Abstract
Neurodegeneration in familial amyotrophic lateral sclerosis (ALS) is associated with enhanced redox stress caused by dominant mutations in superoxide dismutase–1 (SOD1). SOD1 is a cytosolic enzyme that facilitates the conversion of superoxide (O2•–) to H2O2. Here we demonstrate that SOD1 is not just a catabolic enzyme, but can also directly regulate NADPH oxidase–dependent (Nox-dependent) O2•– production by binding Rac1 and inhibiting its GTPase activity. Oxidation of Rac1 by H2O2 uncoupled SOD1 binding in a reversible fashion, producing a self-regulating redox sensor for Nox-derived O2•– production. This process of redox-sensitive uncoupling of SOD1 from Rac1 was defective in SOD1 ALS mutants, leading to enhanced Rac1/Nox activation in transgenic mouse tissues and cell lines expressing ALS SOD1 mutants. Glial cell toxicity associated with expression of SOD1 mutants in culture was significantly attenuated by treatment with the Nox inhibitor apocynin. Treatment of ALS mice with apocynin also significantly increased their average life span. This redox sensor mechanism may explain the gain-of-function seen with certain SOD1 mutations associated with ALS and defines new therapeutic targets.
Introduction
Superoxide dismutase–1 (SOD1) is a ubiquitously expressed cytosolic enzyme that facilitates the conversion of superoxide (O2•–) to H2O2 (1). Inherited dominant mutations in SOD1 lead to progressive motor neuron loss in the cerebral cortex, brain stem, and spinal cord in adult-onset amyotrophic lateral sclerosis (ALS) (2). Oxidative stress and inflammation are thought to play major roles in the pathogenesis of ALS, and several sources of ROS — including mitochondria, NADPH oxidase (Nox), and the SOD1 mutant itself — have been proposed to elevate redox stress in ALS (3–5). For example, studies in SOD1G93A transgenic ALS mice have recently demonstrated that deletion of Nox2, and to a lesser extent Nox1, can significantly slow disease progression and improve survival of ALS mice (3, 5). Although these studies implicate multiple Nox genes in the pathology of ALS, it remains unclear whether enhanced Nox-dependent redox stress in ALS is a primary or secondary event of disease caused by mutant SOD1. Currently it is thought that enhanced Nox2-dependent redox stress in ALS is a secondary event associated with microgliosis during later stages of the inflammatory processes. Recent studies using controlled expression of mutant SOD1 in motor neurons and glia have also demonstrated that these 2 cell types contribute to different phases of ALS disease progression — motor neurons in early phases of disease onset and microglia in later phase disease progression (6). These findings implicate primary defects in microglial and neuronal functions as a consequence of mutant SOD1 expression.
Seven known Nox catalytic subunits exist: Nox1, Nox2gp91phox, Nox3, Nox4, Nox5, Duox1, and Duox2 (7, 8). NADPH oxidases generate O2•– by transferring an electron from NADPH to molecular oxygen. The most widely characterized Nox is phagocytic gp91phox (Nox2), which is also expressed in microglia (3) and a variety of other nonphagocytic cell types. Rac1 is a central activator of Nox2 and Nox1, along with several other subunits that can act in a cell-specific fashion to promote Nox complex activation (p40phox, p47phox, p67phox, NoxO1, and NoxA1) (7, 8). Despite the finding that Nox2 contributes to inflammatory components of ALS disease (3, 5), the mechanism(s) by which mutations in SOD1 lead to dysregulation of O2•– production by Nox2 remain poorly understood.
In the present study, we investigated whether primary defects in Nox regulation are caused by ALS-associated mutations in SOD1. Our findings demonstrate that SOD1 controlled GTP hydrolysis by Rac1 through a redox-dependent interaction uncoupled by an H2O2-mediated modification to Rac1. In this context, when SOD1 was bound to Rac1 under reducing conditions, GTP hydrolysis by Rac1 was inhibited. However, in oxidizing conditions, SOD1 dissociated from Rac1 and no longer inhibited intrinsic GTP hydrolysis by Rac1. ALS mutant forms of SOD1 more strongly associated with Rac1 and were less sensitive to redox uncoupling. These SOD1/Rac1 interactions created a redox-dependent sensor for activation of Rac1-dependent NADPH oxidases and the production of O2•– by endomembranes. Importantly, this regulatory circuit was disrupted by familial ALS–associated mutations in SOD1. Hence, we propose a model by which SOD1 can directly regulate Nox activation by acting as a redox sensor for Rac1 inactivation. Such findings may help to explain the gain of function found in familial ALS SOD1 mutations associated with enhanced redox stress.
Results
ALS-associated SOD1 mutations activate cellular Nox activity.
We hypothesized that ALS mutant SOD1, but not WT SOD1, directly dysregulates production of Nox-derived O2•–. Indeed, analysis of transgenic mice overexpressing SOD1WT or SOD1G93A demonstrated that only the mutant form of SOD1 enhanced NADPH-dependent O2•– production in brain and spinal cord endomembranes and tissue (Figure 1, A and B), and that this was inhibited by the flavoprotein inhibitor diphenyleneiodionium chloride (DPI), but not the mitochondrial complex I inhibitor rotenone (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI34060DS1). Interestingly, the liver, an organ that does not demonstrate notable pathology in ALS, also demonstrated similar SOD1 mutant–associated increases in Nox activity (Figure 1A).
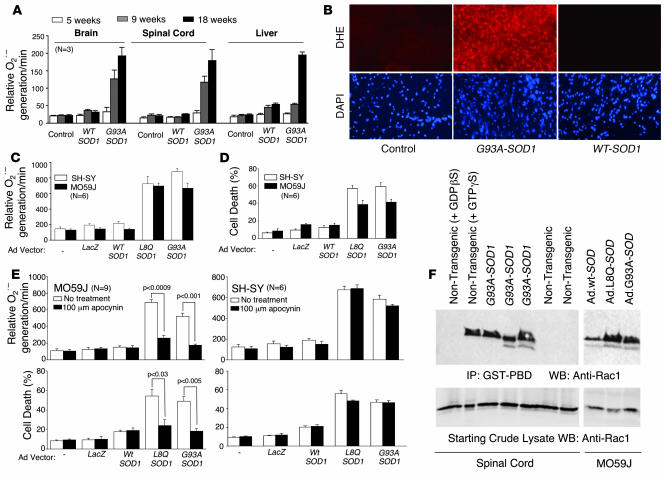
Figure 1. ALS-associated SOD1 mutations activate cellular Nox activity.
(A) Rate of NADPH-dependent O2•– production by total endomembranes isolated from the brain, spinal cord, and liver of nontransgenic or transgenic mice overexpressing SOD1WT or SOD1G93A (mean ± SEM; n = 3 per group). (B) DHE fluorescent detection of O2•– in lumbar spinal cord sections from nontransgenic and transgenic mice overexpressing SOD1WT or SOD1G93A. DAPI staining shows cell nuclei in each section. (C) Rate of NADPH-dependent O2•– production in total endomembranes isolated from SH-SY (neuronal) or MO59J (glial) cells at 48 hours following infection with adenoviral vectors expressing LacZ, SOD1WT, SOD1L8Q, or SOD1G93A. (D) Cell death was quantified in SH-SY and MO59J cells using trypan-blue exclusion at 72 hours after infection with the indicated adenoviral vectors. (E) Using conditions specified in C and D, the rate of NADPH-dependent O2•– production and cell death was assessed in the presence or absence of the Nox inhibitor apocynin (100 μM). Values are mean ± SEM (n = 6 per group). (F) Assessment of GTP-bound Rac1 (activated form) in spinal cord lysates from 2 nontransgenic or 3 SOD1G93A transgenic mice (120 days old) and from MO59J cells overexpressing WT or mutant SOD1 proteins (at 36 hours after adenoviral infection). The 2 left lanes are controls for the Rac activation assay, in which nontransgenic spinal cord lysates were preincubated with the indicated non-hydrolysable guanine nucleotide analogs.
To evaluate whether mutant SOD1 proteins enhance Nox activity directly in the absence of disease-associated inflammatory processes seen in vivo in ALS mice, we expressed WT, L8Q, and G93A forms of SOD1 in both MO59J glial cells and SH-SY neuronal cells using recombinant adenoviruses. Overexpression only of the mutant SOD1 proteins enhanced NADPH-dependent O2•– production in endomembranes from both glial and neuronal cell types (Figure 1C) and significantly increased cell death (Figure 1D). These findings implicate a gain of function in SOD1 mutants that leads to enhanced Nox activation and cellular injury. Apocynin, a known inhibitor of NADPH oxidases such as Nox2 (9), abrogated SOD1 mutant–facilitated NADPH-dependent O2•– production only in glial cells, with a corresponding increase in cell viability (Figure 1E). In contrast, apocynin did not inhibit NADPH-dependent O2•– production in SH-SY neuronal cells induced by mutant SOD1 expression, nor did it protect from associated cellular injury (Figure 1E). These findings suggest that dysregulated ROS production in SOD1 mutant–expressing neuronal cells likely occurs through a pathway distinct from that in glial cells.
