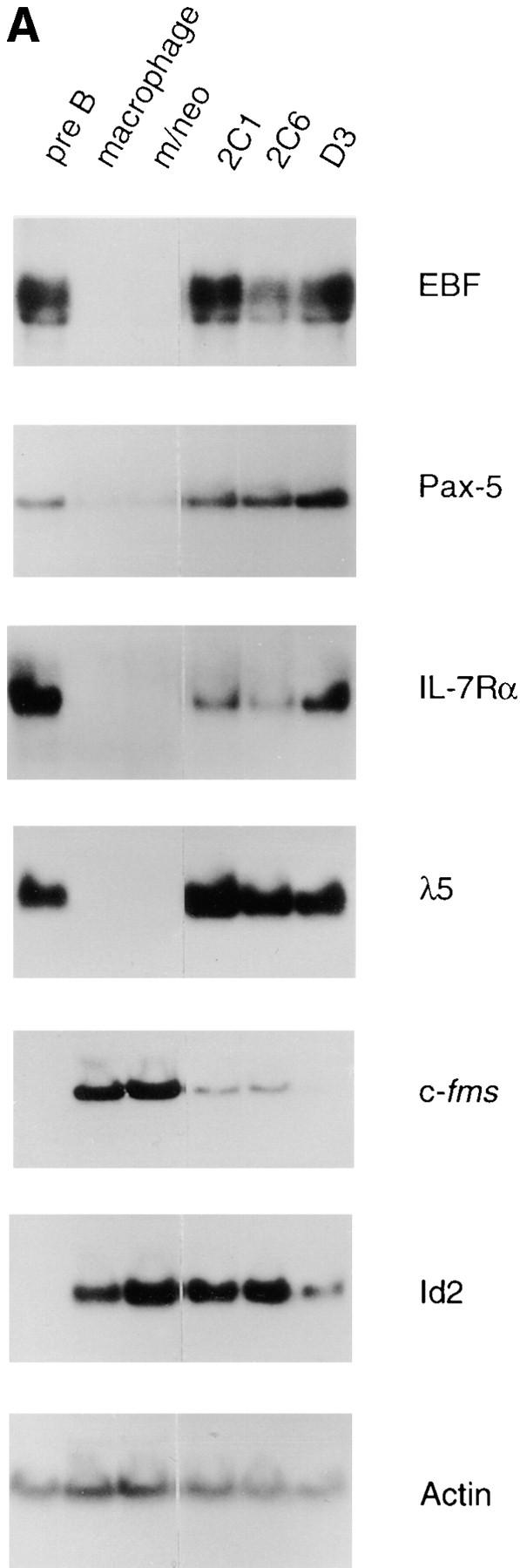
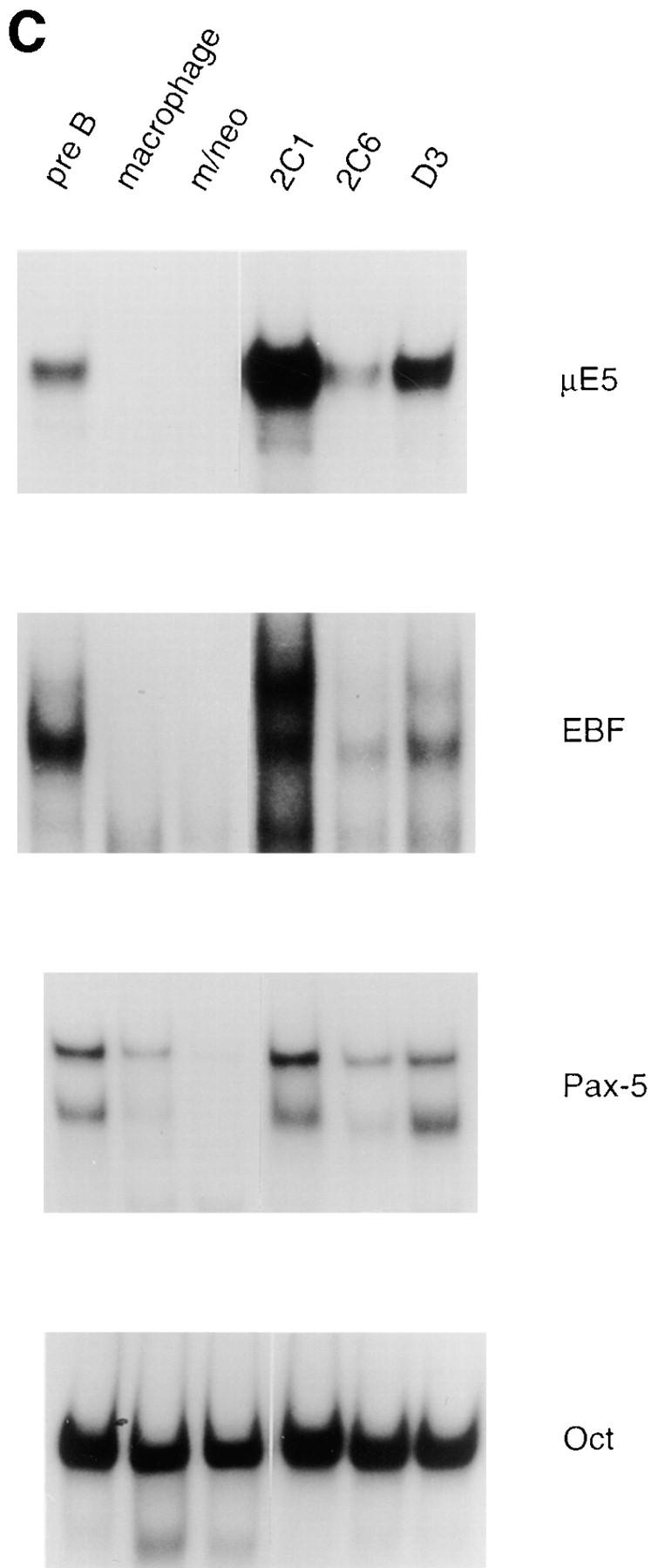
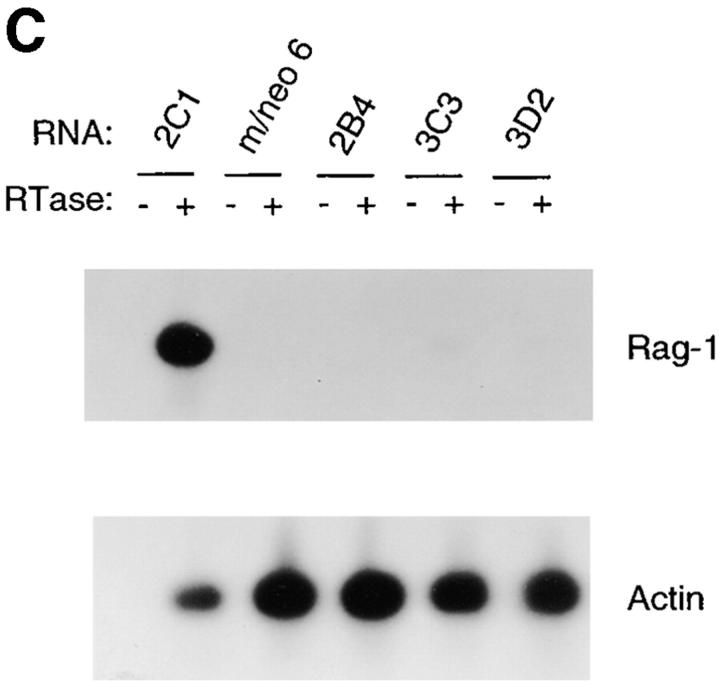
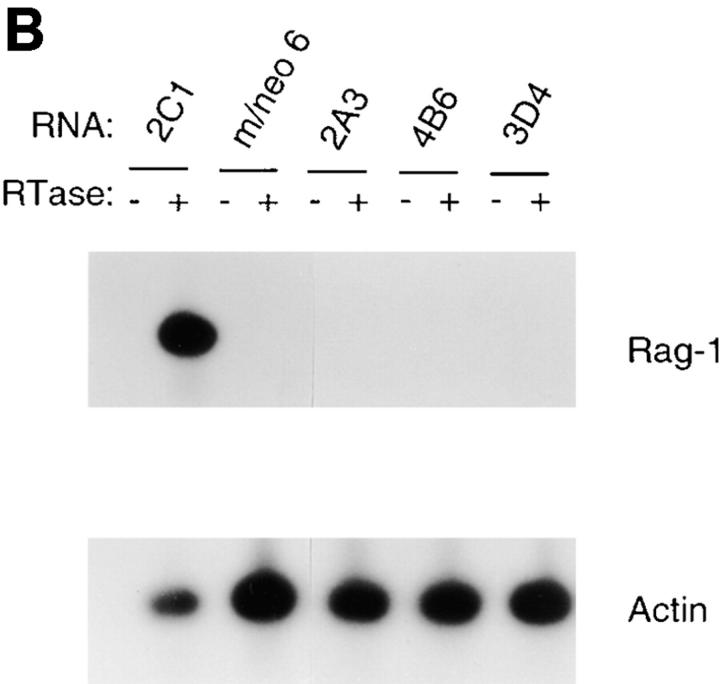
Increased expression of B lineage–associated genes in E12- expressing macrophages. (A) Northern blot analysis of EBF, Pax-5, IL7Rα, λ5, c-fms, Id2, and actin mRNA expression. 10 μg of total RNA extracted from the 70Z/3 pre-B, macrophage, m/neo, and the E12-expressing macrophage clones 2C1, 2C6, and D3 was electrophoresed through an 0.8% agarose gel and transferred to nylon membrane. The blots were probed sequentially with 32P-labeled cDNA probes for the indicated genes. (B) RT-PCR analysis of Rag-1 and Ikaros RNA expression. 2 μg of total RNA was reverse transcribed using an oligo dT15 primer in the presence (+) or absence (−) of RT. 100 ng of cDNA was used for PCR amplification with primers specific for Rag-1 (25 cycles), the COOH-terminal domain of Ikaros (25 cycles), or actin (13 cycles; C) μE5, EBF, Pax-5, and Oct DNA binding activity in E12-expressing macrophage cell lines. 10 μg of nuclear extract from each of the indicated cell lines was incubated with a 32P-labeled oligonucleotide probe containing the binding site for E2A (μE5), EBF, Pax-5, and Oct.
