Abstract
Qa-1b binds a peptide (AMAPRTLLL), referred to as Qdm (for Qa-1 determinant modifier), derived from the signal sequence of murine class Ia molecules. This peptide binds with high affinity and accounts for almost all of the peptides associated with this molecule. Human histocompatibility leukocyte antigen (HLA)-E, a homologue of Qa-1b, binds similar peptides derived from human class Ia molecules and interacts with CD94/NKG2 receptors on natural killer cells. We used surface plasmon resonance to determine the ability of Qa-1b to bind related ligands representing peptides derived from the leaders of class I molecules from several mammalian species. All of the peptides reported to bind HLA-E bound readily to Qa-1b. In addition, peptides derived from leader segments of different mammals also bound to Qa-1b, indicating a conservation of this “Qdm-like” epitope throughout mammalian evolution. We have attempted to define a minimal peptide on a polyglycine backbone that binds Qa-1b. Our previous findings showed that P2 and P9 are important but not sufficient for binding to Qa-1b. Although a minimum peptide (GMGGGGLLL) bound Qa-1b, its interaction was relatively weak, as were peptides sharing five or six residues with Qdm, indicating that multiple native residues are required for a strong interaction. This finding is consistent with the observation that this molecule preferentially binds this single ligand.
Keywords: major histocompatibility complex class Ib, Qa-1b, surface plasmon resonance, peptide, binding
Most MHC class I molecules are capable of binding a large array of individual peptides (1). In contrast, the murine class Ib molecule, Qa-1b, predominantly binds a single species (2, 3). We refer to this peptide as Qdm (for Qa-1 determinant modifier; reference 2), and it is derived from amino acids 3–11 of class Ia D-region–encoded molecules. HLA-E, which differs from Qa-1b in 55 of 181 residues in the α1 and α2 domains, binds leader peptides from human class Ia molecules that are very similar to the murine class Ia leader peptide bound by Qa-1b (4). HLA-E and Qa-1b, unlike other class Ia molecules, have serines rather than the conserved residues threonine and tryptophan at positions 143 and 147 in the “F” pocket, respectively. In the “B” pocket, HLA-E and Qa-1b also share the key residues methionine and alanine at positions 45 and 67, respectively. The HLA-E crystal structure reveals that side chains of five of the nine amino acids of the bound peptide protrude into the pockets of the HLA-E groove (5). Based on this structure of HLA-E, it would be predicted that only a few substitutions in the native Qdm peptide would be tolerated for binding to Qa-1b. This use of multiple anchors would also account for our previous finding that the Qdm peptide binds to Qa-1b with a very high affinity (6). Here, we test this issue by examining the ability of class I leader– derived peptides from several mammalian species to bind Qa-1b and define a minimum Qa-1b binding peptide. Using surface plasmon resonance (SPR), we find that Qa-1b binds class I leader peptides from almost all species tested. Unlike most class Ia molecules, the binding of peptide to Qa-1b requires the retention of multiple amino acids from the native Qdm peptide sequence. The fact that this single peptide dominates the occupancy of Qa-1b/HLA-E may also be related to the functional properties of these molecules, since recent data show that HLA-E interacts with CD94/NKG2 receptors on NK cells to deliver an inhibitory signal (7, 8).
Materials and Methods
Cells.
Drosophila melanogaster cells (S2 cells) cultured at room temperature in Schneider's medium (Life Technologies, Inc., Gaithersburg, MD) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) were cotransfected with pRMHa-3/Qa-1b truncated (12 μg), pRMHa-3/β2-microglobulin (β2m) murine (12 μg), and phshsneo (1μg) using the calcium-phosphate precipitation method (9).
Cloning Soluble Qa-1b for the Production of Soluble Molecules.