Apocynin is thought to inhibit NADPH oxidases by interfering with recruitment of p47phox to the Nox complex (10). Three known Nox catalytic subunits have been shown to be regulated by p47phox in vitro (Nox1, Nox2, and Nox3) (7, 11) and these Nox isoforms are also regulated by the small GTPase Rac (8). However, thus far only Nox2 has been confirmed to be regulated by p47phox in vivo. To this end, we hypothesized that mutant SOD1 expression in apocynin-responsive MO59J glial cells enhances Rac1 activation. Indeed, both SOD1G93A and SOD1L8Q mutant proteins elevated Rac1-GTP levels in MO59J cells compared with overexpression of SOD1WT, as determined by Pak1 pulldown assays (Figure 1F). More importantly, spinal cords from ALS transgenic mice overexpressing the SOD1G93A mutant demonstrated an even greater enhancement in Rac1-GTP levels (i.e., activated Rac1) compared with nontransgenic age-matched controls (Figure 1F). These findings of enhanced Rac1 activation by SOD1 mutant expression led us to hypothesize that SOD1 directly interacts with Rac1 and/or other Nox complex components to stabilize the activated form of this complex. In support of this hypothesis are previous findings that SOD1 and Rac1 both recruit to Nox2-active early endosomes following cytokine stimulation (12).
Rac1 binds to SOD1 in a redox-dependent manner.
To investigate potential binding partners for SOD1 that were associated with the Nox complex, we performed IP experiments for p47phox, p67phox, p22phox, and Rac1 — 4 regulators of the Nox2 complex — from brain tissue lysates followed by Western blotting for SOD1 and found that only Rac1 effectively pulled down SOD1 (Figure 2A and data not shown). To determine whether Rac1/SOD1 interactions occur in other tissue types, we conducted co-IP experiments from several mouse organs including brain, liver, kidney, and heart. Indeed, IP of Rac1 pulled down SOD1 from each of these organs in SOD1+/+ mice, but not SOD1–/– mice (Figure 2A). The amount of SOD1 associated with Rac1 was noticeably highest in the brain and lowest in the heart. To test whether this interaction was direct, we used in vitro pulldown assays with purified proteins. Immobilized His-tagged Rac1 clearly associated with SOD1 when Rac1 was preloaded with GDPβS, but not when Rac1 was preloaded with GTPγS or in the absence of nucleotide (Figure 2B). In contrast, the related Rho GTPase Cdc42 did not associate with SOD1 (Figure 2B). These results suggested that the GDP-bound form of Rac1 associates with SOD1.
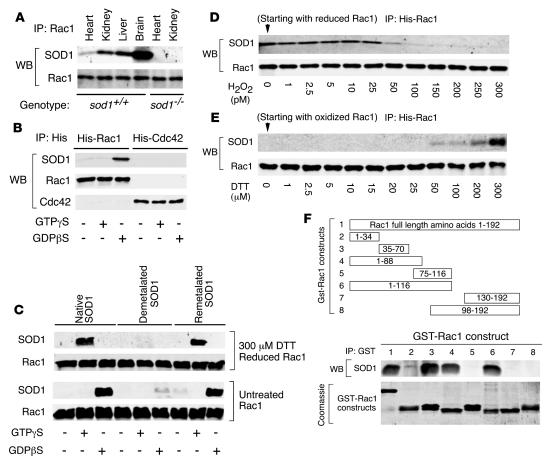
Figure 2. Rac1 binds to SOD1 in a redox-dependent manner.
(A) Rac1 IP from heart, kidney, liver, and/or brain tissue of SOD1+/+ or SOD1–/– mice followed by Western blotting (WB) for SOD1 and Rac1. (B) In vitro IP of purified His-tagged Rac1 and Cdc42 in the presence of purified bovine SOD1 followed by Western blot for SOD1, Rac1, and Cdc42. The His-tagged GTPases were preloaded with the indicated nucleotide analogs prior to incubation with SOD1. (C) In vitro IP of purified His-tagged Rac1 in the presence of purified native, demetalated, or remetalated bovine SOD1 followed by Western blot for SOD1 and Rac1. The His-tagged Rac1 was preloaded with the indicated nucleotide analogs prior to incubation with SOD1. Additionally, untreated His-tagged Rac1 and 300 μM DTT prereduced His-tagged Rac1 were used for in vitro pulldown assays with each of the 3 forms of SOD1. (D) His-tagged Rac1 was prereduced (300 μM DTT), loaded with GTPγS, and treated with the indicated concentrations of H2O2 before performing pulldown assays with SOD1. (E) The indicated concentrations of DTT were added to the 300 pM H2O2-treated His-tagged Rac1 sample in D, and pulldown assays were performed with SOD1. (F) Schematic of GST-Rac1 deletion mutants used to define the SOD1 binding domain and in vitro IP of various GST-tagged Rac1 deletion mutants in the presence of purified bovine SOD1. GST-Rac1 fusion construct numbers above correspond with lane numbers below. Top lanes are Western blot for SOD1 following IP of GST; bottom lanes are Coomassie-stained gel of the purified fusion peptides used for IP.
Given that Rac1 can regulate O2•– production through certain NADPH oxidases (13, 14) and that SOD1 dismutates O2•– to H2O2, we hypothesized that SOD1 enzymatic activity is fundamentally important for interactions with Rac1. Cu binding at the active site of SOD1 is necessary for its enzymatic activity, and a specific Cu chaperone is required for the loading of SOD1 with Cu in vivo (15). Using in vitro pulldown assays, we asked whether the interaction between Rac1 and SOD1 was redox regulated and whether the metal content of SOD1 affected this interaction. Interestingly, reduction of Rac1 with DTT switched the nucleotide preference required for binding to SOD1 (Figure 2C). Nonreduced bacterially expressed Rac1 most efficiently bound to SOD1 in the presence of GDPβS, as shown in Figure 2B. In contrast, reduced Rac1 bound to SOD1 when loaded with GTPγS but not GDPβS (Figure 2C). Furthermore, only metalated (i.e., native) and remetalated forms of SOD1 bound to Rac1, while demetalated (i.e., enzymatically inactive) SOD1 failed to bind Rac1 (Figure 2C and Supplemental Figure 2A). In contrast, neither reduced Cdc42-GTPγS nor Cdc42-GDPβS bound SOD1 (Supplemental Figure 2B). These findings demonstrate that SOD1 can indeed bind Rac1-GTP under reducing conditions and suggest that the redox state of Rac1 influences its affinity for SOD1 in GTP versus GDP bound states.
Intrigued by these results, we sought to directly test whether sequential reduction and oxidation of GTP-bound Rac1 cycles Rac1 into SOD1 bound and unbound states, respectively. To this end, we exposed Rac1 (reduced with DTT and preloaded with GTPγS) to different concentrations of H2O2 and evaluated its ability to associate with SOD1 after removing excess H2O2 and found that H2O2 concentrations as low as 50 pM caused a significant decrease in the binding affinity of Rac1 for SOD1 (Figure 2D). To exclude the possibility of H2O2-mediated irreversible damage to Rac1 protein, we repeated the same experiment adding back different concentrations of DTT to oxidized Rac1 exposed to 300 pM H2O2. Indeed, H2O2-mediated inhibition of Rac1/SOD1 binding was reversed by treatment of Rac1 with 50–300 μM DTT (Figure 2E). These in vitro association data demonstrate that Rac1/SOD1 binding is redox regulated and can cycle between bound and unbound states depending on the redox state of Rac1.
To determine the domain of Rac1 that associated with SOD1, we constructed GST-tagged deletion mutants of Rac1 (Figure 2F) and conducted in vitro pulldown assays. SOD1 most efficiently bound a region of Rac1 contained within amino acids 35–70 (Figure 2F). This region of Rac1 spans several domains important for nucleotide binding (i.e., switch I, G2, switch II, and G3 domains; Supplemental Figure 2C; refs. 16–18). Binding of SOD1 to this region on Rac1 is also consistent with the observed differences in binding between SOD1 and GTPγS- versus GDPβS-bound Rac1. Interestingly, Rac1 guanine-nucleotide exchange factor (GEF) Tiam1 binds to a region of Rac1 that spans the interacting domain with SOD1 (19). In addition, the switch regions on 2 related Rho GTPases, RhoA and Cdc42, are involved in binding to RhoGAP (20, 21). Therefore, we hypothesized that SOD1 influences Rac1 activity by acting as a GEF or GAP. However, SOD1 did not significantly affect GTP loading on Rac1 in vitro (Supplemental Figure 2D) and hence appeared not to act as a GEF.
SOD1 regulates Rac1/Nox2 activation, and this function is dysregulated in certain SOD1 ALS mutants.
We next evaluated whether SOD1 serves as a GAP to stimulate GTP hydrolysis by Rac1. Such a hypothesis was consistent with elevated Rac1-GTP levels in mutant SOD1–expressing glial cells and spinal cord (Figure 1F). However, SOD1 significantly inhibited the GTPase activity of Rac1 and also prevented p29Rho-GAP from activating GTP hydrolysis by Rac1 (Figure 3A). However, E. coli SOD, a homolog of mammalian MnSOD/SOD2, did not alter Rac1 GTPase activity (Figure 3B), nor did it associate with Rac1 in vitro (data not shown). The ability of SOD1 to inhibit GTP hydrolysis was also specific for Rac1 and was not observed with the closely related small GTPase Cdc42 (Figure 3C). Furthermore, demetalated (i.e., enzymatically inactive) SOD1, which did not associate with Rac1 (Figure 2C), also did not inhibit Rac1 GTPase activity (Supplemental Figure 2E). Given that Rac1 has exceptionally high intrinsic GTPase activity (22), these findings are consistent with SOD1 acting to stabilize Rac1-GTP by inhibiting its GTPase activity.