The absence of a μE5-binding complex in the macrophage line could result from a decrease in the amount of E2A proteins or an increase in the expression of an Id protein, which have been shown previously to be expressed in myeloid cells (39, 40, 41, 42, 43). Preliminary RT-PCR analysis and Northern blot assays demonstrated that mRNA encoding Id1, Id3, and Id4 were not expressed in 70Z/3 macrophages; however, Id2 mRNA could be detected (data not shown). To determine whether Id2 protein was present in the macrophage or pre-B lines, cells were labeled with [35S]methionine and Id2 was immunoprecipitated from total cellular extracts using a polyclonal anti-Id2 antibody. Id2 could be immunoprecipitated from the macrophage, but not the pre-B cell extracts (Fig. 1 B). No Id2 was immunoprecipitated with a nonspecific control antiserum (data not shown). Western blot analysis demonstrated that the amount of E12 and E47 protein was decreased at least 10-fold in nuclear extracts from the macrophage line as compared with the pre-B cell line (Fig. 1 C). These data indicate that the decrease in μE5 binding activity observed in the macrophage nuclear extract can be attributed to a decrease in the expression of E2A proteins and an increase in expression of Id2.
Ectopic Expression of E12 in the 70Z/3 Macrophage Cell Line Leads to a Decrease in Mac-1, an Increase in Surface μ, and Induction of κ Light Chain in Response to Mitogen.
The increased level of Id2 and decreased level of the E2A proteins in the macrophage line are consistent with the hypothesis that E2A is a major contributor to the B lineage phenotype. To test this hypothesis directly we stably transfected the macrophage line with constructs encoding human E12 or neomycin under the control of the β-actin promoter (pHβAPneo; reference 54). Greater than 25 neomycin- expressing macrophage clones have been examined, all of which resemble the parental macrophage clone. In contrast, greater than 15 E12-expressing macrophage clones have been isolated all of which have demonstrated a similar phenotype to the three representative E12-expressing macrophage clones, 2C1, 2C6, and D3 described here. Each of the E12-expressing macrophage clones expressed levels of μE5 binding activity comparable to, or higher than, that of the 70Z/3 pre-B cell line (see Fig. 3 C). All of the μE5 binding activity in these clones was supershifted with an antibody specific for human E2A or E12 and flow cytometric analysis indicated that essentially all of the cells in each clone expressed the human E12 protein (data not shown).
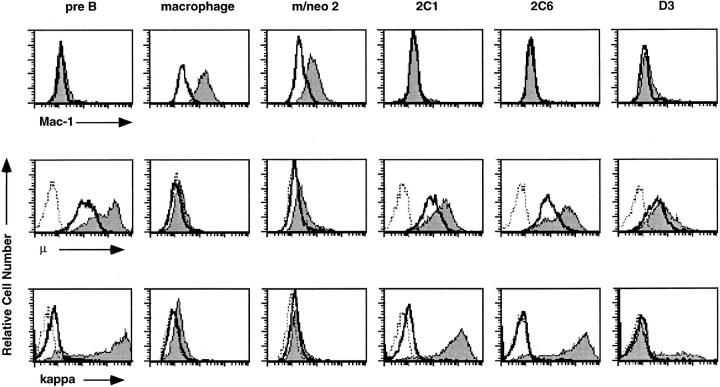
A number of B lineage- or macrophage-associated traits have been identified that distinguish the 70Z/3 pre-B and macrophage cell lines (56, 64). The macrophage line is irregular in shape and adherent to plastic whereas the majority of pre-B cells are round and nonadherent. Interestingly, the E12-expressing macrophage clones were less adherent and more rounded than the parental macrophage line. Approximately 40–50% of the cells in each of the E12- expressing lines were nonadherent, whereas <15% of the cells in the parental line, or the neomycin-expressing clones, were nonadherent (data not shown). The macrophage and pre-B cell lines also differ in the expression of the myeloid adhesion protein Mac-1 (CD11b/CD18). Mac-1 expression can be detected on the surface of the macrophage but not the pre-B cell line (64). Surprisingly, FACS® analysis revealed that the E12-expressing macrophage lines ceased to express Mac-1 (Fig. 2, top). By contrast, all the neomycin-expressing macrophage clones expressed Mac-1 (Fig. 2 and data not shown).
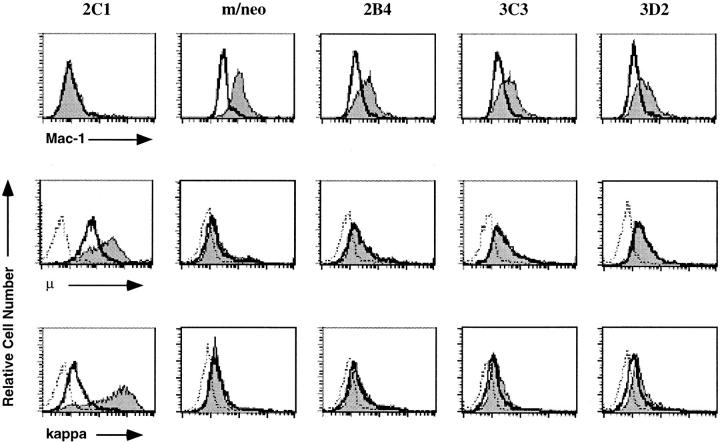
Figure 2.
E12-expressing macrophage clones cease to express Mac-1 and respond to LPS by induction of κ light chain and μ. The 70Z/3 pre-B, macrophage, m/neo, and the E12-expressing macrophage cell lines, 2C1, 2C6, and D3 were examined by flow cytometry for the expression of the myeloid associated marker Mac-1 (top). The shaded histogram represents specific Mac-1 staining; open histograms represent staining in the presence of a nonspecific control antibody. Flow cytometric analysis of μ (middle) or κ (bottom) expression on the indicated cell lines after incubation for 36 h in the presence (shaded histogram) or absence (open histogram) of LPS. The histogram outlined with a dashed line represents staining in the presence of an isotype control antibody.
The 70Z/3 pre-B and macrophage cell lines also demonstrate distinct responses to the bacterial mitogen lipopolysaccharide (LPS; reference 64). The pre-B cell line responds to LPS by induction of its rearranged κ light chain and upregulation of the level of surface μ (65). In contrast, the macrophage line does not induce κ or μ in response to LPS (56). Remarkably, each E12-expressing macrophage clone was shown to induce κ in response to LPS by flow cytometry (Fig. 2, bottom). Greater than 90% of the cells in the 2C1 and 2C6 clones and 30% of the D3 cells were κ inducible. A similar proportion of cells in each clone demonstrated induction of μ (Fig. 2, middle). Furthermore, expression of μ was detected on the surface of these cells before LPS stimulation, a characteristic of the pre-B cell line but not the parental or neomycin-expressing macrophage lines.
We have observed a degree of variability in the ability of independent clones to induce κ that is not correlated with the level of expression of E12. Interestingly, a similar degree of variability (ranging from 20 to 80%) has been observed in subclones of the parental 70Z/3 pre-B line (Paige, C.J., personal communication). This finding suggests that the variation in κ induction observed in our E12-expressing macrophage clones is likely due to an inherent variation in the response to LPS in the parental line. Regardless, we have found that expression of E12, but not neomycin, in this macrophage line was sufficient to restore the ability of some cells in each clone to induce κ light chain and μ in response to LPS. Moreover, these alterations in cell surface phenotype suggest that the E12- expressing macrophage clones were losing the characteristics of the macrophage lineage and acquiring those associated with pre-B lymphocytes.
