Abstract
The generation of a productive “in-frame” T cell receptor β (TCR β), immunoglobulin (Ig) heavy (H) or Ig light (L) chain variable region gene can result in the cessation of rearrangement of the alternate allele, a process referred to as allelic exclusion. This process ensures that most αβ T cells express a single TCR β chain and most B cells express single IgH and IgL chains. Assembly of TCR α and TCR γ chain variable region genes exhibit allelic inclusion and αβ and γδ T cells can express two TCR α or TCR γ chains, respectively. However, it was not known whether assembly of TCR δ variable regions genes is regulated in the context of allelic exclusion. To address this issue, we have analyzed TCR δ rearrangements in a panel of mouse splenic γδ T cell hybridomas. We find that, similar to TCR α and γ variable region genes, assembly of TCR δ variable region genes exhibits properties of allelic inclusion. These findings are discussed in the context of γδ T cell development and regulation of rearrangement of TCR δ genes.
Keywords: T cells, γδ T cells, T cell receptor rearrangement, allelic exclusion, T cell receptor δ
Lymphocyte antigen receptor variable region genes are assembled during development from component variable (V),1 diversity (D), and joining (J) gene segments in the case of the TCR β and δ chain genes and the Ig heavy (H) chain gene or from V and J gene segments in the case of TCR α and γ chain genes and Ig light (L) chain genes (1, 2). Productive rearrangement of TCR β or IgH chain variable region genes results in cessation of further V to DJ rearrangements on the alternate allele, a process referred to as allelic exclusion (2, 3). “Functional” rearrangement of IgL κ or λ L chain genes (i.e., rearrangements which generate an IgL chain that can pair with a pre-existing IgH chain) also lead to cessation of further IgL chain rearrangements resulting in both allelic and IgL chain isotype exclusion (4). In contrast, TCR α and TCR γ chain variable region gene assembly does not exhibit properties of allelic exclusion (3, 5–7). Consequently αβ and γδ T cells can express two TCR α or γ chains, respectively (6, 7).
Several models have been proposed to account for allelic exclusion. One model proposed that the probability of a productive rearrangement is low making it unlikely that an individual cell could have two productive rearrangements (8). However, it is now known that the probability of a productive rearrangement can be as high as 33% (9). Another model proposed that the probability of two complete V(D)J rearrangements in any one cell was low. However, a significant percentage of peripheral B and T cells have two IgH or TCR β V(D)J rearrangements, respectively, arguing against this model (3, 10). It has been proposed that IgH chain allelic exclusion occurs due to a toxic effect of expressing two IgH chains (11). However, the recent demonstration that B cell development proceeds normally in mice that express two IgH transgenes essentially rules out this model (12). An early model, based on analyses of rearrangement patterns in cell lines, proposed that allelic exclusion is regulated and that expression of a productively rearranged IgH or IgL chain prevents further rearrangements at the IgH and IgL chain loci, respectively (4, 13, 14). This regulated model was supported by studies demonstrating that expression of IgH or IgL transgenes resulted in a block in endogenous IgH or IgL chain gene rearrangement, respectively (15–18). Studies of TCR β transgenic mice have supported an analogous model by which the TCR β transgene feeds back to block endogenous TCR β rearrangements (19). In addition, it has recently been demonstrated that expression of IgH or TCR β chains as pre-B or pre-T cell receptors, respectively, is required for allelic exclusion (20–23).
T cells can be divided into two distinct lineages based on expression of either αβ or γδ TCRs. The genes that encode the TCR β and TCR γ chains lie in distinct loci, whereas the genes that encode the TCR δ and TCR α chains lie in a single locus (TCR α/δ locus; Fig. 1; references 24, 25). In the adult thymus TCR β rearrangements are initiated at the CD4−/CD8− (double negative, DN) stage of thymocyte development and are ordered with Dβ to Jβ rearrangement occurring on both alleles before Vβ to DJβ rearrangement (3, 26, 27). Once a productive V(D)Jβ rearrangement is made and a TCR β chain expressed, cells proceed to the CD4+/CD8+ (double positive, DP) stage of development and further Vβ to DJβ rearrangements cease (3, 26, 27). As a result, many αβ T cells have DJβ rearrangements on a single allele (28). Vα to Jα rearrangements are initiated at the DP stage. However, unlike the TCR β locus, expression of a TCR α chain does not result in cessation of Vα to Jα rearrangements (3, 26, 27). This process continues on both alleles, and Vα to Jα rearrangements can result in the deletion of previously assembled productive VJα rearrangements (29). It has been proposed that the downregulation of recombinase activating gene (RAG) gene expression may ultimately be responsible for termination of Vα to Jα rearrangement (30).