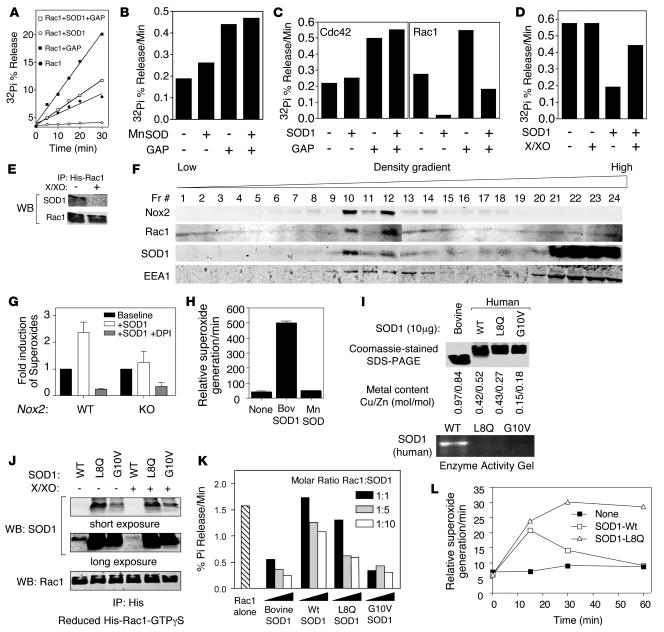
Figure 3. SOD1 activates Rac1 and Nox2, a function dysregulated in certain SOD1 ALS mutants.
(A and B) Rac1 GTPase assays were performed in the presence or absence of purified (A) bovine SOD1 or (B) E. coli MnSOD and/or p29-GAP. His-tagged Rac1 was preloaded with γP32-GTP, and aliquots of the reaction were analyzed at various time points by TLC for GTP hydrolysis by assessing percent 32Pi released from Rac1. (C) GTPase assay for Rac1 and Cdc42 in the presence or absence of bovine SOD1 and/or p29-GAP. (D) Rac1 GTPase assay in the presence or absence of bovine SOD1 and/or X/XO (100 mU; final concentration 100 μM). (E) Pulldown assays of GTPγS-loaded His-tagged Rac1 in the presence of bovine SOD1 with or without 15-minute exposure to X/XO-derived ROS. Data are representative of at least 3 independent experiments. (F) Endosomes were isolated from PMDFs using Iodixanol density gradient fractionation, and fractions were characterized by Western blot for Nox2gp91phox, Rac1, SOD1, and EEA1. (G) Lucigenin assays were used to assess the rate of NADPH-dependent O2•– production in fraction 10 vesicles from Nox2 WT and KO PMDFs in the presence or absence of 2.5 μM bovine SOD1 and/or 10 μM DPI, a general Nox inhibitor (n = 6). (H) Vesicular fractions from PMEFs heterozygous for a gene disruption of mouse SOD1 were assessed for NADPH-dependent O2•– production in the presence or absence of exogenously added 2.5 μM bovine SOD1 or 2.5 μM E. coli MnSOD (n = 3). (I) Coomassie-stained SDS-PAGE of purified bacterially expressed human SOD1 proteins. Bovine SOD1 was used as a reference control and migrates faster than human SOD1 (12). The Cu/Zn content of each SOD1 protein is shown. SOD activity gel of the bacterial purified SOD1 proteins is shown at bottom. (J) In vitro IP of purified prereduced His-tagged Rac1-GTPγS in the presence of the indicated human SOD1 proteins. Rac1/SOD1 complexes were then divided into 2 parts; 1 half was treated with X/XO-derived ROS for 15 minutes at room temperature prior to IP of the His-tag. Following IP, Western blots for SOD1 and Rac1 were performed. Long and short exposures of the SOD1 blot are shown to demonstrate enhanced binding of each of the mutant forms of SOD1 to Rac1. (K) Rac1 GTPase assays were performed using native bovine SOD1 or the indicated bacterially expressed purified human SOD1 proteins. The molar ratio of Rac1/SOD1 is indicated. (L) Time course of NADPH-dependent O2•– generation by isolated PMEF endosomes in the presence or absence of 1 μM human SOD1WT or SOD1L8Q.
Because the binding of Rac1 to SOD1 was controlled by the redox state of Rac1, we hypothesized that the ability of SOD1 to regulate Rac1 GTPase activity might also be redox regulated. To directly evaluate whether ROS alter the ability of SOD1 to inhibit GTP hydrolysis by Rac1, we performed GTPase assays in the presence of a xanthine/xanthine oxidase (X/XO) O2•– generating system. Given that Rac1 regulates O2•– production by certain NADPH oxidases, such a question was potentially relevant to processes that regulate ROS production in vivo. Interestingly, SOD1 lost its ability to inhibit GTP hydrolysis by Rac1 in the presence of this ROS-generating system (Figure 3D). However, the levels of ROS generated under our experimental conditions did not affect the intrinsic Rac1 GTPase activity in the absence of SOD1 (Figure 3D). IP of Rac1-GTPγS/SOD1 complexes using the GTPase assay conditions demonstrated that exposure to X/XO-derived ROS dissociated SOD1 from Rac1 (Figure 3E). These findings demonstrated that ROS alter the ability of SOD1 to regulate Rac1 GTPase activity by controlling physical interactions between these 2 proteins. These findings are consistent with the ability of H2O2 to disrupt SOD1/Rac1 interactions (Figure 2D).
Rac1 and Rac2 are well recognized for their ability to regulate cellular O2•– through its interactions with Nox1 and Nox2gp91phox (7). Rac2 is a hematopoietic-specific protein and regulates Nox2gp91phox in neutrophils (23). Rac1, on the other hand, is a ubiquitous protein that can regulate Nox1 (11, 24) as well as Nox2gp91phox activity in human monocytes (25) and myeloid lineage cells such as microglia (26). These interactions have placed Rac1 central to a number of ROS-regulated cellular processes controlled by O2•– and/or H2O2, the dismutated product of O2•– (13, 27–30). Interestingly, we recently demonstrated that SOD1 is recruited to the surface of endosomes that produce lumenal Nox2-dependent O2•– following IL-1β activation (12). This led us to hypothesize that SOD1 activates Rac1/Nox2 complexes in the endosomal compartment to produce O2•– by inhibiting the GTPase activity of Rac1. To this end, we isolated unstimulated Nox2-containing endosomes from primary mouse dermal fibroblasts (PMDFs) and tested whether supplementing SOD1 would activate NADPH-dependent O2•– generation by this compartment. To confirm that endosomal O2•– was indeed derived from Nox2, we used PMDFs isolated from Nox2gp91phox KO mice or WT control littermates. Iodixanol density gradient separation of vesicular fractions from WT heavy mitochondrial supernatant demonstrated 2 predominant peak fractions containing Nox2gp91phox, Rac1, and SOD1 proteins (fractions 10 and 12) that overlapped with a small peak in NADPH-dependent O2•– production and the early endosomal marker EEA1 (Figure 3F). The addition of purified bovine SOD1 to these isolated endosomes led to a significant enhancement in their ability to produce NADPH-dependent O2•– (Figure 3G). This enhancement in endosomal O2•– was sensitive to DPI, a Nox inhibitor, and was not observed in Nox2gp91phox KO PMDFs (Figure 3G), suggesting that the O2•– was indeed derived from Nox2. These results were reproduced in endosomes isolated from WT primary mouse embryonic fibroblasts (PMEFs), in which the production of NADPH-dependent O2•– was induced by the addition of bovine SOD1, but not E. coli SOD (Figure 3H), despite the equal capacity of both enzymes to dismutate O2•– (Supplemental Figure 3).
Our findings demonstrating that SOD1 can regulate Rac1/Nox2 activation in a redox-dependent fashion provided insights into SOD1 function. However, it remained unclear how ALS-associated mutations in SOD1 dysregulated Rac1 activation and Nox activity. To this end, we generated and purified WT human SOD1 and 2 human ALS mutant SOD1 proteins (L8Q and G10V) from bacteria as GST fusions and cleaved the GST prior to analysis (Figure 3I). Of note, generation of the SOD1G93A GST fusion was unsuccessful, likely because of toxicity of this fusion in bacteria. However, in contrast to previous studies demonstrating that SOD1G93A has normal enzymatic activity (31), SOD1L8Q and SOD1G10V proteins purified from E. coli lacked dismutase activity compared with SOD1WT prepared under the same conditions (Figure 3I). Interestingly, both human SOD1L8Q and SOD1G10V mutant proteins had enhanced ability to bind Rac1 when compared with human SOD1WT (Figure 3J). Unlike human SOD1WT, binding of these SOD1 mutants to Rac1 was not disrupted by X/XO-derived ROS (Figure 3J), suggesting that the redox regulation of SOD1/Rac1 interactions is altered by L8Q and G10V mutations in SOD1. Importantly, human SOD1L8Q and SOD1G10V mutant proteins also demonstrated enhanced ability to inhibit Rac1-GTP hydrolysis compared with human SOD1WT (Figure 3K). Reduced metalation of bacterial-derived human SOD1WT led to decreased effectiveness for inhibiting Rac1-GTPase activity compared with purified bovine SOD1 (Figure 3I), which was likely the result of the reduced binding affinity to Rac1 shown in Figure 2C. However, the extent of metalation appeared to have less of an effect on the ability of bacterial-derived human mutant SOD1 proteins to bind Rac1 and inhibit GTPase activity.
