Abstract
The potential involvement of early growth response (Egr)-1, a zinc-finger transcription factor belonging to the immediate-early genes, in positive/negative selection of thymocytes has been implicated by its expression in the population of CD4+CD8+ double positive (DP) cells undergoing selection. To further investigate this possibility, transgenic mice overexpressing Egr-1 in thymocytes were bred with a transgenic mouse line expressing a T cell receptor (TCR) recognizing the H-Y male antigen in the context of H-2b class I major histocompatibility complex (MHC) molecules. In Egr-1/TCR H-Y double-transgenic mice, efficient positive selection of H-Y CD8+ T cells occurred, even in mice on either a nonselecting H-2d background or a β2-microglobulin (β2m)-deficient background in which the expression of class I MHC heavy chains is extremely low; no positive selection was observed on a Kb−/−Db−/−β2m−/− background where class I MHC expression is entirely absent. Similarly, when the Egr-1 transgene was introduced into a class II MHC–restricted TCR transgenic mouse line, Egr-1/TCR double-transgenic mice revealed increased numbers of CD4+ T cells selected by class II MHC, as well as significant numbers of CD8+ T cells selected by class I MHC (for which the transgenic TCR might have weak affinity). Thus, Egr-1 overexpression allows positive selection of thymocytes via TCR–MHC interactions of unusually low avidity, possibly by lowering the threshold of avidity required for positive selection. Supporting this possibility, increased numbers of alloreactive T cells were positively selected in Egr-1 transgenic mice, resulting in a strikingly enhanced response against allo-MHC. These results suggest that expression of Egr-1 and/or its target gene(s) may directly influence the thresholds required for thymocyte selection.
Keywords: Egr-1, positive selection, T cell, avidity, transgenic mouse
Positive/negative selection of thymocytes at the CD4+CD8+ double positive (DP)1 stage is the key checkpoint for thymocyte maturation, at which their fate—whether they develop further to CD4+ or CD8+ single positive (SP) cells or die by clonal deletion—is decided (1, 2). Accumulating evidence indicates that the avidity between T cell receptors (TCRs) expressed on thymocytes and MHC/antigen-peptide complexes displayed on the surface of APCs appears to define the type of intracellular signals generated, which promote thymocytes to be either positively selected (further developing to the SP stage) or negatively selected (clonally deleted); avidity is dependent on TCR–MHC affinity and the expression levels of both complexes (3, 4). It is believed that there are thresholds in the strength of TCR–MHC avidity that determine the nature and consequences of subsequent TCR signaling (3, 4). When thymocytes and APCs interact with high avidity, cells are negatively selected through the apoptotic pathway, whereas thymocytes are promoted to the SP cell stage when the avidity is moderate but sufficient. If the avidity is too low, cells cannot undergo either type of selection, resulting in their death as “neglected cells.” However, the molecular mechanism that defines these thresholds for selection events is unclear.
Signals generated during positive/negative selection must be differentially controlled; hence, rapidly responding transcriptional regulators able to elicit a cascade of changes in gene expression should be important. Immediate-early genes, expression of which is rapidly induced after cell-surface receptor ligation without de novo protein synthesis, are strong candidates for such a rapid response mediator (5). Shao et al. reported recently that the expression of one of these immediate-early genes, early growth response (Egr)-1, a zinc-finger transcription factor rapidly induced by TCR ligation, appears to correlate with selection events, as it is expressed at much lower levels in DP cells from mutant mice deficient for both MHC class I and class II molecules than in DP cells from wild-type mice. In addition, Egr-1 expression in the mutant DP cells was able to be upregulated by anti-CD3 mAb ligation (6).
We recently generated transgenic mouse lines overexpressing Egr-1 in thymocytes under the control of the lck-proximal promoter (7), and showed that in Egr-1 transgenic mice on a recombination-activating gene (RAG)- deficient background, thymocytes bypassed the block at the CD25+CD44− DN stage and matured to the immature single positive (ISP) cell stage. Here, the effect of Egr-1 expression on positive/negative selection is extensively analyzed by breeding Egr-1 transgenic mice with various TCR transgenic mice, as well as by evaluating positive selection of alloreactive T cells in Egr-1 transgenic mice. This report provides interesting insight into what is actually required for positive selection and what transcriptional pathways may be involved in the selection events.
Materials and Methods
Mice.
All mice used here were maintained in the specific pathogen–free facility of the Basel Institute for Immunology. Matched sets of littermates with transgene-positive and -negative genotypes were used in all experiments.
Serological Reagents and Flow Cytometry.
Reagents used for staining T cells and subsets thereof were as described (7–9, and references therein). Thymus, lymph node, and spleen cells were stained with saturating levels of mAbs and analyzed using a FACSCalibur® cytometer (Becton Dickinson, San Jose, CA).
Cell Survival Assay.
H-Y CD8+ SP cells were sorted after staining thymocytes for CD4, CD8, T3.70, and peanut agglutinin receptor (PNAr). 5 × 105 cells from each type of mice were cultured in triplicate in 96-well flat-bottomed microculture plates in complete DMEM culture medium including 10% FCS. 24, 48, 72, 96, and 120 h after starting the culture, the number of living cells was evaluated by trypan blue exclusion and plotted as a survival curve. H-Y CD8+ SP cells from all types of mice showed a similar number of living cells at each time point.