Figure 1.
Schematic of the mouse TCR α/δ locus. Shown are the Vα/Vδ gene segments, the Dδ and Jδ gene segments, the TCR δ enhancer (Eδ), the TCR δ constant region gene (Cδ), and the Vδ5 gene segment. This is followed by the Jα gene segments, the TCR α constant region gene (Cα), and the TCR α enhancer (Eα). Also shown are probes 1 and 3 through 6 and the approximate position of the BglII (B2) sites. The schematic is not drawn to scale.
Several notable differences exist between the developmental regulation of assembly of αβ and γδ TCR variable region genes. Assembly of TCR γ and TCR δ variable region genes occurs at the DN stage of thymocyte development (31). It is not known whether rearrangement of these genes is concurrent or sequential. In addition, assembly of TCR γ genes does not appear to exhibit allelic exclusion (7). Similar to the TCR β locus, assembly of TCR δ variable region genes does not proceed to completion on all alleles. However, unlike the TCR β locus, TCR δ variable region gene assembly does not appear to be ordered, since incomplete DDδ, DJδ and VDδ rearrangements have been described (32, 33). It is unresolved whether productive TCR δ rearrangements lead to termination of further TCR δ rearrangements (allelic exclusion) or whether TCR δ rearrangements are limited by factors independent of the formation of productive rearrangements. To address this issue, we have analyzed TCR δ rearrangements in a panel of T cell hybridomas derived from splenic γδ T cells. We find the percentage of cells with two in-frame V(D)Jδ rearrangements is similar to that predicted in the absence of allelic exclusion. These findings are discussed in the context of γδ T cell development.
Materials and Methods
Isolation of γδ T Cells and Production of γδ T Cell Hybridomas.
Whole spleen cell suspensions from C57BL6 × CBA mice were incubated in DMEM-15 containing 40 U recombinant human IL-2/ml (PharMingen, San Diego, CA) on plates that had been coated with 10 μg/ml rat anti–hamster Ig (PharMingen) followed by 10 μg/ml of an anti-TCR δ chain mAb (GL4; PharMingen). Cultures were maintained for 6 d and the resulting cells were >90% pure γδ T cells as determined by flow cytometry (data not shown). Hybridomas were produced by fusing these γδ T cells to the thymoma BW-1100.129.237 using a fusion protocol that has been described elsewhere (34, 35).
Flow Cytometry.
Single cell suspensions were prepared from thymus, spleen and lymph nodes as previously described (36). Hybridomas were stained with FITC-conjugated anti-TCR β chain (H57-597) and PE-conjugated anti-TCR δ chain (GL3) monoclonal antibodies from PharMingen and were analyzed by a FACScan® (Becton Dickinson & Co., Sparks, MD).
Genomic DNA Analysis.
Genomic DNA was isolated and Southern blotting carried out as previously described using Zetaprobe membranes (Bio-Rad Laboratories, Hercules, CA) and probes generated by random hexamer priming (Boehringer Mannheim Corp., Indianapolis, IN) using α-[32P]dCTP (34, 35). Probe 1 is a 600-bp HindIII fragment (39). Probe 3 is a 550-bp Msp1 to Nde1 fragment and probe 4 a 1-kb Nde1-Xba1 fragment from pTAE-7 (40). Probe 5 is a 350-bp PCR product generated as described elsewhere (41). Probe 6 is a 1.5-kb EcoRI Cδ cDNA fragment (42).
PCR and Sequence Analysis.
PCR reactions were carried out using 200 ng of genomic DNA isolated from hybridomas and 2.5 U AmpliTaq polymerase (Perkin-Elmer Corp., Norwalk, CT). PCR conditions were: 92°C for 1 min 30 s, 62°C for 2 min 30 s, 72°C for 1 min 30 s cycled 30 times. PCR products were subcloned into pT7blue (Novagen Inc., Madison, WI) before sequencing on a ABI Prism 377 DNA sequencer (Perkin-Elmer Corp.). The Vδ nomenclature of Arden et al. (43) is used. The Vδ and Jδ primers used to PCR VDJδ joins were as follows: Vδ primers: ADV7S (Vδ7/Vδ6), TCACCCTGGACTGTTCATAT; ADV11S5, ATTTTACGACCACCATGAGG; ADV17S2 (Vδ9), ATGCTGATTCTAAGCCTGCT; DV2S8 (Vδ8), AGCAGGTGAGACAAAGTCC; DV4S8, ACGATAGAGTGCAACTACTCA; DV6S2 (Vδ3), ATGGGATGTGTGAGTGGAAT; V10S7 (Vδ7), TGAAGAGGCTGCTGTGCTC; DV101S1 (Vδ1), ATGCTTTGGAGATGTCCAGT; DV102S1(Vδ2), ATGGGGATGTTCCTTCAAGT; DV104S1(Vδ4), CAGGTGGCAGAATCAGCAA; DV105S1(Vδ5), ATGATTGTTGCCGCGACCC; Jδ primers: Jδ1, AGAGTCCAAGAATCATCTACG; Jδ2, CTTCTGTGTACTACTTTTATTTTC.