Based on the above results, we hypothesized that certain ALS mutations in SOD1 might dysregulate Nox2 activation in the endosomal compartment by virtue of their more persistent and redox-insensitive activation of Rac1. To this end, we tested the time course of NADPH-dependent O2•– production by isolated endosomal fractions following the addition of human SOD1WT or SOD1L8Q proteins. As previously observed for bovine SOD1WT (Figure 3H), human SOD1WT activated the production of NADPH-dependent O2•– by isolated PMEF endosomes (Figure 3L). This activation in O2•– production peaked by 15 minutes and returned to baseline by 1 hour. Such transient activation was consistent with Nox-derived ROS inhibiting SOD1/Rac1 interactions and activating GTP hydrolysis by Rac1, leading to a self-regulated reduction in Nox activation. In contrast, adding SOD1L8Q to PMEF endosomes gave rise to persistent NADPH-dependent O2•– production, to 1 hour (Figure 3L). Collectively, these results suggest that the SOD1L8Q ALS mutant is dysregulated in its ability to activate Nox2 by virtue of altered redox-sensitive interactions with Rac1. Such findings are consistent with elevated Nox activity in spinal cords and brains of ALS mice.
Treatment with the Nox inhibitor apocynin increases lifespan and slows disease progression in ALS mice.
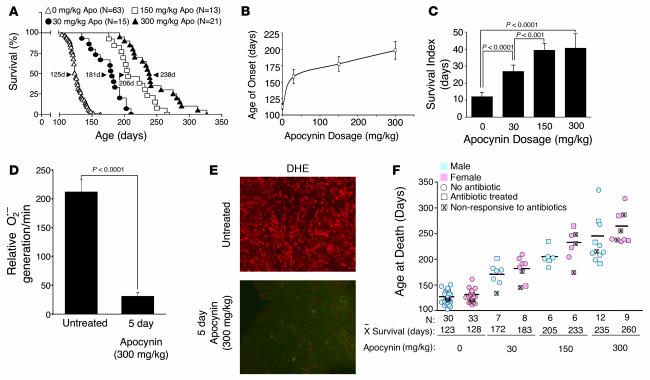
Our finding that certain ALS-associated SOD1 mutations lead to primary defects in Nox activation suggests that inhibiting Nox activation may be therapeutically beneficial in ALS. Indeed, 2 recent studies have demonstrated that deletion of Nox2 can enhance survival and delay disease onset in ALS mice (3, 5). In light of these findings, we tested whether apocynin administration in the drinking water from 2 weeks of age would prolong survival of hemizygous SOD1G93A mice. As shown in Figure 4A, apocynin treatment increased survival of ALS mice in a dose-dependent fashion. At the highest dose, 300 mg/kg, 50% survival times were increased from 125 to 238 days. Both the highest and lowest doses also significantly increased the number of motor neurons in the lumbar spinal cord at 120 days (Supplemental Figure 4, A and B), and there was a clear apocynin dose response in delaying age of disease onset. Treatment with apocynin also significantly increased the survival index of ALS mice (assessed as time to death from first signs of symptoms) using 2 confirmatory methods for onset of disease: weight loss (Figure 4, B and C) and gait (Supplemental Figure 4, C and D). Cumulatively, these findings demonstrated that long-term apocynin treatment significantly delayed disease progression in ALS mice. To confirm that apocynin treatment inhibited Nox activity in vivo, we also treated terminal-stage ALS mice for 5 days with apocynin in the water and evaluated Nox activity and ROS production in the spinal cord by lucigenin and dihydroethidium (DHE) assays, respectively. These studies demonstrated that apocynin treatment effectively inhibited NADPH-dependent O2•– production in vivo (Figure 4, D and E) at later stages of disease associated with microgliosis and increased Nox2 expression (3). The cellular level of increased DHE staining in the spinal cord also suggests that multiple cell types in addition to microglia may have enhanced redox stress. This notion is supported by the fact that multiple Nox genes (i.e., Nox1 and Nox2) have been shown to influence progression of disease in SOD1G93A transgenic mice (5).
Figure 4. Treatment with the Nox inhibitor apocynin increases lifespan and slows disease progression in mice hemizygous for the SOD1G93A transgene.
(A) Kaplan-Meier survival curve for mice treated with indicated doses of apocynin in their water beginning at 14 days of age. n is shown along with median survival time (arrowhead) for each group. Survival differences were significant in all between-group comparisons (log-rank test). (B) Age of disease onset, as determined by a 10% weight loss from peak body weight, for the various doses of apocynin. (C) Dose affect of apocynin treatment on survival index, measured as time from disease onset (as determined by weight loss) until clinical death. (D) Rate of NADPH-dependent O2•– production in total endomembranes isolated from lumbar spinal cords of end-stage SOD1G93Atransgenic mice (~120 days of age) that were either untreated or treated with apocynin (300 mg/kg) in the drinking water for 5 days prior to harvesting spinal cords (n = 5 per group). (E) DHE fluorescence was assessed in lumbar spinal cord sections from 2 mice evaluated in D. (F) Survival data of male and female mice at the indicated apocynin dose. Mice treated for eye infections with antibiotics are marked as squares; those unsuccessfully treated that died from eye infections are marked by “X” within the square. Circles denote animals that never contracted eye infection. n and mean survival time is indicated for each group. Data in B–D are mean ± SEM.
One unexpected finding was that ALS mice with prolonged survival developed eye infections that, if left untreated, led to rapid death without the normal course of motor abnormalities. This was similarly observed in ALS mice on the Nox2-deficient background (5). Treatment of these eye infections with systemic antibiotics led to resolution in approximately 50% of cases (Figure 4F). Importantly, treatment of ALS mice with antibiotics did not increase survival in the absence of apocynin, and non-ALS mice treated with apocynin did not develop eye disease (data not shown).
Discussion
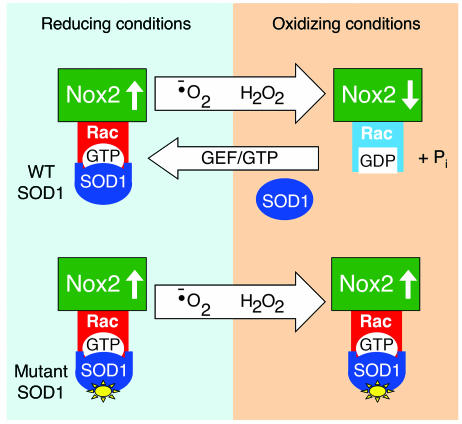
Our findings demonstrate that SOD1, an enzyme that ubiquitously directs the conversion of O2•– to H2O2 in cells, has the ability to control Rac1/Nox2 activation through redox-dependent physical interactions with Rac1. Based on our findings, we propose a redox sensor model by which SOD1 can regulate Nox2-dependent O2•– production through its ROS-sensitive control of Rac-GTP hydrolysis (Figure 5). Upon stimulation, activated Rac-GTP is recruited to the assembling membrane-associated Nox2 complex along with SOD1. Under the reducing conditions of the cytoplasm, SOD1 efficiently binds to Rac-GTP and inhibits intrinsic and/or GAP-facilitated GTPase activity, thereby maintaining Rac in the active state and consequently increasing the production of Nox2-derived O2•–. Local accumulation of H2O2, by either spontaneous or SOD1-facilitated dismutation of O2•–, leads to the dissociation of SOD1 from Rac-GTP and inactivation of Rac through GTP hydrolysis. Since Rac-GDP cannot support Nox2 activation, this event leads to inactivation of the Nox2 complex and reduction in ROS production. It is this redox-sensitive uncoupling of SOD1 from Rac that appears to be dysfunctional in certain ALS mutants of SOD1, leading to hyperactivation of Nox-derived O2•– by endomembranes.
Figure 5. Redox-sensor model for SOD1-mediated regulation of Nox2 ROS production through Rac.
Under reducing conditions SOD1 is bound to Rac-GTP and stabilizes Rac activation by inhibiting intrinsic and GAP-mediated GTP hydrolysis. Increased Rac-GTP levels lead to activation of Nox2 and production of O2•–. O2•– generated by the Nox2 complex is converted to H2O2 by SOD1 or through spontaneous dismutation. As the local concentration of H2O2 rises, oxidation of Rac leads to the dissociation of SOD1. With SOD1 no longer bound to Rac-GTP, hydrolysis to Rac-GDP occurs more quickly, leading to inactivation of the Nox2 complex. SOD1 can then recycle to repeat the process as Rac/Nox2 is reactivated. Through this mechanism, we propose that SOD1 can sense the local concentration of ROS at sites of Rac/Nox2 complex activation and control the activity of the complex. In certain ALS mutants of SOD1, redox-dependent dissociation of SOD1 from Rac1 is impaired, leading to sustained activation of Rac1-GTP and higher levels of Nox2 activation.