Expression of E12 in the 70Z/3 Macrophage Line Results in the Induction of EBF and Multiple B Lineage–associated Genes.
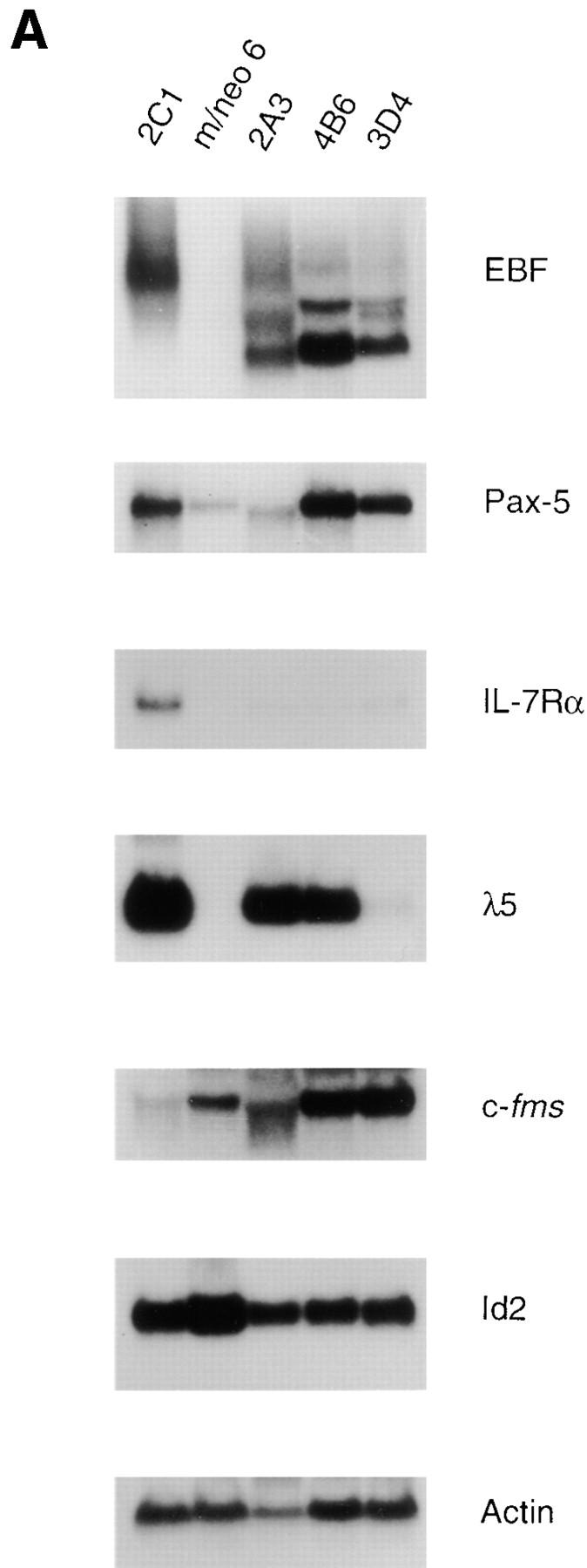
To further investigate the effects of E12 in the 70Z/3 macrophage line we examined the expression of a number of B lineage genes by Northern blot analysis. High levels of mRNA encoding EBF, λ5, and IL7Rα were evident in all of the E12-expressing macrophage clones (Fig. 3 A) By contrast, neither the parental macrophage, nor the neomycin clones, expressed significant levels of these mRNAs (Fig. 3 A). In addition, Pax-5 transcripts were induced significantly in the E12-expressing lines, although low levels of Pax-5 can be detected in the parental macrophage line indicating that E12 is not required for Pax-5 expression (Fig. 3 A). None of these genes have been shown previously to be induced upon expression of E12, or E47, in non-B lineage cells. However, Rag-1 and Rag-2 were upregulated after expression of E47 in a pre-T cell line (66, 67). Therefore, we also examined our clones for the expression of mRNA encoding Rag-1 and Rag-2 using a sensitive RT-PCR assay. Neither the 70Z/3 pre-B nor macrophage lines expressed Rag-1 by this analysis (Fig. 3 B). In contrast, each of the E12-expressing macrophage clones expressed significant levels of Rag-1 mRNA (Fig. 3 B). None of the lines tested demonstrated detectable expression of Rag-2 mRNA (data not shown). By comparison, each of the lines expressed a similar amount of the Ikaros gene, as determined by PCR primers that recognize a COOH-terminal portion of the gene common to all Ikaros isoforms (55; Fig. 3 B).
To determine whether the increase in mRNA observed in the E12-expressing macrophage lines translated into an increase in protein for EBF and Pax-5 we tested nuclear extracts prepared from each of these clones for binding to specific DNA probes by EMSA. Each E12-expressing macrophage clone demonstrated an increase in binding to the EBF probe as compared with the parental or neomycin- expressing macrophage clones (Fig. 3 C). The E12-expressing clones generally demonstrated two or three specific complexes binding to the EBF probe which could also be detected in extracts from the pre-B cell line with a longer exposure. These slower migrating complexes are presumed to be higher order multimers as has been suggested previously (45). Nuclear extracts from the E12-expressing clones were also examined for the presence of proteins able to bind to a Pax-5–binding site. Two of the clones, 2C1 and D3, demonstrated an increase in Pax-5 binding compared with the parental macrophage line. However, the 2C6 line demonstrated binding comparable to that observed in the parental macrophage, but greater than that observed in the neomycin-expressing macrophage clone (Fig. 3 C). We have found a significant variation in the amount of Pax-5 binding between individual neomycin-expressing macrophage clones (compare the macrophage extract to the neomycin-expressing macrophage extract in Fig. 3 C). However, none of the neomycin-expressing clones have demonstrated an increase in binding to the Pax-5 probe compared with the parental macrophage line (data not shown). The source of this variability remains to be elucidated. However, at the present time it is difficult to determine whether the level of Pax-5 binding in the 2C6 clone is increased from that in the individual cell that was transfected. Regardless, the mRNA encoding EBF and Pax-5 is clearly upregulated in all E12-expressing macrophage clones.
We also examined the expression of genes associated with the myeloid lineage including the receptor for macrophage colony-stimulating factor (c-fms), and Id2 by Northern blot analysis. The level of c-fms mRNA was reduced 3- to 20-fold in the E12-expressing macrophage clones compared with the levels detected in the parental and neomycin-expressing clones, although it was significantly greater than that detected in the pre-B line (Fig. 3 A and data not shown). By contrast, all of the E12-expressing clones contained levels of Id2 mRNA that were similar to, or slightly reduced compared with that observed in the parental and neomycin-expressing macrophage lines. Taken together, our data demonstrate that expression of E12 in this macrophage line was sufficient to induce a number of B lineage–associated genes and is, furthermore, not inconsistent with the expression of some myeloid genes such as c-fms. The observation that Id2 is not negatively regulated by E12 is consistent with our identification of both Id2 and E12 (and E47) mRNA in bipotent B cell/macrophage progenitors (data not shown).
Induction of the B Lineage Phenotype in the Macrophage Line Requires the Transactivation Domains of E12.
The capacity of E12 to activate transcription directly is less well established than that for E47. E12 is not generally a major component of E-box–binding complexes, a finding that is thought to be due to the presence of an inhibitory domain which lowers the affinity of E12 for its DNA-binding site (68). It is possible that E12 may function to sequester Id from other bHLH proteins, thereby allowing those factors to activate transcription. Alternatively, the bHLH domain of E12 may interact with other proteins that alter its transactivation potential, as has been demonstrated for the bHLH domain of MyoD (69, 70). To gain insight into the mechanism by which E12 regulates the expression of EBF and other B lineage–associated genes we created stable macrophage lines expressing the E12 bHLH domain, but lacking the transactivation domains (71, 72). All 15 neomycin-resistant clones tested expressed levels of the E12 bHLH protein comparable to, or higher than, that of our E12-expressing macrophage clones as determined by FACS® and Western blot analysis (data not shown). Three of these clones 2B4, 3C3, and 3D2 were chosen for further analysis.
The bHLH-expressing macrophage lines differed dramatically from the E12-expressing macrophage lines. The bHLH-expressing lines remained adherent to plastic and irregular in shape similar to the parental macrophage line (data not shown). In addition, Mac-1 expression could be detected on these cells by flow cytometry (Fig. 4, top). Moreover, we found that the bHLH-expressing macrophage lines failed to induce μ or κ in response to LPS (Fig. 4, middle and bottom). Each of the bHLH-expressing clones demonstrated high levels of μE5 binding activity as measured by EMSA (Fig. 5 A), indicating that the failure to activate B lineage traits is not due to an inability to dimerize or bind DNA. Therefore, our data indicate that the bHLH domain alone, in the absence of the NH2-terminal transactivation domains, is unable to induce these B lineage characteristics in the 70Z/3 macrophage line.