Southern blotting of PCR products was carried out with internal oligos to Jδ1 (CGACAAACTCGTCTTTGG) or Jδ2 (CTCCTGGGACACCCGACAGA).
Theoretical Determination of In-Frame Rearrangement Percentages.
All mature γδ T cells must have at least one productive VDJδ rearrangement. If the probability that a VDJδ rearrangement will be in-frame equals P then the probability that a VDJδ rearrangement will be out of frame will be (1 − P). If there is an equal chance of a rearrangement in each of the three reading frames then P = 1/3. In the absence of allelic exclusion and in cells with two VDJδ rearrangements, the percentage of cells with two in-frame TCR δ rearrangements will be equal to the probability that the cell will have two in-frame rearrangements divided by the probability that the cell will have at least one in-frame rearrangement.
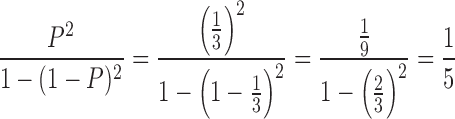 |
Results
Generation of γδ T Cell Hybridomas.
Splenic γδ T cell hybridomas were generated from C57BL6 × CBA F1 mice by stimulating unfractionated spleen cells with plate-bound anti-TCR δ antibody (GL3) as described in Materials and Methods section. The resulting cell population was >90% pure γδ T cells as determined by flow cytometry (data not shown). These cells were fused to the BW-1100.129.237 (BW) thymoma which is incapable of producing TCR δ, β, or α chains (35). T cell hybridomas generated by fusion of a γδ T cell to BW were identified by flow cytometric analysis of cell surface TCR δ expression (data not shown). Only those hybridomas that expressed TCR δ were chosen for further analysis.
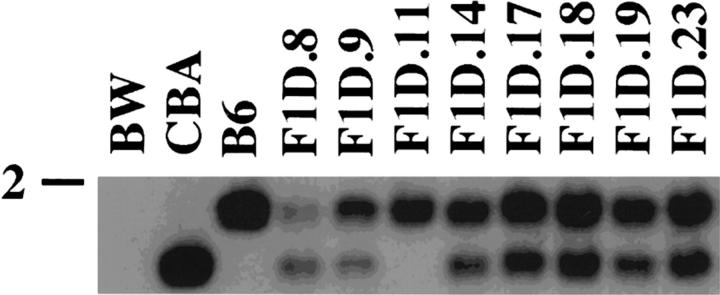
To ensure that both TCR δ alleles were present in the resulting panel of γδ T cell hybridomas genomic DNA isolated from these hybridomas was assayed by Southern blot analyses using TCR δ restriction fragment length polymorphisms that exist between C57BL6 and CBA mice (44). Genomic DNA isolated from γδ T cell hybridomas was digested with HindIII and subjected to Southern blot analysis using probe 6 which is directed against the TCR δ constant region gene (Cδ; Figs. 1 and 2). Probe 6 hybridizing bands are not found in BW as Cδ has been deleted on both alleles due to Vα to Jα rearrangements (Fig. 2). Using probe 6, distinct size bands are generated by the C57BL6 and CBA TCR δ alleles, and hybridomas that had lost either allele (for example F1D.11) were excluded from further analysis (Fig. 2).
Figure 2.
Analysis for the presence of the CBA and C57BL6 TCR δ alleles. Genomic DNA from the hybridoma fusion partner BW 1100.129.237 (BW), CBA kidney (CBA), C57BL6 kidney (B6), or γδ T cell hybridomas (F1D) was digested with HindIII and subjected to Southern blot analysis using probe 6 (Fig. 1). The 2-kb marker is shown.
To determine whether the γδ T cell hybridomas chosen for analysis were clonal, genomic DNA was subjected to Southern blot analysis using probe 4 to detect rearrangements to Jδ1 (Fig. 3 a), probe 5 to detect rearrangements to Jδ2 (Fig. 3 b) or a probe that detects rearrangements to Jβ2 (data not shown). Hybridomas that were oligoclonal on the basis of having three or more TCR δ or TCR β rearrangements were excluded from further analysis. The resulting 27 γδ T cell hybridomas that satisfied the above criteria were characterized further.