The finding that SOD1 functions to regulate Rac1-dependent O2•– production by NADPH oxidases in a redox-dependent fashion has important implications for ALS and the development of targeted antioxidant therapies such as apocynin. Several antioxidant therapies directed at clearance of ROS have been previously evaluated in clinical trials and case studies for ALS (32–34). However, these studies have failed to enhance survival. The therapeutic effect of long-term apocynin administration to SOD1G93A transgenic mice resembled or exceeded that of Nox2 or Nox1 deletion in the same ALS mouse model (5). The robust therapeutic effect of apocynin in preventing progression of ALS in this model suggests that inhibiting ROS production, as opposed to scavenging ROS, may be significantly more protective in ALS. The ability of picomolar quantities of H2O2 to liberate SOD1 from Rac-GTP and allow for GTP hydrolysis to occur suggests that the mechanism of in vivo regulation of Nox may be exquisitely sensitive to small changes in cellular ROS. This mechanism may allow Rac1 to sense and regulate changes in cellular O2•– through SOD1 enzymatic conversion to H2O2. Such spatial regulation may be a key aspect of SOD1 function as a redox sensor and may explain the lack of therapeutic benefit from general ROS scavengers compared with direct inhibition of dysregulated Nox complexes with apocynin.
When considering the mechanism of the therapeutic effect of apocynin to inhibit Nox activation, it is worth noting several other potential aspects of this molecule that might also contribute to its neuroprotective effects in ALS mice. Apocynin can also inhibit other relevant enzymes that participate in cellular redox balance. For example, inhibition of Nox activity by apocynin has been linked to changes in the intracellular reduced/oxidized glutathione ratio and NF-κB–dependent cyclooxygenase-2 synthesis (35). Additionally, apocynin has been shown to alter the abundance of arachidonic acid–derived inflammatory mediators (36). Whether these apocynin-dependent effects are all linked to alterations in Nox activation remains to be fully investigated. There are also several factors that make apocynin appealing for clinical applications. Apocynin itself is inactive and only becomes active after the combined action of ROS and peroxidase activity (37). These characteristics theoretically minimize the off-target effects and makes the active metabolite largely available at sites of oxidative stress.
There are at least 2 key points worth considering about potential applications of apocynin to treat ALS in humans. First, it is unclear whether Nox dysregulation occurs in sporadic forms of ALS (not involving SOD1 mutations) that account for 90%–95% of cases. Second, it is also unclear at what point during the progression of ALS apocynin treatment could provide therapeutic benefit. To begin to address the latter question, we initiated apocynin treatment of ALS mice at 14, 60, and 80 days of age and evaluated survival (Supplemental Figure 5). In a relatively small cohort of sibling ALS mice, there was a significant benefit to treatment at all time points, although early intervention led to maximal therapeutic benefit. These findings suggest that inhibition of Nox activity during early phases of disease is important to slowing disease progression. Hence, early genetic diagnosis of patients with familial ALS would be critical for potential clinical applications with apocynin.
The appreciation that SOD1 is both a catabolic enzyme and regulatory protein involved in cellular ROS metabolism has potential broad-ranging implications for cell biology and the redox-dependent regulation of Rac1. The regulation of ROS production by Rac1 is important for many cellular processes involved in signal transduction (27), actin cytoskeletal rearrangements (28, 38, 39), and cell migration (29), proliferation (13, 29, 40, 41), and differentiation (30). In the context of redox-dependent signal transduction, it has become increasingly recognized that the localized production of O2•– and H2O2 at discrete subcellular domains appears to be a major regulatory aspect of pathways that depend on Nox-derived ROS (12, 42). Some of these pathways also involve activation and recruitment of Rac1 to specific membrane domains. Hence, further investigation into the role SOD1 plays in regulating the redox environment at discrete subcellular domains may uncover new layers of complexity in redox signaling in health, disease, and aging.
Methods
Cell culture and adenoviral infections
SH-SY and M059J cell lines were purchased from ATCC and cultured according to the manufacturer’s instructions. Cells were infected with replication defected adenoviral vectors containing human WT or mutant SOD1 at 1,000 particles/cell when cells reached 70% confluency. Cells were harvested at 36, 48, and 72 hours after infection for Rac activation assays, Nox activity assays, and cell death assays, respectively. Recombinant adenoviruses expressing SOD1WT or LacZ have been previously described (43). L8Q and G93A mutations in SOD1 were introduced into the pAd.CMVlinkwtSOD1 proviral vector using the Gene Editor in vitro site-directed mutagenesis system (Promega). Viruses were then generated as previously described (44).
Trypan blue exclusion assay
Cells were trypsinized and resuspended by gentle pipetting 72 hours after adenoviral infection. A 0.4% trypan blue solution (50 μl) was added to 200 μl cell suspension for a final trypan blue concentration of 0.08%. The cell suspension was then incubated for 2 minutes at room temperature, and the number of stained cells was counted using a hemocytometer. The percentage of dead cells was calculated as the number of stained cells divided by the total number of cells.
In situ DHE staining for O2•–
Mice were euthanized with an overdose of sodium pentobarbital. The lumbar spinal cord was removed immediately and embedded in OTC as 4-mm-long segments. From each group, 30-μm sections were immediately generated within hours of embedding and stained in DHE as follows. Sections were rinsed in PBS containing 100 μM rotenone to inhibit mitochondrial respiration prior to incubation with 1 μM DHE (Invitrogen) for 5 minutes in the dark and the continued presence of 100 μM rotenone. Sections were then rinsed in PBS and coverslipped with Vectashield mounting media containing DAPI. DHE fluorescence was detected using a rhodamine emission filter. Microscope/camera exposure settings were kept constant between samples within an individual experiment, and control and experimental animals were always processed in parallel.
IP and Western blotting
SOD1-null mice (Sod1tm1Leb) were purchased from Jackson Laboratories (45). Tissue lysates from WT and SOD1 KO littermates were generated by homogenization in ice-cold PBS followed by the addition of an equal volume of 2× lysis buffer containing 40 mM Tris-HCl (pH 7.4), 300 mM NaCl, 2% Triton X-100, 100 mM NaF, 80 mM β-glycerophosphate, 10 mM EDTA, and a protease inhibitor cocktail tablet. Protein concentrations were measured by the Bradford assay. IP of Rac1 proteins was performed by incubating 600 μg total protein with 4 μg primary anti-Rac1 antibody (Upstate Cell Signaling Solutions) in 500 μl lysis buffer, followed by rotating for 2 hours at 4°C. Protein A-dynabeads, washed twice with lysis buffer, were then added and rotated overnight at 4°C, followed by magnetic removal of the IP complexes. Beads were washed 4 times with lysis buffer. Pellets were then resuspended in SDS-PAGE reducing loading buffer and incubated at 98°C for 5 minutes before separation by SDS-PAGE. The nitrocellulose membranes bearing the transferred proteins were blocked overnight at 4°C in blocking buffer containing 4% w/v nonfat dried milk and 0.3% Tween 20 in PBS and incubated with primary antibodies to SOD1 (The Binding Site Limited) and Rac1 (Santa Cruz Biotechnology Inc.) and then with infrared dye–conjugated secondary antibodies. Protein bands were detected by the Odyssey infrared imaging system (LI-COR Biotechnology).
Pulldown assays with His-tagged proteins
Dynabeads talon for His-tagged proteins were washed with potassium phosphate buffer (PPHB) containing 100 mM KH2PO4, 10 mM NaCl, 0.25 mM MgCl2, and 100 nM CaCl2. His-tagged Rac1 or Cdc42 (25 pmol) were incubated with 100 μl beads in PPHB at room temperature for 30 minutes with intermittent gentle agitation. The GTPases were either used directly or preloaded with 1 mM GTPγS or GDPβS in GTP binding buffer (50 mM HEPES (pH 7.6), 150 mM NaCl, and 0.1 mM EDTA) for 10 minutes at room temperature followed by the addition of MgCl2 to 10 mM final concentration and wash in PPHB. Samples treated with H2O2 were resuspended in 1 ml PPHB containing the indicated H2O2 concentration for 30 minutes at room temperature and then washed 4 times with PPHB (1 ml each wash). SOD1 (250 pmol) in 10 μl PPHB was then added to each tube of beads, and samples were incubated at room temperature for 30 minutes with intermittent gentle agitation. Beads were then washed 3 times with PPHB to remove unbound SOD1. The fourth wash was carried out in 50 mM Tris (pH 6.8). Proteins were eluted using 20 μl 125 mM imidazole, and samples were then mixed with SDS-PAGE reducing loading buffer and separated by SDS-PAGE for Western blotting.