Figure 4.
E12 bHLH-expressing macrophage clones express Mac-1 but fail to respond to LPS by induction of κ light chain and μ. The E12-expressing 2C1 line, m/neo, and three E12 bHLH-expressing macrophage lines 2B4, 3C3, and 3D2 were examined by flow cytometry for the expression of the myeloid associated marker Mac-1 (top). The shaded histogram represents specific Mac-1 staining, open histograms represent staining in the presence of a nonspecific control antibody. Flow cytometric analysis of μ (middle) or κ (bottom) expression on the indicated cell lines after incubation in the presence (shaded histogram) or absence (open histogram) of LPS for 36 h. The histogram outlined with a dashed line represents staining in the presence of an isotype control antibody.
Figure 5.
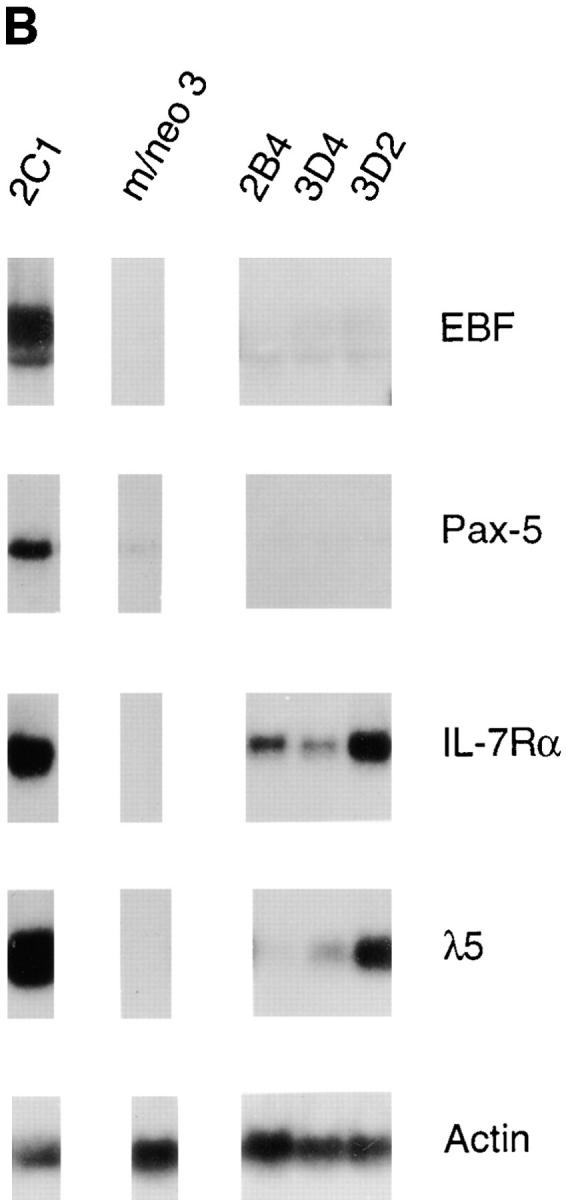
The transactivation domains of E12 are required for activation of most B lineage genes. (A) EMSA of μE5-binding complexes in 70Z/3 pre-B, macrophage and three E12 bHLH-expressing macrophage clones 2B4, 3C3 and 3D2 (which lack the transactivation domains). All lanes are from the same gel, however the pre-B and macrophage panel was exposed for threefold longer than the 2B4, 3C3 and 3D2 lanes. The position of the full-length E2A and E12 bHLH-binding complexes are indicated with an arrow on the left side of the figure. (B) Northern blot analysis of EBF, Pax-5, IL7Rα, λ5, and actin expression in E12 bHLH-expressing macrophage clones. 10 μg of total RNA from each of the cell lines was electrophoresed through an 0.8% agarose gel and transferred to nylon membrane. For comparison an E12-expressing (2C1) and neomycin expressing (m/neo3) macrophage clone run on the same gel, are shown. The blots were probed sequentially with 32P-labeled cDNA probes for the indicated genes. The blot was probed with actin to demonstrate that similar amounts of RNA were loaded in each lane. (C) RT-PCR analysis of Rag-1 and actin RNA expression in the E12 bHLH-expressing macrophage clones. 2 μg of total RNA was reverse-transcribed using an oligo dT15 primer in the presence (+) or absence (−) of RT. 100 ng of cDNA was used for PCR amplification with primers specific for Rag-1 (25 cycles) or actin (13 cycles).
To determine whether the transactivation functions of E12 were required for the induction of other B lineage genes RNA was prepared from the bHLH-expressing macrophage lines and examined by Northern blot or RT-PCR analysis. Remarkably, neither EBF nor Rag-1 mRNA were detected in the bHLH-expressing clones (Fig. 5, B and C). In addition, the level of Pax-5 mRNA remained low in all bHLH clones, similar to the macrophage line (Fig. 5 B). Therefore, the induction of EBF, Pax-5, and Rag-1 in the E12-expressing macrophage cell lines also is dependent on the transactivation functions of E12.
Surprisingly, expression of IL7Rα and λ5 were detected in the bHLH-expressing macrophage clones (Fig. 5 B). This result was unexpected and may indicate that E12 functions by distinct mechanisms to regulate the expression of different genes. Consistent with this notion, the DNA binding domain of both MyoD and Pax-5 have been shown to be sufficient to activate expression of a subset of MyoD and Pax-5 target genes (73, 74). However, in our experiments the level of mRNA detected for either IL7Rα or λ5 was highly variable and generally much lower than that observed in the E12-expressing clones. This observation suggests that the presence of the E12 transactivation domains facilitates expression of these genes. In this regard it should be noted that some E47:E12 bHLH heterodimer was detected in these cells which may provide a low level of transactivation capacity (Fig. 5 A and data not shown). Therefore, although the bHLH domain alone may be sufficient for activation of IL7Rα and λ5, it is also conceivable that the threshold for activation of these genes is lower than for other B lineage targets and therefore effected by low levels of mixed heterodimers. Further studies will be required to determine the mechanism by which the bHLH domain activates the IL7-Rα and λ5 genes.
Ectopic Expression of EBF in the 70Z/3 Macrophage Line Leads to the Activation of a Subset of E12 Inducible Traits.
Both E2A and EBF are obligatory for the proper development of B lineage cells (18–20). Our data suggest that, in the 70Z/3 macrophage line, one of the transcriptional targets of E12 is EBF. This raises the question as to how much of the B lineage phenotype induced by E12 in this macrophage line is due to the activity of EBF. To address this question, we created stable macrophage clones expressing the rat Olf/EBF gene under the control of the β-actin promoter. We identified five clones that expressed Olf/EBF mRNA of the predicted size (2.1 kb) from the rat Olf/EBF expression vector by Northern blot analysis. Three of these clones, 2A3, 4B6, and 3D2 were selected for further analysis.
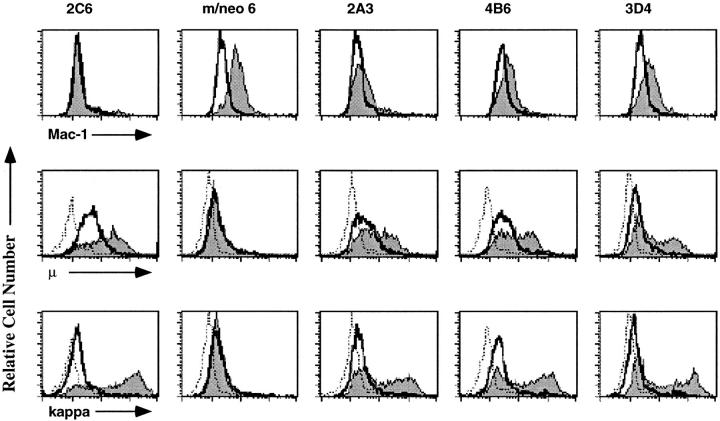
The EBF-expressing macrophage clones were found to exhibit a decrease in plastic adherence such that 50–70% of the cells were nonadherent, similar to the phenotype of the E12-expressing clones (data not shown). However, unlike the E12-expressing lines, the myeloid associated marker Mac-1 could be detected on the EBF-expressing macrophages, although the levels of Mac-1 have been decreasing gradually with time in culture (Fig. 6, top). To our surprise, these EBF-expressing clones did demonstrate an increase in the level of μ as determined by flow cytometric analysis (Fig. 6, middle). Moreover, some cells in each clone demonstrated the capacity to induce κ light chain and μ in response to LPS (Fig. 6, bottom and middle). These observations suggest that indeed, EBF alone is able to induce some of the B lineage traits observed in the E12-expressing macrophage lines.