Figure 3.
Analysis of rearrangements to Jδ1 and Jδ2. Genomic DNA samples described in the legend to Fig. 1 were digested with either BglII (a) or HindIII (b) and subjected to Southern blot analysis using probes 1, 3, and 4 (a) or probe 5 (b). Shown are the 9-, 6- and 4-kb molecular mass markers.
TCR δ Allele Configurations in γδ T Cell Hybridomas.
To determine the extent of TCR δ rearrangement in the panel of γδ T cell hybridomas, hybridoma genomic DNA was digested with BglII and subjected to Southern blot analysis using probe 4 (Figs. 1 and 3 a, data not shown). None of the hybridomas exhibited germline size bands, demonstrating that most TCR δ alleles are rearranged in splenic γδ T cells (Fig. 3 a, data not shown). In addition, most hybridomas gave two bands with probe 4, showing that most TCR δ rearrangements in splenic γδ T cells use the Jδ1 gene segment. F1D.58 exhibited single nongermline bands with probes 4 and 5, demonstrating that it had undergone rearrangements to Jδ1 and Jδ2 (Fig. 3 b). All other hybridomas exhibited germline bands from the C57BL6 and CBA TCR δ alleles using probe 5, showing that there is minimal rearrangement to the Jδ2 gene segment in splenic γδ T cell hybridomas analyzed here (Fig. 3 b, data not shown).
To assay for incomplete TCR δ rearrangements, BglII-digested hybridoma genomic DNA was probed with probes 1 and 3 (Figs. 1 and 3 a, data not shown). Probe 1 hybridizing bands of similar size to probe 4 hybridizing bands would be generated by alleles that have undergone Dδ1 to Dδ2 or Dδ1 to Jδ1 rearrangements. Hybridomas F1D.19, 45, 51, 55, 71, and 72 all exhibit a 4.5-kb BglII band with probes 1 and 4 (Fig. 3 a, Table 1, data not shown), whereas hybridoma F1D.58 yields a 5.5-kb BglII band with probes 1 and 4 (Fig. 3 a and Table 1). To determine which hybridomas had undergone a Dδ1 to Dδ2 rearrangement, BglII-digested DNA was assayed with a probe (probe 3) to the region between Dδ1 and Dδ2 which will be deleted upon Dδ1 to Dδ2 rearrangement (Fig. 1). F1D.58 has a 5.5-kb BglII band that hybridizes to probe 3, demonstrating that one of the alleles in this hybridoma has undergone a Dδ1 to Dδ2 rearrangement (Fig. 3 a, Table 1). The hybridomas that yielded a 4.5-kb BglII band with probes 1 and 4 do not have probe 3 hybridizing bands, demonstrating that they have undergone Dδ1 to Jδ1 rearrangements (Fig. 3 a).
Table 1.
γδ T Cell Hybridomas with One Complete VDJδ Rearrangement
| Hybridoma | V(D)Jδ rearrangement | Incomplete rearrangement | ||
|---|---|---|---|---|
| F1D.19 | DV104S1-Jδ1 | D(D) Jδ1 | ||
| F1D.45 | DV7S-Jδ1 | D(D) Jδ1 | ||
| F1D.51 | DV104S1-Jδ1 | D(D) Jδ1 | ||
| F1D.55 | DV104S1-Jδ1 | D(D) Jδ1 | ||
| F1D.58 | DV101S1-Jδ2 | DD | ||
| F1D.68 | DV104S1-Jδ1 | DV6S2-(D)D | ||
| F1D.71 | DV104S1-Jδ1 | D(D) Jδ1 | ||
| F1D.72 | DV7S-Jδ1 | D(D) Jδ1 |
The Vδ gene segments utilized were determined by PCR as described in Materials and Methods section. The Dδ2 gene segment is indicated in brackets in some rearrangements as it can not be determined from this analysis whether it is utilized. The Vδ gene segment nomenclature of Arden et al. (43) is used.
The DV105S1 (Vδ5) gene segment rearranges by inversion and, therefore, a nongermline probe 1 hybridizing band should be generated by the reciprocal product of a DV105S1 to Dδ1 rearrangement. Furthermore, this band would likely be of a different size than the probe 4 hybridizing band generated by the same rearrangement. Hybridomas F1D.17, 23, 32, and 61 all have 9-kb BglII probe 1 hybridizing bands (Fig. 3 a, data not shown). None of these hybridomas has a 9-kb probe 4 hybridizing BglII band, and each was found to have a DV105S1 to Jδ1 rearrangement by PCR analysis (Table 2). Finally, Vδ to Dδ rearrangements by Vδ gene segments other than DV105S1 will result in loss of probe 1 hybridizing bands and generation of a non-germline probe 3 hybridizing band that should be similar in size to the band generated by probe 4 when probing BglII-digested DNA. In this regard F1D.68 has a 3.5-kb BglII band that hybridizes to probes 3 and 4 and was found to have a Vδ to Dδ2 rearrangement by PCR analysis (Fig. 3 a, Table 1).