Purification of CD8+ T Cells.
CD8+ T cells were purified by incubating spleen cells for 30 min on ice with 10% (vol/vol) culture supernatant of hybridomas RL172 (IgM antibody to CD4) and M5/114 (antibody to class II MHC), washed, and then incubated for an additional 1 h at 37°C in 10% Lo-Tox complement (Cedarlane Labs Ltd., Hornby, Ontario, Canada). Dead cells were excluded by Ficoll gradation. Living cells were washed twice and resuspended in complete DMEM.
T Cell Proliferation Assay.
Purified CD8+ T cells (5 × 105/ well) were cultured in the presence of various concentrations of either immobilized anti-CD3 mAb (KT-3; provided by Dr. C. Benoist, Strasbourg, France) or Con A (Sigma Chemical Co., St. Louis, MO). The KT-3 mAb was immobilized by preincubation of wells at 4°C overnight. All cultures were performed for 3 d, and proliferation was assessed by [3H]thymidine incorporation in the last 16 h of culture.
In Vivo and In Vitro Recognition of H-Y Antigen.
This is a modification of a similar experiment described elsewhere (10). In brief, purified CD8+ T cells including 2 × 106 H-Y CD8+ T cells calculated by the percentage of T3.70+ population in the CD8+ cells after fluorocytometric analysis, were suspended in 200 μl of PBS, and injected intravenously into 6–8-wk-old male and female Cα-deficient mice. In one experiment, cells from each type of donor mouse were injected into two pairs of male and female recipient mice. 4 d after injection, spleen cell suspension from each recipient was stained for CD8 and T3.70, and analyzed by FACScan® (Becton Dickinson). Absolute number of H-Y CD8+ T cells was calculated from spleen cell number and percentage of CD8+T3.70+ cell population. For the in vitro proliferation assay, purified CD8+ T cells from each type of mouse, including various numbers of H-Y CD8+ T cells, were stimulated by 8 × 105/ well of 3,000-rad irradiated spleen cells from male or female B6 mice in complete DMEM in 96-well microculture plates. Cultures were performed for 5 d, and proliferation was assessed by [3H]thymidine incorporation in the last 16 h of culture.
MLRs.
Responders (B cell–depleted lymph node cells) were cultured with stimulators (3,000-rad irradiated spleen cells) at various ratios for 5 d in complete DMEM in flat-bottomed 96-well microculture plates. Proliferation was assessed by [3H]thymidine incorporation in the last 16 h of culture.
Bone Marrow Chimeras.
To construct bone marrow chimeras, recipient mice were 10-Gy (from a cobalt source) irradiated. 24 h later, they were injected intravenously with 107 bone marrow cells from which mature T and B cells had been depleted by treatment with anti-CD4 (RL172) and anti-CD8 (31M) mAbs plus Lo-Tox complement (Cedarlane Labs Ltd.). After grafting, mice were rested for 5 wk to allow reconstitution before analysis.
Results
Efficient Positive Selection of H-Y CD8+ T Cells by Low Avidity TCR–MHC Interactions in the Presence of Egr-1 Overexpression.
As described in our previous report, Egr-1 transgenic mice (Egr-1–TG) exhibit transgene expression at levels far exceeding endogenous Egr-1, when analyzed by Northern blotting using thymic RNA from transgenic and negative littermate mice (7). As observed for many other transgenes driven by the lck-proximal promoter (11), Egr-1 transgene expression was detected in a wide range of developmental stages of thymocytes of Egr-1–TG, including DP cells, when analyzed by the reverse transcription PCR method (12) using RNA from sorted cells from each population (data not shown). No obvious transgene expression was detected either in spleen or lymph node by Northern blot analysis, indicating thymocyte-specific transgene expression (7).
To assess the possible effect of Egr-1 expression on thymocyte selection, Egr-1–TG were cross-bred with transgenic mice expressing a TCR recognizing the H-Y male antigen in the context of H-2b class I, Db molecules. It is well-established that thymocytes expressing H-Y–specific αβ TCRs are positively selected in female H-2b/b mice and negatively selected in male H-2b/b mice, but do not undergo either positive or negative selection in female or male mice on an H-2d/d background (8, 13). Fig. 1 a shows CD4/CD8 profiles of mature thymocytes bearing the H-Y TCR defined as T3.70 (mAb against H-Y TCR α chain)high, PNArlow. As shown in the upper panels, in H-2b/b females, both Egr-1-TG with an H-Y TCR transgene (Egr/H-Y) and Egr-1–TG-negative littermate mice with an H-Y TCR transgene (NL/H-Y) harbored mature H-Y CD8+ SP thymocytes. The CD8+ SP cells in these profiles do not contain immature CD8 SPs (ISPs), which exhibit high levels of PNAr (7). However, in H-Y mice, since expression levels of transgenic H-Y TCR in DN cells (which are PNArlow) are as high as those in mature SP cells (8), a significant proportion of DN cells as well as mature SP cells are plotted in these profiles. The numbers of total thymocytes were slightly larger in Egr/H-Y mice than in NL/H-Y mice (Table 1). However, the absolute numbers of mature H-Y CD8+ SP cells were ∼1.7 times higher in Egr/H-Y than in NL/H-Y mice (Table 1). In line with this, the percentages of T cells bearing the transgenic α chain (T3.70+) in the DP and the total mature CD8+ populations were larger in Egr/H-Y than in NL/H-Y mice (Table 1). Thus, H-Y CD8+ T cells appear to be more efficiently positively selected in the presence of Egr-1 overexpression. More surprisingly, significant numbers of mature H-Y CD8+ SP thymocytes were also observed in female Egr/H-Y mice on either a nonselecting H-2d/d background or a β2m-deficient (β2m0) H-2b/b background (14, 15), in which class I MHC heavy chains are expressed at extremely low levels (Fig. 1 a, middle and bottom; absolute numbers of both total thymocytes and mature H-Y CD8+ SP cells are shown in Table 1). Very few (<5 × 103) mature H-Y CD8+ SP cells were detected in NL/H-Y mice on both H-2d/d and β2m0 backgrounds.