Figure 6.
EBF-expressing macrophage lines express Mac-1 and respond to LPS by induction of κ light chain and μ. The 2C6, m/neo 6 (a neomycin-expressing macrophage), and 3 EBF-expressing macrophage cell lines, 2A3, 4B6, and 3D4 were examined by flow cytometry for the expression of the myeloid-associated marker Mac-1 (top). The shaded histogram represents specific Mac-1 staining, open histograms represent staining in the presence of a nonspecific control antibody. Flow cytometric analysis of μ (middle) or κ (bottom) expression on the indicated cell lines after incubation for 36 h in the presence (shaded histogram) or absence (open histogram) of LPS. The histogram outlined with a dashed line represents staining in the presence of an isotype control antibody. The 70Z/3 pre-B, macrophage, m/neo, and the E12-expressing macrophage cell lines, 2C1, 2C6, and D3 were examined by flow cytometry for the expression of the myeloid associated marker Mac-1 (top).
To further investigate the ability of EBF to induce B lineage–associated traits we examined the expression of B lineage genes by Northern blot analysis. As expected, the mRNA encoding Olf/EBF in each of the transfected clones was 2.1 kb, as compared with 5.2–5.8 kb for the endogenous gene. The larger transcripts that hybridize to the Olf/EBF probe appear to be transcripts that fail to terminate at the polyadenylation sequences 3′ of the Olf/ EBF gene in the pHβAPneo expression vector (data not shown). Overall, the level of Olf/EBF mRNA expressed in these clones was found to be as high, or higher than, that in the 70Z/3 pre-B– or E12-expressing macrophage clones (note that the decreased level of actin mRNA for the 2A3 line; Fig. 7 A). Remarkably, each of the EBF-expressing macrophage clones demonstrated an increase in the expression of Pax-5 and λ5 mRNA, although λ5 was expressed at a lower level than that seen with E12 (Fig. 7 A). In contrast, EBF did not induce a significant increase in IL7Rα (Fig. 7 A) or Rag-1, as determined by RT-PCR analysis (Fig. 7 B). Therefore, expression of EBF in the 70Z/3 macrophage line results in the activation of a subset of E12-inducible genes.
Figure 7.
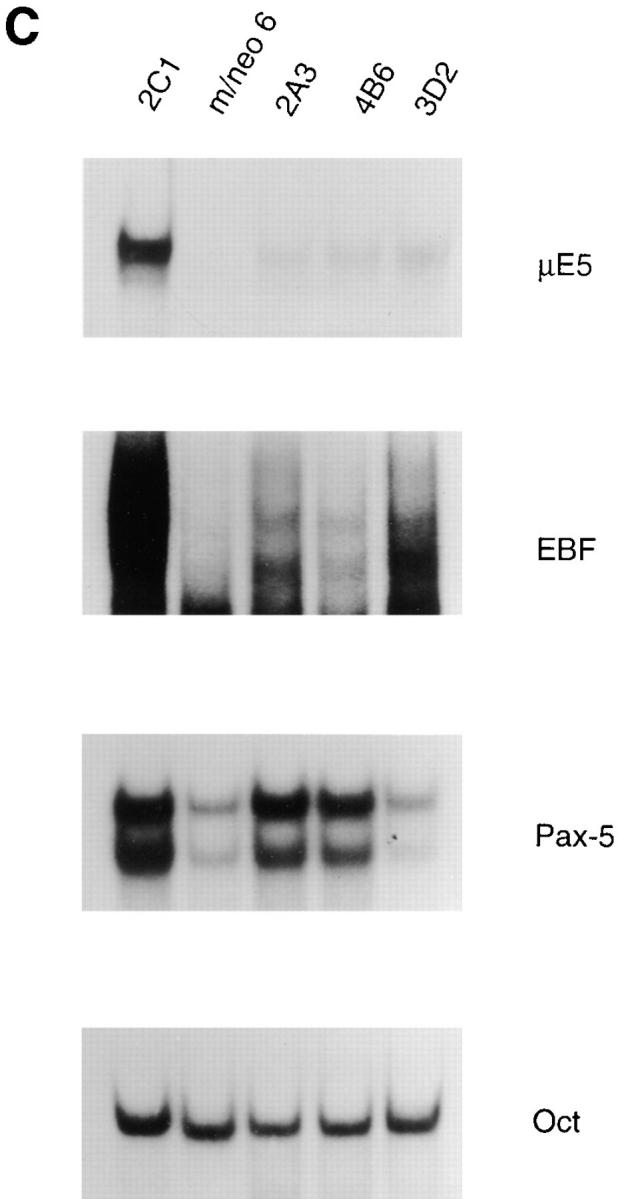
Expression of a subset of B lineage genes in EBF-expressing macrophage cell lines. (A) Northern blot analysis of EBF, Pax-5, IL7Rα, λ5, c-fms, Id2, and actin mRNA expression. 10 μg of total RNA extracted from the EBF-expressing macrophage cell lines 2A3, 4B6, and 3D4 or from a neomycin- or E12-expressing macrophage clone 2C1 was electrophoresed through an 0.8% agarose gel and transferred to nylon membrane. The blots were probed sequentially with 32P-labeled cDNA probes for the indicated genes. (B) RT-PCR analysis of Rag-1 and actin RNA expression. 2 μg of total RNA was reverse-transcribed using an oligo dT15 primer in the presence (+) or absence (−) of RT. 100 ng of cDNA was used for PCR amplification with primers specific for Rag-1 (25 cycles), or actin (13 cycles). (C) μE5, EBF, Pax-5, and Oct DNA binding activity in EBF- expressing macrophage cell lines. 10 μg of nuclear extract from each of the indicated cell lines was incubated with a 32P-labeled oligonucleotide probe containing the binding site for E2A (μE5), EBF, Pax-5, and Oct.
We examined the ability of EBF to induce E2A by EMSA. Nuclear extracts from the EBF-expressing macrophages did not demonstrate an increase in the level of μE5-binding complexes (Fig. 7 C). The slight increase in μE5-binding complex was within the range observed in our panel of 25 neomycin-expressing clones and was therefore unlikely to be due to the expression of EBF (data not shown). In addition, no differences in the amount of E12 or E47 protein were detected by Western blot analysis of nuclear extracts prepared from EBF- or neomycin-expressing macrophage clones (data not shown). In contrast, an increase in binding to the Pax-5 probe could be detected in two of the EBF-expressing lines consistent with the increase in Pax-5 mRNA (Fig. 7 C). Surprisingly, the amount of EBF oligo shifted by EMSA was significantly lower than that in the 2C1 line (Fig. 7 C). We estimate that the amount of EBF binding to the EBF probe by EMSA is between two- and sixfold lower than that observed in the 70Z/3 pre-B line (compare 2C1 to 70Z/3 pre-B in Fig. 3 C). This estimate is consistent with the level of expression observed by others after transfection of EBF into Ba/F3 cells (47). However, we have found it difficult to quantify the amount of EBF since binding to the EBF oligo, in our EMSA, does not appear to be linearly related to the concentration of EBF in the extract. In the absence of an EBF-specific antibody we were unable to determine whether all of the cells in each clone expressed EBF, however, the phenotype of the EBF-expressing macrophage clones suggests that sufficient levels of EBF are expressed to mediate gene activation in these cells. At the present time we can not exclude the possibility that an increase in the amount of EBF may have led to activation of additional B lineage genes.
We also examined the EBF-expressing macrophage clones for mRNA encoding the myeloid associated genes c-fms and Id2, by Northern blot analysis. Unlike the E12-expressing clones, the EBF clones did not show a decrease in the expression of c-fms (Fig. 7 A). We did detect a two- to threefold decrease in the level of Id2 expression; however, this was also observed in a few of our neomycin transfected macrophage clones suggesting that this decrease was not the result of EBF expression directly (data not shown). The pattern of B lineage–associated traits detected in the E12- or EBF-expressing macrophage lines is summarized in Table 1. Taken together, our data indicate that, in the context of this macrophage cell line, EBF was able to regulate a subset of E12-inducible traits including expression of λ5, Pax-5, and responsiveness of κ light chain to LPS. These observations suggest that one of the critical functions of E12 may be to activate EBF and together these factors coordinately regulate multiple B lineage–associated genes.
Table 1.
Summary of B Lineage Traits in 70Z/3 Pre-B, Macrophage and E12- or EBF-Expressing Macrophage Cell Lines
| Cell lines | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pre-B | Macrophage | Macrophages expressing | ||||||||
| E12 | bHLH | EBF | ||||||||
| E2A | +++ | − | +++≳ | − | − | |||||
| EBF | +++ | − | +++ | − | +++≳ | |||||
| Pax-5 | +++ | + | +++ | + | +++ | |||||
| λ5 | +++ | − | +++ | ++ | ++ | |||||
| IL7Rα | +++ | − | +++ | ++ | − | |||||
| Rag-1 | − | − | +++ | − | − | |||||
| κ Induction | +++ | − | +++ | − | ++ | |||||
Expression due to transfected construct.