Total mRNA isolated from spleen cells of a C57BL/6 mouse (RNA STAT-60; Tel-Test, Inc., Friendswood, TX) was the template in the synthesis of first strand cDNA with reverse transcriptase (SuperScript II RT; Life Technologies, Inc.) that used oligo(dT)12–18 as a primer. Qa-1b cDNA was synthesized by PCR with oligonucleotides 5′-GTGAGGATGTTGCTTTTTGCCC and 5′-TCATGCCTTCTGAGGCCAGTC. The truncated Qa-1b (consisting of the leader, α1, α2, and α3 domains with an attached [His]6-tag) cDNA was cloned into the modified vector pRMHa-3 (9). sH2-M3 was a gift from Dr. Johann Deisenhofer (University of Texas Southwestern Medical Center at Dallas).
Production of Soluble Qa-1b.
Soluble (s)Qa-1b from the supernatant of stably transfected Drosophila cells was concentrated 10-fold, loaded onto a C10/10 column packed with 6 ml of Ni-Nta agarose (QIAGEN Inc., Chatsworth, CA) and eluted with 150 mM imidazole (pH 7.4). The protein was further purified by ion exchange chromatography (Mono Q; Amersham Pharmacia Biotech, Inc., Piscataway, NJ).
SPR.
All binding experiments were performed on a Biacore 2000 (Biacore International AB, Uppsala, Sweden) at 25°C. Cysteine-substituted analogue peptides of Qdm were immobilized to the biosensor surface (Sensor Chip CM5; Biacore International AB) using an approach similar to that described by Khilko et al. (10). The peptides were immobilized via thioether coupling to the biosensor flow cell, and Qa-1b was run over it in the soluble phase. In brief, upon activation of the surface with N-hydroxylsuccinimide (NHS)-N-ethyl-N′(dimethylaminopropyl)carbodiimide (EDC), amino groups were generated by a 10-min injection of 1 M ethylenediamine (pH 8.5; Sigma Chemical Co., St. Louis, MO). This was followed by a 4-min introduction of reactive maleimido groups from 50 mM sulfo-SMCC (Pierce Chemical Co., Rockford, IL) in 25 mM sodium bicarbonate, pH 8.5. The cysteine-substituted peptide analogue QdmC5 (200 μM in 10 mM sodium acetate, pH 5.0, except in Fig. 1, B and C, where the QdmC5 concentration was 500 μM) was run over the biosensor surface for 10 min. Unreacted maleimido groups were inactivated by a 2-min exposure to 0.1 M sodium hydroxide. All immobilization steps were performed using a flow rate of 5 μl/min, except the step in which cysteine-substituted peptides were run at 2 μl/ min. The flow rate for peptide binding experiments was 1 μl/min.