Figure 1.

(a) Thymocyte suspensions from female Egr/H-Y and NL/H-Y mice on H-2b/b, H-2d/d, or β2m0 background were stained for CD4, CD8, H-Y transgenic α chain (T3.70), and PNAr, and analyzed by flow cytometry. CD4/CD8 plots of T3.70high, PNArlow cells are shown. The averages of the absolute numbers of mature H-Y CD8+ SP cells (T3.70high, PNArlow) of each type of mice are indicated in each panel. Five pairs of female Egr/TG and NL/H-Y mice of each background were examined. (b) CD4/CD8 plots of T3.70high, PNArlow thymocytes from female I0β2m0 recipient mice into which female β2m0 Egr/H-Y bone marrow cells (BM) or female β2m0 NL/H-Y bone marrow cells were transplanted (left panels). Right, The same profile of female β2m0 recipient mice into which female β2m0 Egr/H-Y bone marrow cells were grafted. The averages of the absolute numbers of mature H-Y CD8+ SP cells (T3.70high, PNArlow) from two sets of transfer experiments are indicated in each panel. (c) CD4/CD8 plots of T3.70high, PNArlow thymocytes from β2m0 or β2m0II0 Egr/H-Y mice. The averages of the absolute numbers of mature H-Y CD8+ SP cells (T3.70high, PNArlow) of each type of mouse are indicated in each panel. Three pairs of female β2m0 and β2m0II0 Egr/H-Y mice were examined.
Table 1.
Positive Selection of H-Y CD8 T Cells
| Mice |
Absolute number (× 10−7) | % of T3.70+ cells in: | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | H-Y CD8+ | DP | Total CD8+ | |||||||
| Thymus | NL/H-Y (b/b) | 7.1 ± 0.2 | 1.4 ± 0.3 | 33 ± 2.5 | 82 ± 3.4 | |||||
| Egr/H-Y (b/b) | 8.0 ± 0.2 | 2.6 ± 0.2 | 62 ± 6.2 | 90 ± 2.4 | ||||||
| Egr/H-Y (d/d) | 7.0 ± 0.4 | 1.0 ± 0.1 | ND | ND | ||||||
| Egr/H-Y (β2m0) | 7.2 ± 0.1 | 0.9 ± 0.2 | ND | ND | ||||||
| Spleen | NL/H-Y (b/b) | 0.9 ± 0.1 | 52 ± 5.3 | |||||||
| Egr/H-Y (b/b) | 1.5 ± 0.3 | 85 ± 3.5 | ||||||||
| Egr/H-Y (d/d) | 1.6 ± 0.4 | ND | ||||||||
| Egr/H-Y (β2m0) | 1.4 ± 0.2 | ND | ||||||||
Absolute numbers of total thymocytes and mature H-Y CD8+ T cells are shown. Percentages of T3.70+ population in DP or total mature CD8+ cells in the thymus and spleen are also presented. Each value represents the mean of data compiled from five mice ± SD.
H-Y CD8+ T cells were observed in peripheral lymphoid tissues in Egr/H-Y mice on H-2b/b, H-2d/d, and β2m0 backgrounds as well as in H-2b/b NL/H-Y female mice (numbers shown in Table 1). As observed in the thymus, the percentage of T cells bearing the transgenic α chain (T3.70+) in splenic CD8+ cells was larger in H-2b/b Egr/H-Y mice than in H-2b/b NL/H-Y mice (Table 1). Peripheral H-Y CD8+ T cells revealed similar expression patterns of several activation and memory markers such as CD25, CD44, and Mel-14 in all types of mice (data not shown).
Thus, in the presence of Egr-1 overexpression, H-Y thymocytes appear to be efficiently positively selected even by H-2d class I molecules and by the extremely low levels of class I expressed in the absence of β2m, and mature to become peripheral H-Y CD8+ T cells.