Discussion
The progression of hematopoietic progenitors through the earliest detectable stage of B cell development requires the activity of the transcription factors encoded by the E2A and EBF genes (18, 19, 20). The absence of B lineage cells in mice that lack these genes has made it difficult to examine the role of each factor in promoting, or maintaining, the B lineage developmental program. In this paper we have described a novel cell line model in which the role of the E2A and EBF proteins in regulating the expression of B lineage traits can be studied. We have shown that the spontaneous loss of B lineage genes from the 70Z/3 pre-B cell line is associated with a loss of E2A and EBF DNA binding activity. Moreover, expression of the E2A protein E12 in this macrophage line results in the induction of a large number of B lineage traits including mRNA encoding EBF, Pax-5, λ5, IL7Rα, and Rag-1, and the ability to activate κ light chain in response to LPS. Our data indicate that activation of EBF may be one of the functions of E12 in regulating the B lineage phenotype in these cells. This hypothesis is supported by the finding that ectopic expression of EBF in the macrophage line results in the activation of a subset of E12-induced genes including Pax-5, λ5, and κ light chain responsiveness to LPS, but not expression of E2A, IL7Rα, or Rag-1. Taken together, our data suggest, that in the hierarchy of transcription factors regulating B cell development, E12 functions upstream of EBF. However, E12 may interact with EBF or Pax-5 to regulate expression of B lineage genes during B cell development.
The ability of E12 to activate most B lineage traits is dependent on the amino-terminal transactivation domains suggesting that E12 does not function by releasing an alternative bHLH protein from inhibition by Id2. Furthermore, the requirement for the transactivation domains suggest that E12 does not function in a manner analogous to the myogenic bHLH protein MyoD. The bHLH domain alone of MyoD has been shown to be sufficient for the activation of muscle-specific genes and the induction of the myogenic program (75). Recent studies have shown that the MyoD bHLH domain is able to interact with a muscle-specific transcription factor, MEF2, and together these proteins demonstrate a transactivation potential that is not found in either protein alone (70). In addition, a novel MyoD domain, NH2-terminal to the basic region, has recently been implicated in the remodeling of chromatin thereby leading to increased activation of the myogenin promoter (73). The bHLH domain of E2A is able to interact with p300/CBP (creb-binding protein) which associates with a histone acetyltransferase activity and could, therefore, mediate chromatin restructuring in a manner analogous to that of MyoD (76, 77). Such a mechanism may contribute the activation of the IL7Rα and λ5 genes observed in the bHLH domain only expressing lines. However, the low level of activation observed in the presence of very high levels of bHLH protein suggest that the transactivation domains of E12 also contribute significantly to expression of these genes (see Fig. 5). Taken together, our data suggest that the transactivation domains of E12, or additional sequences found in the NH2-terminal domain, are required for the activation of the majority of B lineage associated traits in the 70Z/3 macrophage cell line.
Our studies provide the first demonstration of activation of the endogenous EBF gene in response to expression of E12. The finding that EBF is a downstream target of E12 was somewhat surprising given that EBF expression can be detected, at very low levels, in the fetal liver and bone marrow of E2A-deficient mice (18, 29). This discrepancy can be accounted for in at least one of two ways. First, EBF is expressed in adipocytes as well as B lineage cells (45). Therefore, the low level of EBF observed in E2A-deficient mice may be the result of expression in non-B lineage cells. Second, two additional bHLH transcription factors, HEB and E2-2, are expressed in B cell progenitors. These transcription factors bind to the same DNA sequence as the E2A proteins and play an important, although nonessential, role in B lymphopoiesis (78). It is possible that in the absence of E2A, HEB or E2-2 can regulate the expression of EBF. Alternatively, the regulation of EBF by E12 may be unique to this cell line. Although E12 is able to regulate the expression of EBF in the context of this macrophage line it appears that this is not its sole function in regulating B lineage progression since expression of EBF alone does not result in the activation of all of the genes induced by E12.
Ectopic expression of either E12 or EBF lead to an increase in mRNA encoding the B lineage–associated transcription factor Pax-5. Surprisingly, expression of Pax-5 was not completely extinguished during the conversion of 70Z/3 pre-B lymphocytes to the macrophage phenotype indicating that neither factor is required for the continued expression of this gene. Interestingly, it has been suggested that the Pax-5 gene is a target of the IL7R signaling cascade (79), yet we find expression of Pax-5 in the macrophage line and an upregulation of Pax-5 transcripts in EBF-expressing clones that do not express IL7R transcripts. Although expression of the IL7R itself may not be required for Pax-5 expression in these cells, our data do not exclude the possibility that Pax-5 is a target of the transcription factors that are activated by the IL7R signaling cascade. Recent studies have demonstrated that gene regulation by Pax-5 is exquisitely sensitive to the cellular concentration of this protein (80). Accordingly, the threefold decrease in Pax-5 DNA binding activity observed in the macrophage line may have a dramatic effect on the expression of Pax-5 target genes. Pax-5 functions as a repressor of some genes, such as the immunoglobulin J chain, and an activator of others, such as CD19 (81, 82). The affinity of a given Pax-5–binding site, its context within the promoter and the relative concentration of Pax-5 have all been postulated to contribute to the ability of Pax-5 to activate or repress a given gene (80). In our clones it is possible that the low level of Pax-5 observed in the parental macrophage clone is sufficient to synergize with the transfected E12 or EBF to activate genes that may not have been successfully activated by either transcription factor alone. Similarly, the increased level of Pax-5 observed in E12- or EBF-expressing macrophage clones is likely to contribute to the pattern of gene activation in these clones.
The induction of κ light chain in pre-B cell lines has been associated with the activation of at least three distinct DNA binding activities within the κ enhancer, NF-κB, Oct-2, and KBF-A in addition to E2A (83–86). We have found no differences in the ability of 70Z/3 pre-B cells and macrophages to activate any of these factors in response to LPS (Kee, B.L., unpublished data). It was unexpected to find that expression of EBF in the macrophage line led to the ability to induce κ in response to LPS. It is not certain whether this is the result of EBF activity on the κ locus itself, or whether EBF activates a B lineage–associated gene that influences κ expression. However, binding sites for EBF have been identified in the promoters of a number of κ light chain genes within the VκII and VκV families (87). The 5′ flanking region of the κ gene expressed in 70Z/3 pre-B cells does contain a potential EBF-binding site (88). EBF interaction with its binding site in immature B cells has been suggested to lead to inhibition of κ gene expression (87). Therefore, it remains to be determined whether a modification of EBF in response to LPS may relieve this repressive effect or whether κ induction in EBF-expressing clones is the result of a novel EBF-regulated gene. In addition, it remains to be determined whether κ induction proceeds through the same pathway in E12- and EBF-expressing lines since multiple combinations of enhancer binding proteins have been implicated in the regulation of κ transcription (86, 89, 90).
EBF and E47 have been shown previously to synergize to activate the endogenous λ5 and VpreB genes in Ba/F3 cells (47). Our results are consistent with these findings since the level of λ5 is greater in the E12-expressing clones, which also express EBF, than in EBF only–expressing clones. However, expression of EBF alone in this macrophage line led to levels of λ5 which were easily detected by Northern blot analysis. We speculate that the ability of EBF to activate λ5 in this context, but not in Ba/F3 cells, may be due to the presence of other B lineage factors, such as Pax-5, in this cell line. The λ5 promoter contains binding sites for E2A, EBF, Pax-5, and Ikaros proteins and it is possible that any combination of these proteins might be involved in the regulation of this gene. Our data also suggest that in the context of this macrophage line EBF is not absolutely required for λ5 expression since the bHLH domain could activate low levels of λ5 in the absence of EBF.
Expression of E12 in the 70Z/3 macrophage cell line led to the expression of two genes, IL7Rα and Rag-1, which were not induced significantly by expression of EBF. Although EBF expression is not sufficient for the activation of either of these genes, our results to not exclude the possibility that EBF may play a role in their regulation or that higher levels of EBF may have activated these genes. EBF may function in the regulation of Rag genes in combination with E2A and/or Pax-5. However, it seems unlikely that EBF is required for the activation of IL7Rα expression during B cell development since we have found that expression of IL7Rα precedes the expression of EBF by at least two days in cultures of lineage unrestricted B cell progenitors differentiating in vitro (Kee, B.L., unpublished data). In addition, expression of IL7Rα transcripts is less affected in EBF-deficient mice than the expression of a number of additional B lineage associated genes (20). Further studies will be required to determine whether the activity of EBF is required for activation of these genes.