Figure 1.
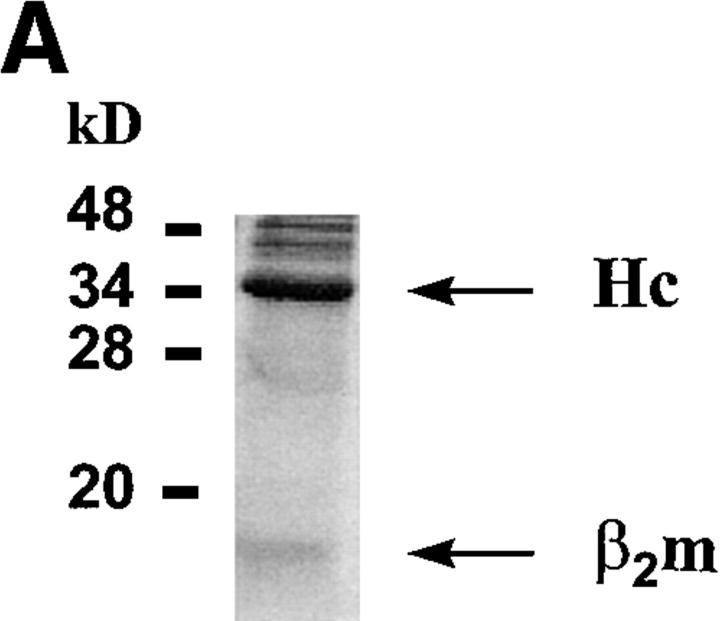
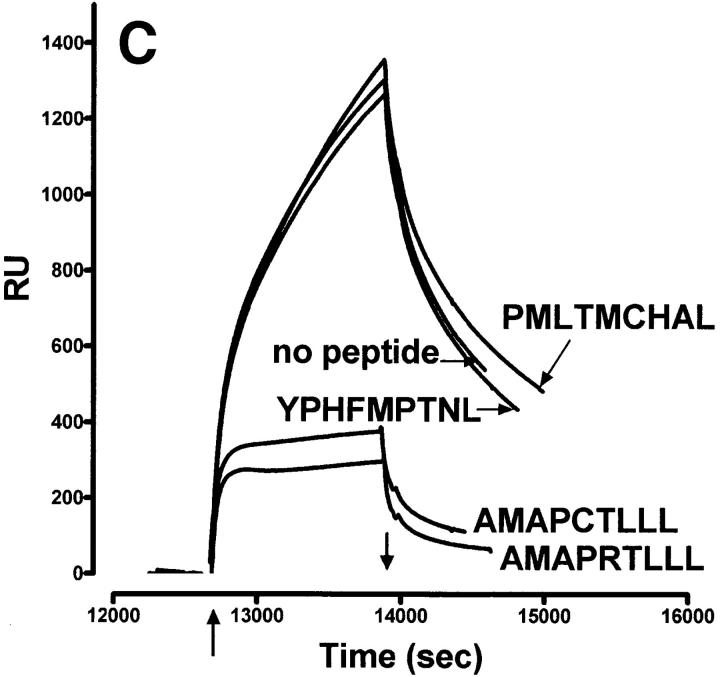
Characteristics of sQa-1b/β2m secreted from transfected D. melanogaster cells and its specific binding to immobilized QdmC5 peptide. (A) After concentration and purification, the supernatant from Qa-1b/β2m-transfected Drosophila cells was resolved on a 15% SDS-PAGE, and stained using Coomassie brilliant blue. Arrows, Qa-1b heavy chain (Hc) and β2m. (B–D) SPR demonstrating binding of sQa-1b/β2m to immobilized QdmC5. Sensorgrams were obtained using injection volumes of 20 μl at a rate of 1 μl/min. Mass increase due to macromolecular binding is measured in resonance units (RU). Arrowheads, Start (↑ ) and end (↓ ) of the injection. (B) Injection of 0.5 μM sQa-1b or sM3. (C) 0.5 μM sQa-1b was run over the chip alone, or in the presence of 20 μM Qdm (AMAPRTLLL), QdmC5 (AMAPCTLLL), or two control peptides, PMLTMCHAL and YPHFMPTNL. (D) 0.5 μM sQa-1b was run at 1 μl/min for 20 min over immobilized QdmC5 in the absence or presence of competitor peptides. Results are presented as relative binding of sQa-1b, where 0 represents binding in the presence of 20 μM Qdm (25 RU), and 1 is the binding in the absence of peptide (263 RU). *, +, and O represent binding in the presence of 20 μM of the entire 24-mer Dd leader, MGAMAPRTL and MAPRTLLLL, respectively.
Peptide Synthesis.
Peptides were synthesized using F-MOC chemistry on a peptide synthesizer (Synergy 432A; Applied Biosystems, Inc., Foster City, CA).
Results
We showed previously that Qa-1b predominantly binds a single peptide species, Qdm (AMAPRTLLL; reference 3). This result precludes our ability to identify anchor residues by conventional techniques. Therefore, the approach used in this investigation was to generate sQa-1b molecules and test their binding ability to a series of related ligands using SPR.
To study antigen binding to the Qa-1b molecule, recombinant sQa-1b/β2m dimers were generated in D. melanogaster (S2) cells following established protocols (11, 12). Truncated Qa-1b molecules secreted by stably transfected cells were purified on Ni-coated beads followed by anion exchange. Both heavy chain and β2m were visible on Coomassie-stained SDS-PAGE (Fig. 1 A).
Binding of Qa-1b to Immobilized Qdm Peptide Is Specific and Concentration Dependent.