Rac1 GTPase assay
Rac1 GTPase assays were performed as previously described (46), with modifications. His-tagged Rac1 or Cdc42 (25 pmol) were incubated with 25 pmol GTP and 2.5 pmol P32-labeled γ-GTP in GTP binding buffer containing 50 mM HEPES (pH 7.6), 150 mM NaCl, and 0.1 mM EDTA for 10 minutes at room temperature and then placed on ice water. A 1-μl aliquot was then taken for thin-layer chromatography (TLC) as time 0 of this GTPase reaction. Three proteins were added in various combinations to each reaction — including bovine SOD1, E. coli SOD1, and/or p29-GAP — as indicated in Results and figure legends. The molar ratio of Rac1 or Cdc42 to p29-GAP was 1:1. The molar ratio of Rac1 or Cdc42 to SOD1 was 1:10. To start the GTPase reaction, an equal volume of 2× GTPase buffer containing 50 mM HEPES (pH 7.6), 150 mM NaCl, 10 mM EDTA, and 10 mM MgCl2 was added to each condition at 15°C. Where indicated, 100 mU xanthine oxidase was incubated in the reaction mixture with a final xanthine concentration of 100 μM. Aliquots (1 μl) were spotted on a TLC plate from each sample at different time points. The TLC was run for 90 minutes at room temperature in 1M acetic acid with 0.8M LiCl running buffer. To quantify GTP hydrolysis, the free phosphate (Pi) bands were cut out along with the corresponding GTP bands. Each was put in liquid scintillation fluid and counted by liquid scintillation spectrometry. The percentage of GTP hydrolyzed was calculated as Pi divided by the sum of Pi and GTP.
Construction of GST-Rac1 and GST-SOD1 fusion proteins
Bacterial expression constructs for WT and deletion mutants GST-Rac1 and/or GST-SOD1 were generated by PCR-mediated cloning into the pGex-2T vector (Amersham Biosciences). All bacterial fusion constructs were confirmed by complete sequencing. ALS mutations L8Q (47) and G10V (48) were introduced into the GST-SOD1 using the Gene Editor in vitro site-directed mutagenesis system (Promega). The primer sequence used to generate the L8Q mutant was 5′-AAGGCCGTGTGCGTGCAGAAGGGCGACGGCCCA-3′. The primer sequence used to generate the G10V mutant was 5′-AAGGCCGTGTGCGTGCTGAAGGTTGACGGCCCA-3′.
Expression and purification of bacterial GST-tagged proteins
The GST-tagged expression constructs were transformed into E. coli using ampicillin selection. Bacterial colonies harboring the WT and mutant constructs were grown in LB medium containing 100 μg/ml ampicillin in 1-l flasks at 37°C to a cell density of A600 = 0.6. Isopropyl-D-thiogalactopyranoside was then added to 1 mM to induce the expression of GST-tagged proteins, and cultures were grown for 6 hours at 37°C. The bacteria were collected by a 4,000 g spin for 15 minutes at 4°C and resuspended in PBS on ice. The bacteria were then lysed on ice by 5 30-seconds sonicator pulses using a virsonic cell disruptor (VirTis Gardiner). The bacterial lysate was then centrifuged at 30,000 g for 30 minutes to pellet debris. The fusion proteins were purified from cellular extracts using glutathione-sepharose beads (Amersham Biosciences) according to the manufacturer’s instructions, and the GST fusion proteins were eluted with 10 mM glutathione, 50 mM Tris-HCl (pH 7.5), and 120 mM NaCl. Protein purity was assessed by Coomassie-stained SDS-PAGE, and protein concentrations were normalized using the Bradford method. It should be noted that GST-Rac1 fusion proteins containing 88 or 116 amino acids of the N terminus of Rac1 consistently migrated faster than their predicted molecular weights in SDS-PAGE, which was likely due to altered folding properties of domains contained within these deletion mutants. The GST-tagged SOD1 proteins were cleaved from GST using a thrombin cleavage capture kit (EMD Bioscences). Following cleavage, SOD1 proteins were separated from the cleaved GST-tag using an FPLC glutathione-sepharose column.
Demetalation of SOD1
Demetalation of purified bovine SOD1 was performed as previously described (1), with modification. Cu and Zn were removed by exposing purified bovine SOD1 to PBS (pH 3.0) plus 2 mM EDTA and stirring for 60 minutes at 4°C. The protein was then dialyzed overnight against 50 mM potassium phosphate (pH 7.4). A fraction of demetalated bovine SOD1 was then remetalated by dialysis against 100 mM sodium acetate (pH 5.5) in the presence of a 40-fold molar excess of Zn, followed by a 4-fold molar excess of Cu. To remove unbound metals, the SOD protein was then dialyzed several times against PBS (pH 7.4). The Cu/Zn content of native, demetalated, and remetalated bovine SOD1 was determined as previously described (49). Briefly, 10 μg SOD1 was mixed with 1 ml assay buffer containing 100 mM sodium borate (pH 7.8), 2% SDS, and 100 μM pyridylazoresorcinol (PAR). The reaction mixture was heated for 20 minutes at 100°C. Zn and Cu levels were calculated as the decrease in 500-nm reading measured on a Shimadzu UV-160 spectrophotometer after the addition of 0.8 mM nitrilotriacetic acid (NTA) and EDTA, respectively. The Zn or Cu content in SOD1 was reported as the molar ratio of Zn or Cu to SOD1. SOD1 enzyme activity gels were performed as we described previously (43). Briefly, 10 μg native, demetalated, or remetalated SOD1 was run on a native 12% polyacrylamide gel. SOD1 activity was determined using nitroblue tetrazolium reduction. Enzymatic activity was defined as the clearance zones in a background of black precipitate.
Subcellular fractionation
Isolation of total endomembranes.
Cells or tissue were lysed in homogenization buffer (50 mM Tris-HCl, 320 mM sucrose, and 1 mM EDTA, pH 7.4) by nitrogen cavitation. Lysate (600 μg) was then centrifuged at 3,000 g to create a postnuclear supernatant devoid of heavy mitochondria (PNS), and the supernatant was subsequently centrifuged at 100,000 g for 1 hour to spin down total endomembranes. The membranes were washed 3 times in homogenization buffer before being resuspended in 150 μl homogenization buffer prior to Nox activity assays.
Iodixanol separation of endosomes.
Buoyant density centrifugation was used for subcellular fractionation and isolation of endosomes containing Nox2, Rac1, and SOD1 activity. Cells were washed twice with ice-cold PBS and scraped into a 1.5-ml microfuge tube using the same buffer. The cells were pelleted and resuspended in homogenization buffer (HMB) containing 0.25 M sucrose, 20 mM HEPES (pH 7.4), 1 mM EDTA, and an EDTA-free protease inhibitor cocktail. The cells were homogenized using nitrogen cavitation in a cell disruption high-pressure chamber (Parr instruments). The pressure was raised to 650 psi for 5 minutes and released suddenly. The homogenate was centrifuged at 3,000 g for 15 minutes to pellet unbroken cells, nuclei, and heavy mitochondria. The heavy mitochondrial supernatant (HMS) was bottom-loaded into an iodixanol discontinuous gradient in a 12.5-ml SW41Ti ultracentrifuge tube using a previously described method with modifications (50, 51). The discontinuous gradient was composed of 1.25 ml HMB without EDTA followed by bottom loading of the following sequential iodixanol steps: 1.0 ml 2.5%, 1.0 ml 5%, 1.5 ml 9%, 1.5 ml 14%, 2.5 ml 19%, 1.5 ml 26%, and finally the HMS in 2 ml 32%. Iodixanol concentrations were prepared fresh using a 50% iodixanol working solution (WS) diluted with HMB without EDTA. The WS was prepared by adding 1 part buffer containing 0.25 M sucrose and 120 mM HEPES (pH 7.4) to 5 parts iodixanol 60% stock solution. The gradients were centrifuged at 100,000 g using an SW41Ti swinging rotor overnight at 4°C. The fractions were collected from the top of the tube using a fraction collector (Labconco) in 500-μl fractions on ice. Endosomal fractions were concentrated by first diluting with buffer and centrifugation at 100,000 g for 1 hour, prior to Western blotting for various proteins. Fractions at the bottom of the gradient containing the sample loaded were not concentrated. The density gradient was designed to optimally separate the following compartments based on previous studies (51–55): fractions 1–5, plasma membrane (density, 1.03–1.05 g/ml); fractions 7–13, endosomal compartment (density, 1.055–1.11 g/ml); fractions 8–10, Golgi apparatus (density, 1.06–1.09 g/ml); fractions 10–13, light endoplasmic reticulum (density, 1.09–1.11 g/ml); fractions 13–18, lysozomes (density, 1.11–1.13 g/ml): fractions 18–21, light mitochondria (density, 1.13–1.15 g/ml); fractions 19–20, heavy endoplasmic reticulum (density, 1.145 g/ml); fractions 21–24, peroxisomes (density, 1.18–1.2 g/ml); and fractions 22–24, cytosolic proteins (density, 1.26 g/ml). Monoclonal antibody anti-Nox2 (54.1) was a kind gift from A.J. Jesaitis (Montana State University, Bozeman, Montana, USA; ref. 56) and W.M. Nauseef (University of Iowa).