Whether positive selection of H-Y CD8+ T cells can occur in the complete absence of class I MHC molecules was assessed by grafting bone marrow cells from female β2m0 Egr/H-Y (H-2b/b) mice into female Kb−/−Db−/− β2m−/− (I0β2m0; H-2b/b) recipient mice, which completely lack class I MHC (16, 17). As shown in Fig. 1 b, no mature H-Y CD8+ SP thymocytes could be observed in the thymi from I0β2m0 recipients into which either β2m0 Egr/H-Y or β2m0 NL/H-Y bone marrow cells were transplanted. In contrast, female β2m0 (H-2b/b) recipients into which β2m0 Egr/H-Y bone marrow cells had been transferred harbored a significant number (6.2 ± 0.6 × 106; n = 2) of mature H-Y CD8+ SP thymocytes. This result strongly indicates that H-Y CD8+ T cells in H-2d/d and β2m0 Egr/H-Y mice were positively selected by H-2d class I molecules or by the extremely low levels of class I expressed in the absence of β2m, but not by other molecules like class II MHC. This was supported by the analysis of β2m, class II MHC doubly deficient (β2m0II0) Egr/H-Y mice, which harbored grossly the same numbers of mature H-Y CD8+ SP thymocytes as β2m0 Egr/H-Y mice (Fig. 1 c).
H-Y CD8+ T cells Selected by Low Avidity TCR–MHC Interactions Are Entirely Functional.
Whether H-Y CD8+ T cells from H-2b/b, H-2d/d, and β2m0 Egr/H-Y mice were functional was determined using matched cells from H-2b/b female NL/H-Y mice as controls. First, purified CD8+ T cells from each type of mouse were stimulated by anti-CD3 mAb or Con A. As shown in Fig. 2, a and b, CD8+ T cells from Egr/H-Y mice on all three backgrounds responded to both anti-CD3 mAb and Con A to a comparable degree as those from H-2b/b female NL/H-Y mice.
Figure 2.
(a and b) CD8+ lymph node T cells were purified from female Egr/H-Y mice (Egr) on H-2b/b, H-2d/d, and β2m0 backgrounds and from female NL/H-Y mice (NL) on H-2b/b by depleting CD4+ cells and class II MHC–positive cells. Cells were stimulated by culture with (a) immobilized anti-CD3 mAb or (b) Con A. In both experiments, proliferation was assessed in duplicate samples by [3H]thymidine incorporation. Data are representative of four independent experiments. (c) Purified CD8+ T cells including 5 × 106 H-Y CD8+ cells in PBS were injected intravenously into male or female Cα−/− recipient mice. 4 d after injection, the numbers of H-Y CD8+ cells in recipient spleens were evaluated and are represented by bars ± SD. Three sets of independent experiments were performed. Numbers of H-Y CD8+ cells in female recipients were identical or less than those of mice with no cells injected [donor (−)], which is indicative of background in the fluorocytometric analysis. (d) Purified CD8+ T cells including the indicated numbers of H-Y CD8+ cells were stimulated by 3,000-rad irradiated spleen cells from male or female B6 mice. Proliferation was assessed by duplicate [3H]thymidine incorporation. Results are representative of three sets of independent experiments; all experiments showed similar results.
Next, to determine whether the H-Y CD8+ T cells specifically recognize the H-Y male antigen, equal numbers of H-Y CD8+ T cells from each type of mouse were injected intravenously into either male or female H-2b/b Cα-deficient mice (which lack T cells; reference 18), and the number of H-Y CD8+ T cells in recipient spleens was evaluated 4 d after injection. As shown in Fig. 2 c, significant numbers of H-Y CD8+ T cells were detected in male recipients into which either Egr/H-Y or NL/H-Y T cells were transplanted. In contrast, no obvious H-Y CD8+ T cells were detected in female recipients. These results suggest that H-Y CD8+ T cells from all types of mice recognized and specifically responded to the male antigen; expansion of the H-Y CD8+ T cells resulted in accumulation of a significant population of those T cells in recipient spleens. Interestingly, the number of H-Y CD8+ T cells in male recipients into which cells from β2m0 Egr/H-Y mice were injected was slightly lower (∼30% less) than those in recipients into which cells from other types of mice were injected. This may be due to rejection by NK cells, since NK cells recognize and kill β2m0 T cell blasts because of their extremely low class I MHC expression (19, 20). Supporting this possibility, the in vitro response of H-Y CD8+ T cells from β2m0 Egr/H-Y mice to irradiated spleen cells from male C57BL/6 (B6: H-2b/b) mice (containing NK cells that are no longer able to kill target cells due to irradiation) was comparable to the response of H-Y CD8+ cells from other types of mice (Fig. 2 d). As observed in in vivo experiments, cells from all types of mice also failed to respond to female spleen cells in vitro.
Thus, the H-Y CD8+ T cells positively selected by low avidity ligands in H-2d/d or β2m0 Egr/H-Y mice were functionally indistinguishable from cells positively selected in NL/H-Y female H-2b/b mice in the absence of the Egr-1 transgene.
More Efficient Negative Selection in Egr-1 Transgenic Mice.
In male H-2b/b thymi, H-Y thymocytes are negatively selected and die through the apoptotic pathway (8). As shown in Fig. 3, both Egr/H-Y and NL/H-Y thymocytes have undergone clonal deletion. Interestingly, even more efficient negative selection appears to occur in Egr/H-Y mice, judging from the lower number of thymocytes in Egr/H-Y mice. However, male H-2d/d and β2m0 Egr/H-Y mice exhibited positive selection of H-Y CD8+ SP cells, and the numbers of mature H-Y CD8+ SP cells were comparable to those of females (data not shown).
Figure 3.
CD4/CD8 profiles of thymocytes from male Egr/H-Y and NL/H-Y mice on an H-2b/b background, gated on T3.70+ cells. Numbers in the profiles indicate the averages of the absolute cell numbers of positive cells within a quadrant, of all mice analyzed. Total thymocyte numbers for both types of mice are shown. Three pairs of male Egr/H-Y and NL/H-Y mice were analyzed.