The 70Z/3 cell line is one of a number of B lineage lines that are capable of losing some of their B cell characteristics and acquiring characteristics of macrophages (56, 58, 59, 60–63, 91). However, 70Z/3 is unique in that it is one of the few lines that undergoes this transition spontaneously in vitro. Like most of the other lineage “switching” cell lines 70Z/3 macrophage cells have never shown reversion to the B lineage phenotype, despite numerous attempts to identify such revertants (64; Kee, B.L., unpublished data). Therefore, it is particularly surprising that expression of a single cDNA (E12 or EBF) can lead to the expression of such a large number of B lineage genes in this macrophage line. In addition, it is remarkable that the loss of E2A and EBF is correlated with the loss of expression of such a large number of B lineage–associated traits. We have examined two other pre-B cell–derived macrophage cell lines, P388D1 and Raw 309, for the expression of E2A and EBF and have found that they also lack expression of both E2A and EBF (Kee, K.L. unpublished data). This finding is consistent with the hypothesis that loss of E2A and EBF are an essential feature of the transition of B lineage cells to the macrophage phenotype. Unfortunately, our attempts to create stable E12 expressing lines from these cells has been unsuccessful, potentially owing to the ability of E2A proteins to inhibit cell growth.
It is also piquant that the loss of the B lineage–associated traits is associated with an increase in macrophage–like characteristics. In general, each of the lineage switching B cell lines has in common the expression of CD5, indicating that they are derived from progenitors which originate during fetal development (reviewed in reference 57). Some B cell progenitors that develop during fetal life have been shown to derive from a bipotent B cell and/or macrophage progenitor (92). This finding is consistent with the hypothesis that the genetic programs regulating the early B lineage or macrophage differentiation programs may diverge by only a few genetic events. Alternatively, macrophages may represent a default pathway that is followed in the absence of lineage specifying genes. Future experiments will be necessary to determine whether E2A proteins are involved in the regulation of cell fate decisions of bipotent B cell/ macrophage progenitors or other lineage unrestricted B cell progenitors.
It is interesting that targeted deletion of the majority of the genes induced by E12 or EBF, such as λ5, IL7Rα, and Rag-1, leads to an arrest in B lymphopoiesis at a later developmental stage than that seen in E2A- or EBF-deficient mice (7, 8, 11, 93–96). Therefore, novel targets of E2A or EBF are likely to be required for progression through the earliest stages of the B lineage pathway. This cell line model provides a useful system by which to identify such essential E2A and EBF target genes.
Acknowledgments
We are indebted to Dr. C.J. Paige for introducing us to the 70Z/3 switching system, for providing the macrophage cell line, communicating unpublished data, and numerous valuable discussions. We also gratefully acknowledge the contributions of Drs. Gretchen Bain and Eben Massari in the form of reagents, discussions, and reading of the manuscript. We thank Dr. R. Reed for the Olf/EBF cDNA and Dr. S. Hedrick for the pHβAPneo expression vector.
Abbreviations used in this paper
- bHLH
basic helix-loop-helix
- c-fms
macrophage colony-stimulating factor
- EBF
early B cell factor
- EMSA
electrophoretic mobility shift assay
- RT
reverse transcriptase
Footnotes
B.L. Kee was supported by a fellowship from the Medical Research Council of Canada and is a Special Fellow of the Leukemia Society of America. This work was funded by the National Institutes of Health (NIH), the Council for Tobacco Research and the Edward Mallinckrodt Jr. Foundation.
References
- 1.Till JE, McCulloch EA. Hemopoietic stem cell differentiation. Biochim Biophys Acta. 1980;605:431–459. doi: 10.1016/0304-419x(80)90009-8. [DOI] [PubMed] [Google Scholar]
- 2.Cumano A, Paige CJ. Enrichment and characterization of uncommitted B-cell precursors from fetal liver at day 12 of gestation. EMBO (Eur Mol Biol Organ) J. 1992;11:593–601. doi: 10.1002/j.1460-2075.1992.tb05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billips LG, Petitte D, Dorshkind K, Narayanan R, Chiu C-P, Landreth KS. Differential roles of stromal cells, interleukin-7, and kit-ligand in the regulation of B lymphopoiesis. Blood. 1992;79:1185–1192. [PubMed] [Google Scholar]
- 4.Yancopoulos GD, Alt FW. Regulation of the assembly and expression of variable-region genes. Annu Rev Immunol. 1986;4:339–368. doi: 10.1146/annurev.iy.04.040186.002011. [DOI] [PubMed] [Google Scholar]
- 5.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 6.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 7.Shinkai Y, Rathburn G, Lam K-P, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, Alt FW. Rag-2 deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 8.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. Rag-1 deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 9.Reth M. Antigen receptors on B lymphocytes. Annu Rev Immunol. 1993;10:98–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- 10.Rolink A, Haasner D, Melchers F, Andersson J. The surrogate light chain in mouse B-cell development. Int Rev Immunol. 1996;13:341–356. doi: 10.3109/08830189609061757. [DOI] [PubMed] [Google Scholar]
- 11.Loffert D, Schaal S, Ehlich A, Hardy RR, Zou Y-R, Muller W, Rajewsky K. Early B-cell development in the mouse: Insights from mutations introduced by gene targeting. Immunol Rev. 1994;137:135–153. doi: 10.1111/j.1600-065x.1994.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 12.Alt FW, Blackwell TK, DePinho RA, Reth MG, Yancopoulos GD. Regulation of genome rearrangement events during lymphocyte differentiation. Immunol Rev. 1986;89:5–30. doi: 10.1111/j.1600-065x.1986.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 13.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robb L, Elwood NJ, Elefanty AG, Kontgen F, Li R, Farnett LD, Begley CG. The SCL gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO (Eur Mol Biol Organ) J. 1996;15:4123–4129. [PMC free article] [PubMed] [Google Scholar]
- 15.Georgopoulos K, Bigby M, Wang J-H, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 16.Scott E, Simon M, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 17.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO (Eur Mol Biol Organ) J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 18.Bain G, Robanus EC, Maandag, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 19.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 20.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 21.Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax-5/BSAP. Cell. 1994;79:901–913. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 22.Nutt SL, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 23.Murre C, McCaw PS, Vaessin H, Caudy M, Jan L, Jan YN, Cabrera CV, Buskin JM, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 24.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD and mycproteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 25.Sen R, Baltimore D. Multiple factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- 26.Ernst P, Smale ST. Combinatorial regulation of transcription II: the mu immunoglobulin heavy chain gene. Immunity. 1995;2:427–438. doi: 10.1016/1074-7613(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 27.Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 29.Bain G, Robanus EC, Maandag, te Riele HPJ, Feeney AJ, Sheehy A, Schlissel M, Shinton SA, Hardy RR, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 30.Lassar A, Davis R, Wright W, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic bHLH proteins requires hetero-oligomerisation with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 31.Lassar, A.B., and H. Weintraub. 1992. The myogenic helix-loop-helix family: regulators of skeletal muscle determination and differentiation. In Transcriptional Regulation. S.L. McKnight and K.R. Yamamoto, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1037–1061.