Due to the SPR limitations in detecting the binding of small molecular weight peptides to immobilized class I molecules, we decided to attach the peptide to the chip. In the following experiments, we used QdmC5 (arginine→ cysteine substitution at position 5), which readily bound to the biosensor chip and in turn was bound by sQa-1b (Fig. 1, B and C). This binding is specific, since sM3 (Fig. 1 B) and sCD1 (not shown) failed to bind. Binding of Qa-1b to immobilized QdmC5 was blocked by adding QdmC5 or Qdm in solution, but not irrelevant control peptides (YPHFMPTNL) or (PMLTMCHAL), the latter of which contains the putative Qa-1b peptide anchors methionine at P2 and leucine at P9 (Fig. 1 C).
Trimming and Extending Qdm at the COOH Terminus Affects Its Binding to Qa-1b.
Since peptides in solution can compete with immobilized QdmC5 for binding to soluble Qa-1b, we used this approach to further analyze the peptide binding characteristics of this molecule. The Qdm nonamer peptide completely blocked binding at concentrations between 200 nM and 20 μM (Fig. 1 D). Extending the Qdm peptide by adding a leucine at position 10 (10-mer) results in decreased binding relative to the nonamer at 20 and 2 μM concentrations, and almost no binding at 200 nM. Trimming the Qdm peptide at the COOH end to an 8-mer gives a similar result. A 7-mer lost virtually all of its binding ability. We also tested the entire 24 amino acid leader of Dd from which the Qdm peptide is derived, and found that it failed to block the binding of Qa-1b to immobilized peptides. Finally, we generated two more nonamers from the leader or Dd. Instead of spanning from residues 3 to 11, these peptides span amino acids 1–9 (MGAMAPRTL) and 4–12 (MAPRTLLLL). They both failed to bind to Qa-1b.
Qa-1b Binds Peptides Derived from the Leader Segment of Human Class I Molecules.
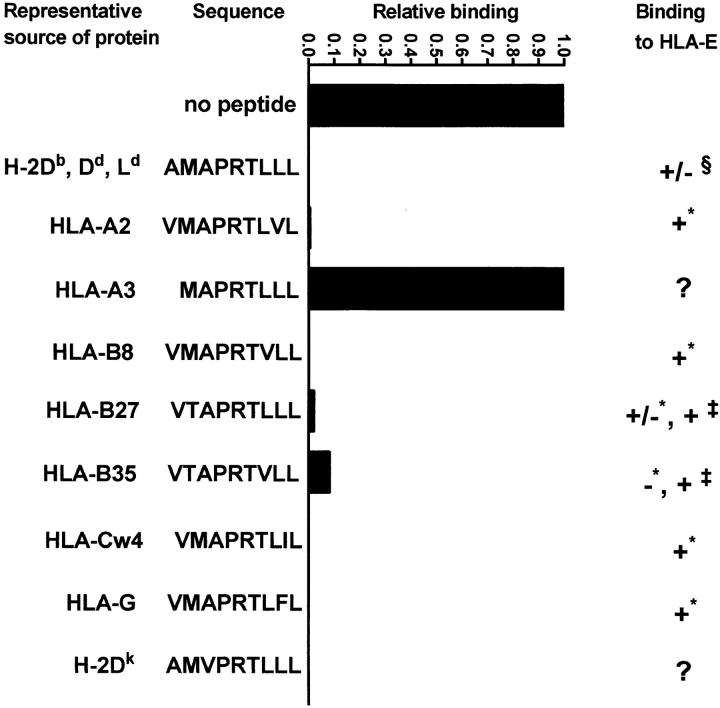
Since HLA-E and Qa-1b share unique features in their peptide binding grooves, we tested whether Qa-1b can bind the same human class I–derived peptides that bind to HLA-E. We found that all of the tested peptides except for the one originating from the leader of HLA-A3 bound to Qa-1b (Fig. 2). All of the peptides that bound to both Qa-1b and HLA-E have very similar sequences that are derived from positions 3–11 of the leader. Peptides with a threonine→ methionine change at P2 (HLA-B27, -35) bound less well, and this was more evident in experiments where the inhibiting peptides were titrated at lower concentrations (data not shown).
Figure 2.