Positive Selection of AND-TCR T Cells in the Presence of Egr-1 Overexpression.
Egr-1 transgenic mice were also bred with another type of transgenic mouse expressing a TCR recognizing the moth cytochrome C peptide in the context of class II MHC I-Ek (called AND mice; reference 9). The AND TCR has a weak affinity for I-Ab class II MHC molecules, and therefore, CD4+ SP thymocytes are positively selected in an H-2b/b background (9, 21). The upper panels of Fig. 4 show the CD4/CD8 profiles of mature thymocytes bearing the AND TCR (defined as transgenic TCR [Vβ3]high, PNArlow) from Egr-1 transgene–positive AND mice (Egr/AND) and Egr-1 transgene–negative AND mice (NL/AND), both of which are on an H-2b/b background. Egr/AND mice contained increased numbers of both total thymocytes (1.1 ± 0.1 × 108 in Egr/AND versus 0.8 ± 0.2 × 108 in NL/AND; n = 4 each) and AND CD4+ SP thymocytes (3.5 ± 0.3 × 107 in Egr/ AND versus 2.2 ± 0.3 × 107 in NL/AND; n = 4 each). This is reminiscent of the increased numbers of H-Y CD8+ SP cells in H-2b/b Egr/H-Y mice and, again, suggests more efficient positive selection in the presence of the Egr-1 transgene. Interestingly, a significant number (8.0 ± 0.6 × 105; n = 4) of mature AND CD8+ SP cells were observed in Egr/AND mice. It is possible that the AND TCR also has a weak affinity for class I MHC (22) which is not normally sufficient to induce signal(s) for positive selection. Egr-1 overexpression might lower the threshold for positive selection, resulting in positive selection of AND cells by class I MHC, creating a population of AND CD8+ SP cells. This hypothesis is well-supported by analysis of Egr/ AND mice on a β2m0 background, where the number of AND CD8+ SP cells strikingly decreased (Fig. 4, middle). However, the difference between H-Y CD8+ SP cells and AND CD8+ SP cells in β2m0 Egr-1 transgenic mice is intriguing; very efficient positive selection of H-Y CD8+ SP cells was observed, while almost no positive selection of AND CD8+ SP cells was achieved, in the β2m0 background. This may be explained by a possible difference in the original affinity of H-Y TCR and AND TCR for class I MHC molecules. The former may have a relatively high affinity, which creates sufficient avidity for positive selection even with extremely low expression levels of class I MHC, when Egr-1 overexpression lowers the threshold. In contrast, the latter may have a low affinity, which contributes to sufficient avidity with normal expression levels of class I MHC in the presence of Egr-1 overexpression, but not with the extremely low levels of class I MHC in β2m0 mice.
Figure 4.
Thymocyte suspensions from Egr/AND and NL/AND mice on either H-2b/b, β2m0, or class II MHC–deficient (IIo) backgrounds were stained for CD4, CD8, AND transgenic β chain (Vβ3), and PNAr, and analyzed by flow cytometry. CD4/CD8 plots of cells gated on Vβ3high, PNArlow are shown. Numbers in the profiles indicate the averages of the absolute cell numbers of positive cells within a quadrant. N.D., No cell detectable.
In a class II MHC–deficient (II0) background, in which “leaky” expression of class II molecules is not detected (23), AND CD4+ cells virtually disappeared in both Egr/ AND and NL/AND mice (Fig. 4, bottom). This is consistent with failed positive selection of H-Y CD8+ T cells on an I0β2m0 background, which entirely lacks class I MHC expression. Deficiency of positive selection in the complete absence of MHC molecules indicates a requirement of TCR-mediated signaling to promote DP cells to the SP stage, even in the presence of Egr-1 overexpression.
Increased Frequency of Alloreactive T Cells in Egr-1 Transgenic Mice.