Lucigenin chemiluminescence assay for NADPH-dependent O2•– production
Nox activities were analyzed by measuring the rate of O2•– generation using a chemiluminescent, lucigenin-based system (57). Lucigenin (5 μM) in 50 μl of each subcellular fraction (when iodixanol fractionation of PNS was used) or 100 μl of endomembranes (from 400 μg PNS) was incubated in the dark at room temperature for 15 minutes. Lucigenin chemiluminescence was measured using a single-tube Luminometer TD20-20 (Turner Designs). The reaction was initiated by the addition of β-NADPH to a final concentration of 100 μM with or without DPI (10 μM) and/or SOD (2.5 μM) as indicated. Lucigenin chemiluminescence was measured over the course of 5 minutes. The initial slope of the luminescence curve (RLU/min) was used to calculate the rate of luminescence product formation and compared between samples as an index of Nox activity. In the absence of NADPH, the luminescence was negligible and did not change over time.
Animal models
All animal experimentation met or exceeded the standards set by the principles and procedures outlined in the NIH guidelines for the care and use of experimental animals. All procedures were approved by the Animal Care and Use Committee at the University of Iowa. All mice were purchased from Jackson Laboratories. These included (a) SOD1 KO mice (strain name, B6;129S7-Sod1tm1Leb/J; stock no. 002972), (b) Nox2 KO mice (strain name, B6.129S6Cybbtm1Din/J; stock no. 002365), (c) transgenic mice overexpressing WT human SOD1 [strain name, B6SJL-Tg(SOD1)2Gur/J; stock no. 002297], control for transgenic mice overexpressing SOD1G93A, and (d) transgenic mice overexpressing a human SOD1G93A mutant transgene [strain name, B6SJL-Tg(SOD1G93A)1Gur/J; stock no. 002726]. Genotypes were confirmed using PCR protocols given by Jackson Laboratories. Only transgenic mice hemizygous for the SOD1WT or SOD1G93A transgenes were used for analysis. The SOD1G93A line of mice contains the high-expressing form of mutant SOD1, and animals develop disease onset at about 100 days of age and die about 25 days later (58). Clinical death was defined as the time when the mice could no longer right themselves within 20 seconds of being placed on their backs or when they lost 20% of their body weight during a 1-week period. If either of these 2 criteria was met, the mice were euthanized. At the time of advanced clinical symptoms, SOD1G93A mice were grueled twice daily and individually housed.
Treatment of ALS mice with apocynin
Only mice hemizygous for the SOD1G93A transgene were used in studies. Apocynin powder was added to hot (~60°C) sterile water at 3 different doses. The water was allowed to cool to room temperature before being given to the mice. The concentrations of apocynin tested were 200 μg/ml, 1 mg/ml, and 2 mg/ml in water. The conversion to mg/kg was estimated based on the average daily water intake of approximately 4 ml/mouse (which was confirmed in 25- to 30-g ALS mice; see also ref. 59), giving rise to dosing of approximately 30, 150, and 300 mg/kg/d. Water was changed every 5–7 days (a time point that demonstrated no detectable decay of apocynin by Electrospray LC-MS). Mice were started on apocynin-treated water at 14 days after birth unless otherwise indicated. The mice in the study were allowed free access to food and water. At the time of advanced clinical symptoms, mice were grueled twice daily with chow soaked in apocynin water or apocynin-free water. Those apocynin-treated SOD1G93A transgenic mice that lived longer than untreated controls were susceptible to superficial eye infections. At first sign of discharge, mice were placed on water or gruel containing antibiotics (0.1 mg/ml gentamycin and 0.1 mg/ml ceftazidime). If the condition worsened, mice were additionally injected subcutaneously with 85 mg/kg enterfloxin (Baytril) once daily for 14 days. To ensure that antibiotic treatment did not alter the course of ALS-like disease, SOD1G93A transgenic mice were put on antibiotic water and monitored for survival (n = 8); no significant changes in survival were seen compared with untreated animals (data not shown).
Physiologic assays for disease
Beginning at 80 days of age, SOD1G93A transgenic mice were evaluated for onset of motor neuron defects and disease symptoms by assessing both weight and stride length weekly. For stride length analysis, front and hind paws of mice were covered in different colored paint to record walking patterns during continuous locomotion. Stride length was measured, and the average stride was calculated from at least 6 consecutive strides (60).
Statistics
Statistical significance for all comparisons with the exception of survival curves was assessed using ANOVA followed by Dunnett’s post-test or Student’s t test. Kaplan-Meier survival curves were generated using Prism software and compared using the log-rank test; resulting P values are 2-tailed. In all statistical analyses, P values less than 0.05 were deemed significant.
Supplementary Material
Acknowledgments
This work was supported by the NIDDK (grant DK067928), the Roy J. Carver Chair in Molecular Medicine (J.F. Engelhardt), and the Center for Gene Therapy Animal Models Core (P30 DK54759). The authors would like to thank Jackie Bickenbach for help isolating the PMDFs. We also gratefully acknowledge Mariah Steele and Ola Awad for editorial assistance and Patricia Yarolem, Sara Nelson, and Andrea Park for technical assistance.
Footnotes
Nonstandard abbreviations used: ALS, amyotrophic lateral sclerosis; DHE, dihydroethidium; DPI, diphenyleneiodionium chloride; Nox, NADPH oxidase; O2•–, superoxide; PMDF, primary mouse dermal fibroblast; PMEF, primary mouse embryonic fibroblast; PPHB, potassium phosphate buffer; SOD1, superoxide dismutase–1; TLC, thin-layer chromatography; X/XO, xanthine/xanthine oxidase.
Conflict of interest: The authors have declared that no conflict of interest existed while scientific results were obtained for the present work. Following completion of the research presented in this article, a relationship with a company was formed to move findings to clinical trials. The authors have no equity in this company.
Citation for this article: J. Clin. Invest. 118:659–670 (2008). doi:10.1172/JCI34060
Maged M. Harraz and Jennifer J. Marden contributed equally to this work.
See the related Commentary beginning on page 474.
References
- 1.McCord J.M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 2.Rosen D.R., et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 3.Wu D.C., Re D.B., Nagai M., Ischiropoulos H., Przedborski S. The inflammatory NADPH oxidase enzyme modulates motor neuron degeneration in amyotrophic lateral sclerosis mice. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12132–12137. doi: 10.1073/pnas.0603670103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruijn L.I., Miller T.M., Cleveland D.W. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 5.Marden J.J., et al. Redox modifier genes in amyotrophic lateral sclerosis in mice. J. Clin. Invest. 2007;117:2913–2919. doi: 10.1172/JCI31265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boillee S., et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 7.Lambeth J.D. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 8.Hordijk P.L. Regulation of NADPH oxidases: the role of Rac proteins. Circ. Res. 2006;98:453–462. doi: 10.1161/01.RES.0000204727.46710.5e. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa S., et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 2004;114:1752–1761. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stolk J., Hiltermann T.J., Dijkman J.H., Verhoeven A.J. Characteristics of the inhibition of NADPH oxidase activation in neutrophils by apocynin, a methoxy-substituted catechol. Am. J. Respir. Cell Mol. Biol. 1994;11:95–102. doi: 10.1165/ajrcmb.11.1.8018341. [DOI] [PubMed] [Google Scholar]
- 11.Ueyama T., Geiszt M., Leto T.L. Involvement of Rac1 in activation of multicomponent Nox1- and Nox3-based NADPH oxidases. Mol. Cell. Biol. 2006;26:2160–2174. doi: 10.1128/MCB.26.6.2160-2174.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q., et al. Nox2 and Rac1 regulate H2O2-dependent recruitment of TRAF6 to endosomal interleukin-1 receptor complexes. Mol. Cell. Biol. 2006;26:140–154. doi: 10.1128/MCB.26.1.140-154.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irani K., et al. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- 14.Abo A., et al. Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1. Nature. 1991;353:668–670. doi: 10.1038/353668a0. [DOI] [PubMed] [Google Scholar]
- 15.Rae T.D., Schmidt P.J., Pufahl R.A., Culotta V.C., O’Halloran T.V. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 16.Hirshberg M., Stockley R.W., Dodson G., Webb M.R. The crystal structure of human rac1, a member of the rho-family complexed with a GTP analogue. Nat. Struct. Biol. 1997;4:147–152. doi: 10.1038/nsb0297-147. [DOI] [PubMed] [Google Scholar]
- 17.Ito Y., et al. Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry. 1997;36:9109–9119. doi: 10.1021/bi970296u. [DOI] [PubMed] [Google Scholar]