Blocking of the binding of sQa-1b to immobilized QdmC5 by peptides derived from leader sequences of human class I molecules. Results are presented as relative binding, where 0 is the binding of sQa-1b in the presence of 20 μM Qdm (32 RU), and 1 is the binding in the absence of peptides (345 RU). 0.5 μM sQa-1b was run over immobilized QdmC5 for 20 min at the rate of 1 μl/min in the presence of 20 μM competitor peptides. For HLA-E binding, + indicates strong binding, − indicates no binding, and +/− indicates weak binding. Data taken from * Table 1 in reference 18, ‡ reference 8, and § reference 4.
MHC Class I Leader–derived Peptides from Various Mammals Bind to Qa-1b.
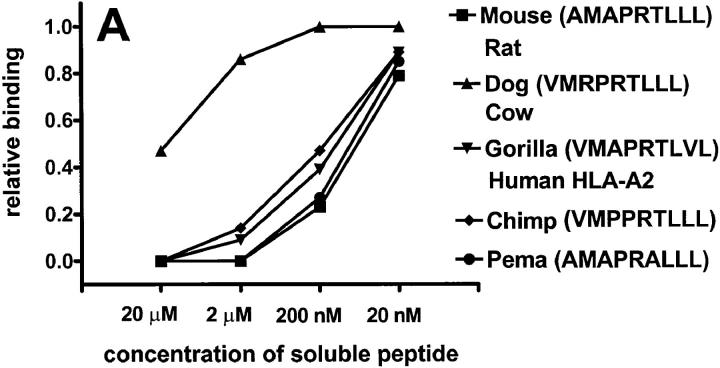
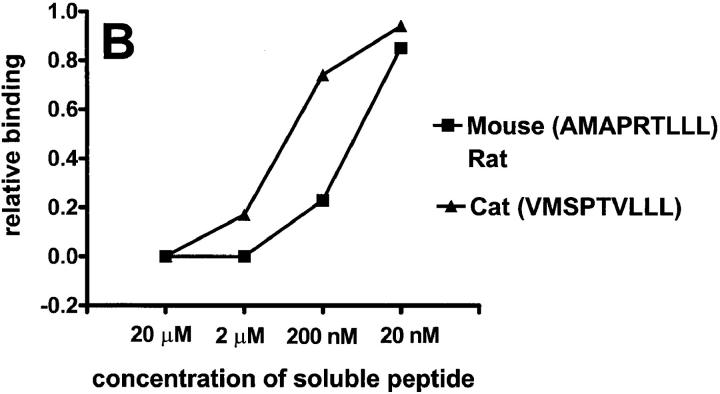
The unusual finding that HLA-E and Qa-1b bind the same set of peptides, all derived from leader sequences of MHC class I molecules, raises the possibility that there is a conservation of similar epitopes in other mammals. In fact, inspection of representative class Ia sequences from a variety of mammalian species reveals a conserved “Qdm-like” epitope (Table 1). We tested the ability of these putative peptides to bind to Qa-1b. Most of the tested peptides, except those from dog or cow class I molecules, bound well to Qa-1b (Fig. 3 A). It is likely that the presence of the positively charged arginine at P3 of the peptide results in weaker binding.
Table 1.
Conservation of “Qdm-like” Epitope in Mammals
| Qdm-like peptides present in class I leaders | Species | Molecule containing sequence | ||
|---|---|---|---|---|
| AMAPRTLLLa | Mouse | D-end molecules | ||
| AMAPRTLLLb | Rat | RT1.A | ||
| AMAPRALLLc | Pema | Pm62 | ||
| VMRPRTLLLd | Cow | BL3-7 | ||
| VMRPRTLLLe | Dog | DLA-6.7B | ||
| VMSPTVLLLf | Cat | FLA24 | ||
| VMAPRTLVLg | Gorilla | GOGO-A0401 | ||
| VMAPRTLVLh | Human | HLA-A2 | ||
| VMPPRTLLLi | Chimpanzee | CHLA A-108 | ||
| MAPRTLLLj | Human | HLA-A3 |
Figure 3.
Putative peptides from leaders of various mammalian class I molecules bind to Qa-1b. Results are presented at relative binding, where 0 is the binding of sQa-1b in the presence of 20 μM Qdm (25 RU in A, 26 RU in B), and 1 is the binding in the absence of blockers (276 RU in A, 230 RU in B). Running buffer was Hepes-buffered saline (HBS) in A and 2% DMSO in HBS in B.