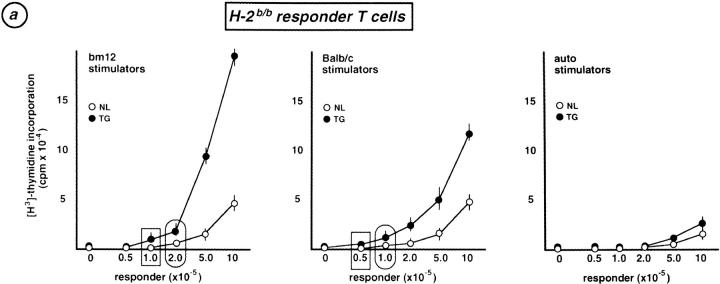
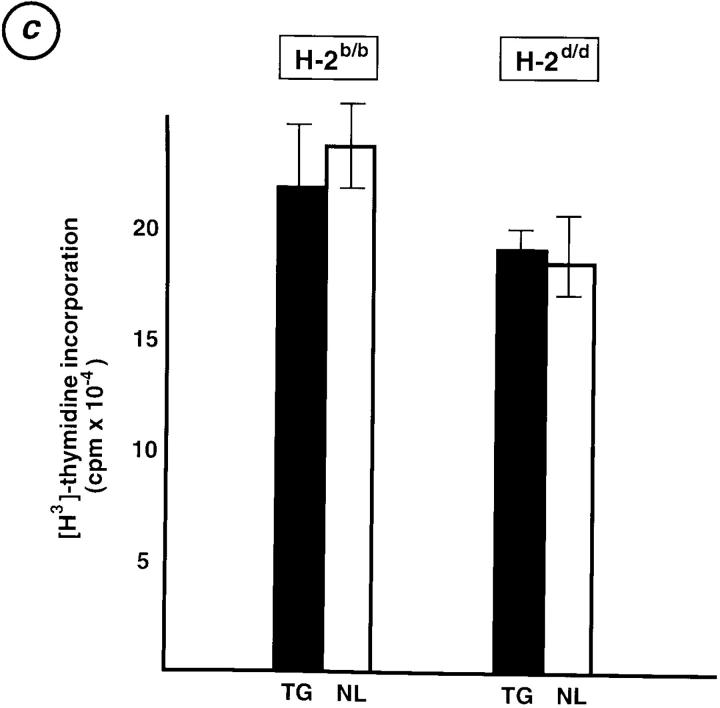
Alloreactive T cells, which recognize allo-MHC complexes, are believed to be positively selected by auto-MHC/antigen-peptide complexes for which the alloreactive TCRs have sufficient, but not too much, avidity (24). Therefore, whether Egr-1 expression might affect the selection of alloreactive T cells was determined using “normal” (no TCR transgenes introduced) Egr-1 transgenic mice. Fig. 5 a demonstrates allo-MLR responses of H-2b/b Egr-1–TG and NL mice to βm12 (H-2b with a mutated class II) and BALB/c (H-2d/d) spleen cells. Transgenic T cells exhibited enhanced responses to both βm12 and BALB/c spleen cells. Similar enhanced allo-responses of H-2d/d transgenic T cells were elicited towards B6 (H-2b/b) spleen cells (Fig. 5 b). These enhanced allo-responses of transgenic T cells appear to be due to an increased frequency of alloreactive T cells, but not to hyperproliferation of each T cell, because fewer transgenic T cells responded to the stimulators, as shown by the limiting dilution of responding cells in Fig. 5, a and b; and transgenic and negative littermate T cells exhibited comparable proliferative responses to Con A (Fig. 5 c). Overall, Egr-1 overexpression appears to allow positive selection of a larger number of alloreactive T cells, some of which originally might not have had sufficient affinity with the auto-MHC to be positively selected in normal mice.
Figure 5.
(a) MLR responders were titrated B cell–depleted lymph node cells from either Egr-1 transgenic (TG) or negative littermate (NL) mice, both with an H-2b/b background, stimulated with 3,000-rad irradiated 5 × 105 spleen cells from βm12 (H-2b with a mutated class II MHC), BALB/c (H-2d), or autologous spleen cells. (b) MLR was performed as above, with H-2d/d responder T cells and B6 (H-2b) stimulator cells. The smallest number of responder cells at which [3H]thymidine incorporation was detected in transgenic T cells (rectangle) and in negative littermate T cells (ellipse) in each panel is indicated. All experiments were repeated with four pairs of Egr-1 transgenic and negative littermate mice. In all panels, each value represents the mean of all results ± SD. (c) 5 × 105 B cell–depleted lymph node cells from transgenic (TG) and negative littermate (NL) mice were stimulated by 1 μg/ml of Con A for 72 h. Proliferation was assessed by duplicate [3H]thymidine incorporation for the last 16 h. Bars represent the mean of all results of three independent experiments ± SD.
Discussion
This is the first description of a transcription factor behaving as a modulator of positive selection; Egr-1 overexpression allows positive selection of thymocytes mediated by low avidity interactions with ligand(s) that are usually not sufficient, perhaps by lowering the threshold of avidity required for positive selection.
One might argue that Egr-1 overexpression induces survival effectors, such as antiapoptotic factors (25–30) in thymocytes. This may result in accumulation of the few leaky mature SP cells, such as the very small number of H-Y CD8+ SP cells that are detectable in both H-2d/d and β2m0 NL/H-Y mice, thus forming a seemingly positively selected cell population. However, this is unlikely for at least three reasons. First, purified H-Y CD8+ SP cells from H-2b/b, H-2d/d, or β2m0 female Egr/H-Y mice and H-2b/b female NL/H-Y mice showed comparable survival curves when cells were simply cultured in vitro (data not shown; see Materials and Methods). Second, I0β2m0 recipient mice into which β2m0 Egr/H-Y bone marrow cells were transplanted did not harbor the significant H-Y CD8+ SP cell population that was seen in H-2d/d or β2m0 female Egr/ H-Y mice, although their thymi contained detectable leaky H-Y CD8+ SP cells. Third, both male H-2b/b Egr/H-Y and NL/H-Y thymocytes exhibited clonal deletion of H-Y cells, indicating no rescue from apoptosis of Egr/H-Y cells undergoing negative selection. On the contrary, even more efficient negative selection appears to occur in Egr/H-Y mice. By these criteria, Egr-1 overexpression does not appear to induce survival effectors for thymocytes. It seems more likely that Egr-1 overexpression allows an unusually low avidity interaction between TCR and MHC/antigen-peptide complexes to initiate positive selection and further development of thymocytes.