- 18.Sprang S.R. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 19.Worthylake D.K., Rossman K.L., Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature. 2000;408:682–688. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
- 20.Rittinger K., Walker P.A., Eccleston J.F., Smerdon S.J., Gamblin S.J. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 21.Rittinger K., et al. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 22.Menard L., et al. Rac1, a low-molecular-mass GTP-binding-protein with high intrinsic GTPase activity and distinct biochemical properties. Eur. J. Biochem. 1992;206:537–546. doi: 10.1111/j.1432-1033.1992.tb16957.x. [DOI] [PubMed] [Google Scholar]
- 23.Roberts A.W., et al. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity. 1999;10:183–196. doi: 10.1016/s1074-7613(00)80019-9. [DOI] [PubMed] [Google Scholar]
- 24.Cheng G., Diebold B.A., Hughes Y., Lambeth J.D. Nox1-dependent reactive oxygen generation is regulated by Rac1. J. Biol. Chem. 2006;281:17718–17726. doi: 10.1074/jbc.M512751200. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X., Carnevale K.A., Cathcart M.K. Human monocytes use Rac1, not Rac2, in the NADPH oxidase complex. J. Biol. Chem. 2003;278:40788–40792. doi: 10.1074/jbc.M302208200. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson B., Koenigsknecht-Talboo J., Grommes C., Lee C.Y., Landreth G. Fibrillar beta-amyloid-stimulated intracellular signaling cascades require Vav for induction of respiratory burst and phagocytosis in monocytes and microglia. J. Biol. Chem. 2006;281:20842–20850. doi: 10.1074/jbc.M600627200. [DOI] [PubMed] [Google Scholar]
- 27.Sulciner D.J., et al. rac1 regulates a cytokine-stimulated, redox-dependent pathway necessary for NF-kappaB activation. Mol. Cell. Biol. 1996;16:7115–7121. doi: 10.1128/mcb.16.12.7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kheradmand F., Werner E., Tremble P., Symons M., Werb Z. Role of Rac1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science. 1998;280:898–902. doi: 10.1126/science.280.5365.898. [DOI] [PubMed] [Google Scholar]
- 29.Yamaoka-Tojo M., et al. IQGAP1, a novel vascular endothelial growth factor receptor binding protein, is involved in reactive oxygen species-dependent endothelial migration and proliferation. Circ. Res. 2004;95:276–283. doi: 10.1161/01.RES.0000136522.58649.60. [DOI] [PubMed] [Google Scholar]
- 30.Puceat M., Travo P., Quinn M.T., Fort P. A dual role of the GTPase Rac in cardiac differentiation of stem cells. Mol. Biol. Cell. 2003;14:2781–2792. doi: 10.1091/mbc.E02-09-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valentine J.S., Hart P.J. Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3617–3622. doi: 10.1073/pnas.0730423100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barber S.C., Mead R.J., Shaw P.J. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochim. Biophys. Acta. 2006;1762:1051–1067. doi: 10.1016/j.bbadis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Galbussera A., et al. Vitamin E intake and quality of life in amyotrophic lateral sclerosis patients: a follow-up case series study. Neurol. Sci. 2006;27:190–193. doi: 10.1007/s10072-006-0668-x. [DOI] [PubMed] [Google Scholar]
- 34.Ferrante K.L., et al. Tolerance of high-dose (3,000 mg/day) coenzyme Q10 in ALS. Neurology. 2005;65:1834–1836. doi: 10.1212/01.wnl.0000187070.35365.d7. [DOI] [PubMed] [Google Scholar]
- 35.Barbieri S.S., et al. Apocynin prevents cyclooxygenase 2 expression in human monocytes through NADPH oxidase and glutathione redox-dependent mechanisms. Free Radic. Biol. Med. 2004;37:156–165. doi: 10.1016/j.freeradbiomed.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Engels F., Renirie B.F., Hart B.A., Labadie R.P., Nijkamp F.P. Effects of apocynin, a drug isolated from the roots of Picrorhiza kurroa, on arachidonic acid metabolism. FEBS Lett. 1992;305:254–256. doi: 10.1016/0014-5793(92)80680-f. [DOI] [PubMed] [Google Scholar]
- 37.Hart B.A., Simons J.M. Metabolic activation of phenols by stimulated neutrophils: a concept for a selective type of anti-inflammatory drug. Biotechnol. Ther. 1992;3:119–135. [PubMed] [Google Scholar]
- 38.Nimnual A.S., Taylor L.J., Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nat. Cell Biol. 2003;5:236–241. doi: 10.1038/ncb938. [DOI] [PubMed] [Google Scholar]
- 39.Lopes N., et al. Thrombospondin 2 regulates cell proliferation induced by Rac1 redox-dependent signaling. Mol. Cell. Biol. 2003;23:5401–5408. doi: 10.1128/MCB.23.15.5401-5408.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pani G., et al. A redox signaling mechanism for density-dependent inhibition of cell growth. . J. Biol. Chem. 2000;275:38891–38899. doi: 10.1074/jbc.M007319200. [DOI] [PubMed] [Google Scholar]
- 41.Moore K.A., et al. Rac1 is required for cell proliferation and G2/M progression. Biochem. J. 1997;326:17–20. doi: 10.1042/bj3260017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ushio-Fukai M. Localizing NADPH oxidase-derived ROS. Sci. STKE. 2006;2006:re8. doi: 10.1126/stke.3492006re8. [DOI] [PubMed] [Google Scholar]
- 43.Zwacka R.M., et al. Redox gene therapy for ischemia/reperfusion injury of the liver reduces AP1 and NF-kappaB activation. Nat. Med. 1998;4:698–704. doi: 10.1038/nm0698-698. [DOI] [PubMed] [Google Scholar]
- 44.Anderson R.D., Haskell R.E., Xia H., Roessler B.J., Davidson B.L. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 2000;7:1034–1038. doi: 10.1038/sj.gt.3301197. [DOI] [PubMed] [Google Scholar]
- 45.Matzuk M.M., Dionne L., Guo Q., Kumar T.R., Lebovitz R.M. Ovarian function in superoxide dismutase 1 and 2 knockout mice. Endocrinology. 1998;139:4008–4011. doi: 10.1210/endo.139.9.6289. [DOI] [PubMed] [Google Scholar]
- 46.Kwon T., Kwon D.Y., Chun J., Kim J.H., Kang S.S. Akt protein kinase inhibits Rac1-GTP binding through phosphorylation at serine 71 of Rac1. J. Biol. Chem. 2000;275:423–428. doi: 10.1074/jbc.275.1.423. [DOI] [PubMed] [Google Scholar]
- 47.Bereznai B., Winkler A., Borasio G.D., Gasser T. A novel SOD1 mutation in an Austrian family with amyotrophic lateral sclerosis. Neuromuscul. Disord. 1997;7:113–116. doi: 10.1016/s0960-8966(96)00419-1. [DOI] [PubMed] [Google Scholar]
- 48.Kim N.H., Kim H.J., Kim M., Lee K.W. A novel SOD1 gene mutation in a Korean family with amyotrophic lateral sclerosis. J. Neurol. Sci. 2003;206:65–69. doi: 10.1016/s0022-510x(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 49.Ghezzo-Schoneich E., Esch S.W., Sharov V.S., Schoneich C. Biological aging does not lead to the accumulation of oxidized Cu,Zn-superoxide dismutase in the liver of F344 rats. Free Radic. Biol. Med. 2001;30:858–864. doi: 10.1016/s0891-5849(01)00473-7. [DOI] [PubMed] [Google Scholar]
- 50.Xia W., et al. Presenilin 1 regulates the processing of beta-amyloid precursor protein C-terminal fragments and the generation of amyloid beta-protein in endoplasmic reticulum and Golgi. Biochemistry. 1998;37:16465–16471. doi: 10.1021/bi9816195. [DOI] [PubMed] [Google Scholar]
- 51.Graham J., Ford T., Rickwood D. The preparation of subcellular organelles from mouse liver in self-generated gradients of iodixanol. Anal. Biochem. 1994;220:367–373. doi: 10.1006/abio.1994.1351. [DOI] [PubMed] [Google Scholar]
- 52.Billington D., Maltby P.J., Jackson A.P., Graham J.M. Dissection of hepatic receptor-mediated endocytic pathways using self-generated gradients of iodixanol (Optiprep). Anal. Biochem. 1998;258:251–258. doi: 10.1006/abio.1998.2561. [DOI] [PubMed] [Google Scholar]
- 53.Graham J.M., Billington D. Iodixanol — a new density gradient medium for the dissection of the endosomal compartment. Z. Gastroenterol. 1996;34(Suppl. 3):76–78. [PubMed] [Google Scholar]
- 54.Graham J.M. Purification of peroxisomes using a density barrier in a swinging-bucket rotor. ScientificWorldJournal. 2002;2:1400–1403. doi: 10.1100/tsw.2002.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plonne D., et al. Separation of the intracellular secretory compartment of rat liver and isolated rat hepatocytes in a single step using self-generating gradients of iodixanol. Anal. Biochem. 1999;276:88–96. doi: 10.1006/abio.1999.4311. [DOI] [PubMed] [Google Scholar]
- 56.Burritt J.B., Quinn M.T., Jutila M.A., Bond C.W., Jesaitis A.J. Topological mapping of neutrophil cytochrome b epitopes with phage-display libraries. J. Biol. Chem. 1995;270:16974–16980. doi: 10.1074/jbc.270.28.16974. [DOI] [PubMed] [Google Scholar]
- 57.Li Y., et al. Validation of lucigenin (bis-N-methylacridinium) as a chemilumigenic probe for detecting superoxide anion radical production by enzymatic and cellular systems. J. Biol. Chem. 1998;273:2015–2023. doi: 10.1074/jbc.273.4.2015. [DOI] [PubMed] [Google Scholar]
- 58.Gurney M.E., et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 59.Bachmanov A.A., Reed D.R., Beauchamp G.K., Tordoff M.G. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav Genet. 2002;32:435–443. doi: 10.1023/a:1020884312053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harper S.Q., et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.