Some of these leader peptides are extremely hydrophobic and not soluble in aqueous solvents. One such peptide is VMSPTVLLL, a Qdm-like epitope derived from the cat class I leader. To circumvent this problem, we diluted the peptide in 2% DMSO, where it remained soluble, and then used 2% DMSO as a running buffer. Under these conditions, we demonstrate that both the murine Qdm peptide and the peptide derived from the cat sequence bind Qa-1b (Fig. 3 B).
The Minimal Requirements for Peptide Binding to Qa-1b.
We next determined the minimum requirement for ligand binding to Qa-1b by synthesizing a number of peptides in which glycines were introduced in different positions (Table 2). We used glycine instead of alanine because the native Qdm sequence contains two alanines. Of the minimal peptides we tested, those with two or three nonglycine residues showed no (GMGGGGGGL, GMGGRGGGL) or very little binding (GMGGGGLGL, GMGGGGGLL) (Table 2). However, a peptide with four of nine native residues GMGGGGLLL blocked >50% of binding of sQa-1b to immobilized QdmC5 peptide at the highest concentration tested (20 μM). However, when this peptide was titrated, we noted relatively little blocking activity at 2 μM and none at 200 nM, in marked contrast to the titration seen when more homologous peptides were tested (Fig. 3 A). This indicates that methionine at P2 and the three COOH-terminal leucines are sufficient for detectable although relatively very weak binding to Qa-1b. Side chains of other amino acids in Qdm also play a role in the overall peptide binding. There is apparently a fine balance in their contribution which is dependent on the neighboring residues, since a peptide with five native residues (GMGGRGLLL) binds better to Qa-1b than a peptide with six native residues (GMGPRGLLL).
Table 2.
Binding of Poly-Gly Peptides to Qa-1b
| Peptide sequence | Qa-1b binding [Inhibitor peptide] | |
|---|---|---|
| 20/2/0.2 μM | ||
| GMGGGGGGL | 0/ND/ND | |
| GMGGRGGGL | 0/ND/ND | |
| GMGGRGGLL | 0/ND/ND | |
| GMGGRGLGL | 0/ND/ND | |
| GMGGRGLLL * | 0.74/ND/ND | |
| GMGPRTLGL | NS‡ | |
| GMGPRGLLL * | 0.59/ND/ND | |
| GMGPRGLGL | 0/ND/ND | |
| GMGGGGLLL | 0.56/0.16/0 | |
| GMGGGGLGL | 0.18/0/0 | |
| GMGGGGGLL | 0.09/0/0 | |
| AMAPRTLLL | 1/1/0.76 |
sQa-1b was run over immobilized QdmC5 for 20 min at the rate of 1 μl/min in the presence of different poly-Gly peptides with HBS as running buffer. Final responses were expressed as a fraction of 1, which was maximal blocking achieved by 20 μM Qdm. A response of 0 indicates that peptide does not bind to Qa-1b. Native residues are shown in bold. Peptides with three or four native residues that showed some blocking at 20 μM were further titrated versus Qdm at 2 μM and 200 nM.
Running buffer was 2% DMSO in HBS.
Peptide GMGPRTLGL was not soluble (NS) in 2% DMSO in HBS.
Discussion
Both Qa-1b (3) and HLA-E (4) bind a similar peptide derived from the leader of class Ia molecules. It appears that these are the major peptides that both of these class I molecules bind. Although Qa-1b and HLA-E share unique residues in their F pocket that are not found in other class I molecules, it is surprising that they bind similar peptides, since they differ considerably in their primary structure. Data presented in this paper show that Qa-1b not only binds peptides derived from the leader of murine MHC molecules, but also binds all of the human class I–derived peptides that were reported to interact with HLA-E, as well as putative class I leader peptides from several other mammalian species. An examination of leader sequences from representative class I alleles from several mammalian species shows a conservation of the Qa-1b/HLA-E binding epitope between positions 3 and 11 of the segment. In fact, among the class I–derived peptides we tested, there is relatively little variability in most of the amino acids. For example, P4 (proline) and P9 (leucine) had no variability, whereas P1, P2, P5, and P7 had a single predominant residue although an alternative amino acid was seen in some peptides. The binding of this array of xenogeneic leader peptides was almost as efficient as the binding of the murine leader itself. It is important to note that leaders of Qa-1b, HLA-E, and their other mammalian homologues are unique and do not contain the conserved Qdm-like epitope. Consequently, the peptide binding grooves of these molecules are not occupied by peptides derived from their own leaders.