An alternative argument is that Egr-1 overexpression itself might be sufficient to promote the DP to SP transition, with no interaction between thymocytes and ligands required. When the Egr-1 transgene was introduced into a RAG-deficient background, thymocytes did overcome the “β selection” beyond the maturation block at the CD25+CD44− DN stage and developed into immature CD8 single positive (ISP) cells without any signaling through the TCR (7). However, this was not the case for positive selection at the DP stage. As we demonstrated in a previous report, although irradiated Egr-1 transgenic mice on a RAG-deficient background developed DP cells expressing the Egr-1 transgene but lacking TCR expression, neither CD4+ nor CD8+ SP cells were observed (7). This suggests that DP cells do require some interaction with ligand(s) to proceed to the SP stage even in the presence of Egr-1 overexpression. This is supported convincingly by the analysis of I0β2m0 recipient mice into which β2m0 Egr/H-Y bone marrow cells were transplanted, in which no positive selection of H-Y CD8+ T cells occurred, as well as of Egr/ AND mice on a class II MHC–deficient background, which harbored no AND CD4+ T cells. Hence, Egr-1 expression appears not to directly initiate transition of DP cells to the SP stage, but may influence the threshold of avidity required for positive selection.
Functions of T cells selected by unusually low avidity TCR–MHC interactions in the presence of Egr-1 overexpression appear normal both in responses to various stimuli and in specificity of antigen-peptide recognition, as demonstrated by the functional analysis of mature H-Y CD8+ T cells in H-2d/d and β2m0 Egr/H-Y mice. However, long-term survival of a naive population of peripheral H-Y CD8+ T cells in H-2d/d Egr/H-Y mice could be affected, as a recent report by Tanchot et al. (16) implicated a requirement of the right MHC for survival of peripheral naive T cells. Therefore, a large proportion of peripheral H-Y CD8+ T cells in H-2d/d Egr/H-Y mice might be newly generated T cells. Tanchot et al. also demonstrated a requirement of only a nonspecific class I MHC for survival of peripheral memory T cells (16). This is consistent with the comparable expression patterns of memory markers such as Mel-14 in the peripheral H-Y CD8+ T cells in H-2d/d Egr/H-Y mice and H-2b/b NL/H-Y mice.
Phenotype in Egr-1 transgenic mice indicates a physiological role of Egr-1 in determining the threshold required for positive selection. Perhaps related to this, Egr-1 overexpression may also affect negative selection. As described above, more efficient negative selection appeared to be achieved by Egr-1 overexpression in male H-2b/b H-Y mice. However, thymocytes express other members of Egr family genes, such as Egr-2, -3, and -4, all of which share a highly conserved DNA-binding domain (5, 6). These members might functionally compensate for each other, and hence no alteration of thymic selection is observed in Egr-1–deficient mice as discussed previously (7, 31). Therefore, a set of genes controlled by a common binding site for Egr genes, including Egr-1, might be critical in defining the overall thresholds required for thymic selection. Further characterization of Egr target genes should shed light on the precise molecular mechanism underlying the positive/negative selection of thymocytes.
Acknowledgments
We thank Drs. S. Gilfillan and K. Campbell for critical reading of the manuscript; E. Wagner, W. Metzger, and R. Zedi for help with the mice; Dr. P. Kisielow for T3.70 mAb; and Dr. C. Benoist for β2m0II0 mice and KT-3 mAb.
Abbreviations used in this paper
- DP
double positive
- Egr-1
early growth response 1
- Egr/H-Y
Egr-1 transgene–positive H-Y transgenic
- I0
class I MHC–deficient
- II0
class II MHC–deficient
- ISP
immature SP
- NL/H-Y
Egr-1 transgene–negative H-Y transgenic
- PNAr
peanut agglutinin receptor
- RAG
recombination-activating gene
- SP
single positive
- TG
transgenic
Footnotes
F.A. Lemonnier is supported by the Association pour la Recherche sur le Cancer. Basel Institute for Immunology was founded and is supported by F. Hoffman-La Roche Ltd. (Basel, Switzerland).