- 32.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 33.Naya FJ, Huang H-P, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai M-J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen C-P, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benezra R. An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell. 1994;79:1057–1067. doi: 10.1016/0092-8674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 36.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 37.Sun X-H, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun X-H. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 39.Jacobs Y, Xin X-Z, Dorshkind K, Nelson C. Pan/E2A expression precedes immunoglobulin heavy-chain expression during B lymphopoiesis in nontransformed cells, and Pan/E2A proteins are not detected in myeloid cells. Mol Cell Biol. 1994;14:4087–4096. doi: 10.1128/mcb.14.6.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xin XQ, Nelson C, Collins C, Dorshkind K. Kinetics of E2A HLH protein expression during myelopoiesis and primary B cell differentiation. J Immunol. 1994;152:16351–16356. [PubMed] [Google Scholar]
- 41.Voronova AF, Lee F. The E2A and tal-1 helix-loop-helix proteins associate in vivoand are modulated by Id proteins during interleukin 6-induced myeloid differentiation. Proc Natl Acad Sci USA. 1994;91:5952–5956. doi: 10.1073/pnas.91.13.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreider BL, Benezra R, Rovera G, Kadesch T. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science. 1992;255:1700–1702. doi: 10.1126/science.1372755. [DOI] [PubMed] [Google Scholar]
- 43.Cooper CL, Brady G, Billia F, Iscove NN, Quesenbery PJ. Expression of the Id family helix-loop-helix regulators during growth and development in the hematopoietic system. Blood. 1997;89:3155–3165. [PubMed] [Google Scholar]
- 44.Wang MW, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 45.Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 46.Hagman J, Gutch MJ, Lin H, Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO (Eur Mol Biol Organ) J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 48.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1477. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bain G, Gruenwald S, Murre C. E2A and E2-2 are subunits of B-cell-specific E2-box DNA-binding proteins. Mol Cell Biol. 1993;13:3522–3529. doi: 10.1128/mcb.13.6.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagman J, Travis A, Grosschedl R. A novel lineage-specific nuclear factor regulates mb-1gene transcription at the early stages of B cell differentiation. EMBO (Eur Mol Biol Organ) J. 1991;10:3409–3417. doi: 10.1002/j.1460-2075.1991.tb04905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer μE5/μE2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 52.Fitzsimmons D, Hodsdon W, Wheat W, Maira S-M, Wasylsk B, Hagman J. Pax-5 (BSAP) recruits ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promotor. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 53.Singh H, Sen R, Baltimore D, Sharp PA. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 54.Gunning P, Leavitt J, Muscat G, Ng SY, Kedes L. A human β-actin expression vector system directs high-level accumulation of anti-sense transcripts. Proc Natl Acad Sci USA. 1987;84:4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale S. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14:711–723. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hara H, Sam M, Maki RA, Wu GE, Paige CJ. Characterization of a 70Z/3 pre-B cell derived macrophage clone. Differential expression of Hox family genes. Int Immunol. 1990;1:691–695. doi: 10.1093/intimm/2.8.691. [DOI] [PubMed] [Google Scholar]
- 57.Borrello MA, Phipps RP. The B/macrophage cell: an elusive link between CD5+B lymphocytes and macrophages. Immunol Today. 1996;17:471–475. doi: 10.1016/0167-5699(96)20031-b. [DOI] [PubMed] [Google Scholar]
- 58.Borzillo GV, Sherr CJ. Early pre-B cell transformation induced by the v-fmsoncogene in long-term mouse bone marrow cultures. Mol Cell Biol. 1989;9:3973–3981. doi: 10.1128/mcb.9.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borzillo GV, Ashmun RA, Sherr CJ. Macrophage lineage switching of murine early pre-B lymphoid cells expressing transduced fms genes. Mol Cell Biol. 1990;10:2703–2714. doi: 10.1128/mcb.10.6.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson WF, Pierce JH, Rudikoff S, Morse HC., III Relationships between B cell and myeloid differentiation. J Exp Med. 1988;168:389–407. doi: 10.1084/jem.168.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klinken SP, Alexander WS, Adams JM. Hemopoietic linage switch: v-raf oncogene converts Eμ-myctransgenic B cells into macrophages. Cell. 1988;53:857–867. doi: 10.1016/s0092-8674(88)90309-1. [DOI] [PubMed] [Google Scholar]
- 62.Strasser A, Elefanty AG, Harris AW, Cory S. Progenitor tumours from Eμ-bcl-2-myctransgenic mice have lymphomyeloid differentiation potential and reveal developmental differences in cell survival. EMBO (Eur Mol Biol Organ) J. 1996;15:3823–3834. [PMC free article] [PubMed] [Google Scholar]
- 63.Principato M, Cleveland JL, Rapp UR, Holmes KL, Pierce JC, Morse HC, III, Klinken SP. Transformation of murine bone marrow cells with combined v-raf-v-myconcogenes yields clonally related mature B cells and macrophages. Mol Cell Biol. 1990;10:3562–3568. doi: 10.1128/mcb.10.7.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka T, Wu GE, Paige CJ. Characterization of the B cell-macrophage transition in 70Z/3 cells. Eur J Immunol. 1994;24:1544–1548. doi: 10.1002/eji.1830240713. [DOI] [PubMed] [Google Scholar]
- 65.Paige CJ, Kincade PW, Ralph P. Murine B cell leukemia line with inducible surface immunoglobulin expression. J Immunol. 1978;121:641–647. [PubMed] [Google Scholar]
- 66.Schlissel M, Voronova A, Baltimore D. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain transcription and rearrangement in a pre-T-cell line. Genes Dev. 1991;5:1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- 67.Choi JK, Shen C-P, Radomska HS, Eckhardt LA, Kadesch T. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO (Eur Mol Biol Organ) J. 1996;15:5014–5021. [PMC free article] [PubMed] [Google Scholar]
- 68.Sun XH, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- 69.Molkentin JD, Olson EM. Defining the regulatory networks for muscle development. Curr Opin Genet Dev. 1996;6:445–453. doi: 10.1016/s0959-437x(96)80066-9. [DOI] [PubMed] [Google Scholar]
- 70.Black BL, Molkentin JD, Olson E. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interactions with MEF2. Mol Cell Biol. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quong MW, Massari ME, Zwart R, Murre C. A new transcriptional activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol. 1993;12:792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Massari ME, Jennings PA, Murre C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiaeand mammalian cells. Mol Cell Biol. 1996;16:121–129. doi: 10.1128/mcb.16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 74.Nutt SL, Morrison AM, Dorfler P, Rolink A, Busslinger M. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO (Eur Mol Biol Organ) J. 1998;17:2319–2333. doi: 10.1093/emboj/17.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tapscott SJ, Davis RL, Thayer MJ, Cheng P-F, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 76.Eckner R, Yao R-P, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 77.Yang X-J, Ogryzko VV, Nishikawa J, Howard BJ, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 78.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2 and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corcoran AE, Riddell A, Krooshoop D, Benkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- 80.Wallin JJ, Gackstetter ER, Koshland ME. Dependence of BSAP repressor and activator functions on BSAP concentration. Science. 1998;279:1961–1964. doi: 10.1126/science.279.5358.1961. [DOI] [PubMed] [Google Scholar]
- 81.Rinkenberger JL, Wallin JJ, Johnson KW, Koshland ME. An interleukin-2 signal relieves BSAP (Pax5)- mediated repression of the immunoglobulin J. chain gene. Immunity. 1998;5:365–376. doi: 10.1016/s1074-7613(00)80263-0. [DOI] [PubMed] [Google Scholar]
- 82.Kozmik Z, Wang S, Dorfler P, Adams B, Busslinger M. The promoter of the CD19 gene is a target for the B cell specific transcription factor BSAP. Mol Cell Biol. 1992;12:2662–2672. doi: 10.1128/mcb.12.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein NF-kappa B by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 84.Staudt LM, Singh H, Sen R, Wirth T, Sharp PA, Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986;323:640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- 85.Nelms K, Hromas R, Van Ness B. Identification of a second inducible DNA-protein interaction in the kappa immunoglobulin enhancer. Nucleic Acid Res. 1990;18:1037–1043. doi: 10.1093/nar/18.4.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shaffer AL, Peng A, Schlissel MS. In vivo occupancy of the κ light chain enhancers in primary pro- and pre-B cells: a model for κ locus activation. Immunity. 1997;6:131–143. doi: 10.1016/s1074-7613(00)80420-3. [DOI] [PubMed] [Google Scholar]
- 87.Sigvardsson M, Akerblad P, Leanderson T. Early B cell factor interact with a subset of κ promoters. J Immunol. 1996;156:3788–3796. [PubMed] [Google Scholar]
- 88.Parslow RG, Blair DL, Murphy WJ, Granner DK. Structure of the 5′ ends of immunoglobulin genes: A novel conserved sequence. Proc Natl Acad Sci USA. 1984;81:2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O'Brien DP, Oltz EM, Van Ness BG. Coordinate transcription and V(D)J recombination of the kappa immunoglobulin light-chain locus: NF-κB-dependent and -independent pathways of activation. Mol Cell Biol. 1997;17:3477–3487. doi: 10.1128/mcb.17.7.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Briskin M, Kuwabara MD, Sigman DS, Wall R. Induction of kappa transcription by interferon-gamma without activation of NF-kappa B. Science. 1988;242:1036–1037. doi: 10.1126/science.3143155. [DOI] [PubMed] [Google Scholar]
- 91.Borrello MA, Phipps RP. Fibroblasts support outgrowth of splenocytes simultaneously expressing B lymphocyte and macrophage characteristics. J Immunol. 1995;155:4155–4161. [PubMed] [Google Scholar]
- 92.Cumano A, Paige CJ, Iscove NN, Brady G. Bipotential precursors of B cells and macrophages in murine fetal liver. Nature. 1992;356:612–615. doi: 10.1038/356612a0. [DOI] [PubMed] [Google Scholar]
- 93.Kitamura D, Roes J, Kuhm R, Rajewsky K. A B cell deficient mouse generated through targeted disruption of the membrane exon of the immunoglobulin μ chain. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 94.Kitamura D, Kudo A, Schaal S, Muller W, Melchers F, Rajewsky K. A critical role of λ5 protein in B cell development. Cell. 1992;69:823–831. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 95.Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1959. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.von Freeden-Jeffrey U, Vieira P, Lucian LA, McNeil T, Burdach SEG, Murray R. Lymphopenia in IL-7 gene deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]