We have attempted to determine the minimum motif required for peptide binding to Qa-1b. Since only one peptide has been eluted from the groove of this molecule, it is not possible to assign anchor residues in the conventional manner. In addition to embedding principal anchors, methionine at P2 and leucine at P9, we needed to introduce two more wild-type residues in the polyglycine chain, leucines at P7 and P8, to observe detectable binding. However, the binding of this pentaglycine analogue was still considerably weaker than that of native Qdm, suggesting that side chains of other residues also contribute to the overall interaction. Although it is possible that binding of the minimal peptide with fewer anchor residues could have been found had we used a backbone other than glycine (13), several other minimal peptides with glycine backbones have been used successfully to identify anchors that participate in binding to class I molecules (14, 15).
Thus, the finding that Qdm requires multiple anchors would explain the dominance of a single peptide in its groove. The data presented here, together with the recent crystal structure of HLA-E bound to its peptide (5), suggest that Qa-1b and HLA-E can only bind Qdm-like peptides with high efficiency. However, it cannot be ruled out that their occupancy by these peptides is a result of a restrictive peptide antigen processing and/or presentation pathway. It is interesting to note that the common ligand that Qa-1b and HLA-E bind is derived from a conserved part of class I leader segments that are expendable in the mature protein and thus would not affect selection for polymorphic peptide binding residues.
Boyson et al. (16) pointed out that a comparison of the rates of synonymous and nonsynonymous nucleotide substitutions in the peptide binding region versus the remainder of the molecule indicates that the peptide binding groove of HLA-E and its homologues in macaques has been conserved for over 36 million years, when the two last shared a common ancestor. Yeager et al. have communicated that, although not orthologous, Qa-1b and HLA-E might have evolved similar functions through convergent evolution at the amino acid sequence level of the peptide binding region (17). Regardless of whether molecular-level convergence or evolutionary conservation of the peptide binding region accounts for the specificity of these grooves, this conservation of specificity suggests a crucial immunological function for these molecules. In this regard, it has recently been shown that HLA-E is a ligand for CD94/ NKG2 receptors on NK cells; interaction of HLA-E with this receptor protects target cells from NK-mediated lysis (7, 8). Although this has not yet been demonstrated for Qa-1b, it is likely that it interacts with its murine CD94/ NKG2 counterpart in a similar manner. Class I molecules in mice could, through Qa-1b, control the activity of NK cells which would be signaled upon interaction with cell surface–expressed Qa-1b. Decreased expression and/or processing of class I molecules would decrease the expression of Qa-1b, which would in turn result in a changed activity level of NK cells.
It is conceivable that occasionally Qa-1b–bound class I–derived peptides could be replaced, or that some of the peptide binding grooves might be initially occupied by other self- or pathogen-derived peptides which would be presented to T cells. Future studies should show whether Qa-1b is recognized by NK cell receptors, and what role peptides play in the response.
Acknowledgments
This work was supported by National Institutes of Health grants P01-AI-37818, R01-AI-34930, and 5-RO1-AI-37942 (to J. Forman), and by a Welch Foundation grant (to C.A. Hasemann).