References
- 1.Benoist C, Mathis D. Positive selection of T cells: fastidious or promiscuous? . Curr Opin Immunol. 1997;9:245–249. doi: 10.1016/s0952-7915(97)80143-4. [DOI] [PubMed] [Google Scholar]
- 2.Marrack P, Kappler J. Positive selection of thymocytes bearing αβ T cell receptors. Curr Opin Immunol. 1997;9:250–255. doi: 10.1016/s0952-7915(97)80144-6. [DOI] [PubMed] [Google Scholar]
- 3.Hogquist KA, Gavin MA, Bevan MJ. Positive selection of CD8+T cells induced by major histocompatibility complex binding peptides in fetal thymic organ culture. J Exp Med. 1993;177:1469–1473. doi: 10.1084/jem.177.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashton-Rickardt PG, van Kaer L, Schumacher TNM, Ploegh HL, Tonegawa S. Peptide contributes to the specificity of positive selection of CD8+T cells in the thymus. Cell. 1993;73:1041–1049. doi: 10.1016/0092-8674(93)90281-t. [DOI] [PubMed] [Google Scholar]
- 5.Kelly K, Siebenlist U. Immediate-early genes induced by antigen receptor stimulation. Curr Opin Immunol. 1995;7:327–332. doi: 10.1016/0952-7915(95)80106-5. [DOI] [PubMed] [Google Scholar]
- 6.Shao H, Kono DH, Chen L-Y, Rubin EM, Kaye J. Induction of the early growth response (Egr) family of transcription factors during thymic selection. J Exp Med. 1997;185:731–744. doi: 10.1084/jem.185.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazaki T. Two distinct steps during thymocyte maturation from CD4−CD8− to CD4+CD8+ distinguished in the early growth response (Egr)-1 transgenic mice with a recombinase-activating gene–deficient background. J Exp Med. 1997;186:877–885. doi: 10.1084/jem.186.6.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+CD8+ thymocytes. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 9.Kaye J, Hsu ML, Sauron ME, Jameson SC, Gascoigne NR, Hedrick SM. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 1989;341:746–749. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- 10.Rocha B, von Boehmer H. Peripheral selection of the T cell repertoire. Science. 1991;251:1225–1228. doi: 10.1126/science.1900951. [DOI] [PubMed] [Google Scholar]
- 11.Calnan BJ, Szychowski S, Chan FKM, Cado D, Winoto A. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995;3:273–282. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 12.Miyazaki T, Dierich A, Benoist C, Mathis D. Independent modes of natural killing distinguished in Lag-3 knockout mice. Science. 1996;272:405–408. doi: 10.1126/science.272.5260.405. [DOI] [PubMed] [Google Scholar]
- 13.von Boehmer H. Developmental biology of T cells in T cell-receptor transgenic mice. Annu Rev Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- 14.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature. 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 15.Bix M, Raulet D. Functionally conformed free class I heavy chains exist on the surface of beta 2-microglobulin negative cells. J Exp Med. 1992;176:829–834. doi: 10.1084/jem.176.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanchot C, Lemonnier FA, Perarnau B, Freitas AA, Rocha B. Differential requirements for survival and proliferation of CD8 naive or memory T cells. Science. 1997;276:2057–2062. doi: 10.1126/science.276.5321.2057. [DOI] [PubMed] [Google Scholar]
- 17.Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Perarnau B. HLA-A2.1–restricted education and cytolytic activity of CD8+ T lymphocytes from β2-microglobulin (β2m) HLA-A2.1 monochain transgenic H-2 Dbβ2m double knockout mice. J Exp Med. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mombaerts P, Clarke AR, Rudnicki MA, Lacomini J, Itohara S, Lafaille JJ, Wang L, Ichikawa Y, Jaenisch R, Hooper MR, Tonegawa S. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 19.Liao N, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency: susceptibility to natural killer (NK) cells and impaired NK activity. Science. 1991;253:199–202. doi: 10.1126/science.1853205. [DOI] [PubMed] [Google Scholar]
- 20.Hoglund P, Ohlen C, Carbone E, Franksson L, Ljunggren HG, Latour A, Koller B, Karre K. Recognition of beta 2-microglobulin-negative (beta 2m−) T-cell blasts by natural killer cells from normal but not from beta 2m− mice: nonresponsiveness controlled by beta 2m− bone marrow in chimeric mice. Proc Natl Acad Sci USA. 1991;88:10332–10336. doi: 10.1073/pnas.88.22.10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaye J, Vasquez NJ, Hedrick SM. Involvement of the same region of the T cell antigen receptor in thymic selection and foreign peptide recognition. J Immunol. 1992;148:3342–3353. [PubMed] [Google Scholar]
- 22.Shimizu T, Takeda S. CD8 T cells from major histocompatibility complex class II-deficient mice respond vigorously to class II molecules in a primary mixed lymphocyte reaction. Eur J Immunol. 1997;27:500–508. doi: 10.1002/eji.1830270222. [DOI] [PubMed] [Google Scholar]
- 23.Cosgrove D, Gray D, Dierich A, Kaufman J, Lemeur M, Benoist C, Mathis D. Mice lacking MHC class II molecules. Cell. 1991;66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- 24.Udaka K, Wiesmuller KH, Kienle S, Jung G, Walden P. Self-MHC-restricted peptides recognized by an alloreactive T lymphocyte clone. J Immunol. 1996;15:670–678. [PubMed] [Google Scholar]
- 25.Strasser A, Harris AW, Cory S. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 1994;67:889–899. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- 26.Strasser A, Harris AW, von Boehmer H, Cory S. Positive and negative selection of T cells in T-cell receptor transgenic mice expressing a bcl-2 transgene. Proc Natl Acad Sci USA. 1994;91:1376–1380. doi: 10.1073/pnas.91.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Field SJ, Tsai F-Y, Kuo F, Zubiaga AM, Kaelin WQ, Jr, Livingston DM, Orkin SH, Greenberg ME. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki L, Jacks T, Bronson R, Goilot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 29.Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 30.Maraskovsky E, O'Reilly LA, Teepe M, Corcoran LM, Peschon JJ, Strasser A. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1−/−mice. Cell. 1997;89:1011–1019. doi: 10.1016/s0092-8674(00)80289-5. [DOI] [PubMed] [Google Scholar]
- 31.Lee SL, Tourtellotte LC, Wesselschmidt RL, Milbrandt J. Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J Biol Chem. 1995;270:9971–9977. doi: 10.1074/jbc.270.17.9971. [DOI] [PubMed] [Google Scholar]