References
- 1.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox AL, Appella E, Engelhard VH. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 2.Aldrich CJ, DeCloux A, Woods AS, Cotter RJ, Soloski MJ, Forman J. Identification of a TAP-dependent leader peptide recognized by alloreactive T cells specific for a class Ib antigen. Cell. 1994;79:649–658. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- 3.DeCloux A, Woods AS, Cotter RJ, Soloski MJ, Forman J. Dominance of a single peptide bound to the class Ib molecule, Qa-1b . J Immunol. 1997;158:2183–2191. [PubMed] [Google Scholar]
- 4.Braud V, Jones EY, McMichael A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at position 2 and 9. Eur J Immunol. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- 5.O'Callaghan CA, Tormo J, Willcox BE, Braud VM, Jakobsen BK, Stuart DI, McMichael AJ, Bell JI, Jones EY. Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E. Mol Cell. 1998;1:531–541. doi: 10.1016/s1097-2765(00)80053-2. [DOI] [PubMed] [Google Scholar]
- 6.Kurepa Z, Forman J. Peptide binding to the class Ib molecule, Qa-1b . J Immunol. 1997;158:3244–3251. [PubMed] [Google Scholar]
- 7.Braud VM, Allan DSJ, O'Callaghan CA, Sodestrom K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. HLA-E binds to natural killer receptors CD94/NKG2A, B and C. Nature. 1998;91:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 8.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA-A)-E complexed with HLA class I signal sequence–derived peptides by CD94/NKG2 confers protection from natural killer cell–mediated lysis. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunch TA, Grinblat Y, Goldstein LSB. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogastercells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khilko SN, Corr M, Boyd LF, Lees A, Inmaa JK, Margulies DH. Direct detection of major histocompatibility complex class I binding to antigenic peptides using surface plasmon resonance. J Biol Chem. 1993;268:15425–15434. [PubMed] [Google Scholar]
- 11.Matsumura M, Saito Y, Jackson MR, Song ES, Peterson PA. In vitro peptide binding to soluble empty class I major histocompatibility complex molecules from transfected Drosophila melanogastercells. J Biol Chem. 1992;267:23589–23595. [PubMed] [Google Scholar]
- 12.Wang CR, Castano AR, Peterson PA, Slaughter C, Lindahl KF, Deisenhofer J. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell. 1995;82:655–664. doi: 10.1016/0092-8674(95)90037-3. [DOI] [PubMed] [Google Scholar]
- 13.Maryanski JL, Verdini AS, Weber PC, Salemme FR, Corradin G. Competitor analogs for defined T cell antigens: peptides incorporating a putative binding motif and polyproline or polyglycine spaces. Cell. 1990;60:63–72. doi: 10.1016/0092-8674(90)90716-r. [DOI] [PubMed] [Google Scholar]
- 14.Parker KC, Bednarek MA, Hull LK, Utz U, Cunningham B, Zweerink HJ, Biddison WE, Coligan JE. Sequence motifs important for peptide binding to the human MHC molecule, HLA-A2. J Immunol. 1992;149:3580–3587. [PubMed] [Google Scholar]
- 15.DiBrino M, Parker KC, Shiloach J, Knierman M, Lukszo J, Turner RV, Biddison WE, Coligan JE. Endogenous peptides bound to HLA-A3 possess a specific combination of anchor residues that permit identification of potential antigenic peptides. Proc Natl Acad Sci USA. 1993;90:1508–1512. doi: 10.1073/pnas.90.4.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyson JE, McAdam SN, Gallimore A, Golos TG, Liu X, Gotch FM, Hughes AL, Watkins DI. The MHC E locus in macaques is polymorphic and is conserved between macaques and humans. Immunogenetics. 1995;41:59–68. doi: 10.1007/BF00182314. [DOI] [PubMed] [Google Scholar]
- 17.Yeager M, Kumar S, Hughes AL. Sequence convergence in the peptide-binding region of primate and rodent MHC class Ib molecules. Mol Biol Evol. 1997;14:1035–1041. doi: 10.1093/oxfordjournals.molbev.a025709. [DOI] [PubMed] [Google Scholar]
- 18.Braud VM, Allan DSJ, Wilson D, McMichael AJ. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol. 1997;8:1–10. doi: 10.1016/s0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 19.Crew MD, Bates LM, Douglass CA, York JL. Expressed Peromyscus maniculatus(Pema) MHC class I genes: evolutionary implications and the identification of a gene encoding a Qa1-like antigen. Immunogenetics. 1996;44:177–185. [PubMed] [Google Scholar]