Table 2.
γδ T Cell Hybridomas with Two Complete VDJδ Rearrangements
| Hybridoma | Rearrangement | In-frame | Rearrangement | In-frame | ||||
|---|---|---|---|---|---|---|---|---|
| F1D.9 | DV107S7-Jδ1 | Y | ADV7S2-Jδ1 | N | ||||
| F1D.17 | ADV7S1-Jδ1 | Y | DV105S1-Jδ1 | N | ||||
| F1D.23 | ADV7S1-Jδ1 | Y | DV105S1-Jδ1 | N | ||||
| F1D.29 | ADV7S1-Jδ1 | Y | DV104S1-Jδ1 | N | ||||
| F1D.32 | DV7S-Jδ1 | Y | DV105S1-Jδ1 | Y | ||||
| F1D.33 | ADV7S1-Jδ1 | Y | ADV17S2-Jδ1 | N | ||||
| F1D.36 | DV104S1-Jδ1 | Y | ND | |||||
| F1D.42 | DV10S7-Jδ1 | Y | ADV17S2-Jδ1 | N | ||||
| F1D.49 | DV7S5-Jδ1 | Y | DV104S1-Jδ1 | Y | ||||
| F1D.57 | DV104S1-Jδ1 | Y | DV7S4-Jδ1 | N | ||||
| F1D.60 | ADV7S2-Jδ1 | Y | DV104S1-Jδ1 | Y | ||||
| F1D.61 | DV105S1-Jδ1 | Y | DV104S1-Jδ1 | Y | ||||
| F1D.64 | ND | Y | DV7S6-Jδ1 | N | ||||
| F1D.67 | DV104S1-Jδ1 | Y | DV7S5-Jδ1 | N | ||||
| F1D.73 | DV104S1-Jδ1 | Y | DV104S1-Jδ1 | Y | ||||
| F1D.75 | ADV7S1-Jδ1 | Y | DV104S1-Jδ1 | Y | ||||
| F1D.83 | DV102S1-Jδ1 | Y | DV7S5-Jδ1 | N | ||||
| F1D.89 | DV104S1-Jδ1 | Y | DV7S5-Jδ1 | N | ||||
| F1D.91 | DV7S6-Jδ1 | Y | ND |
F1D.32 has a rearrangement utilizing a DV7S Vδ gene segment that differs from other known family members by at least five nucleotides (data not shown). This Vδ gene segment may represent a novel DV7S family member or is due to strain differences. The two DV104S1 rearrangements in F1D.73 are distinct as determined by differences in junctional diversity (data not shown). The in-frame TCR δ rearrangement in F1D.64 is presumed as the cell expresses a TCR δ chain.
These Southern blot analyses revealed that, of the 27 γδ T cell hybridomas analyzed, all had complete V(D)J δ rearrangements on one allele (Fig. 3, a and b, Tables 1 and 2. On the other allele, 19 hybridomas also had complete V(D)J δ rearrangements, one had a Dδ1Dδ2 rearrangement, one had a VDδ2 rearrangement and 6 had Dδ1Jδ1 rearrangements (Fig. 3 a, Table 1). In addition, the Jδ2 gene segment was used in only one rearrangement (Fig. 3 b, Table 1).
Analysis of V(D)Jδ Rearrangements in γδ T Cell Hybridomas.
Using primers that should recognize the members of the 11 known mouse Vδ gene families in conjunction with primers that were just downstream of Jδ1 or Jδ2, PCR analysis was carried out on all hybridomas to determine Vδ gene segment usage (Tables 1 and ;reference2. By this analysis none of the hybridomas analyzed gave more than two distinct PCR products (data not shown). PCR products from the 19 hybridomas that had two complete V(D)Jδ rearrangements were cloned and sequenced. None of the V(D)Jδ rearrangements isolated used known Vδ pseudogenes (43). Two distinct rearrangements were isolated from 16 of the 19 hybridomas determined to have two V(D)Jδ rearrangements. All hybridomas had at least one in-frame V(D)Jδ rearrangement except for F1D.64 in which only a single out of frame V(D)Jδ rearrangement was isolated (Table 2). The other allele of this hybridoma must have an in-frame V(D)Jδ rearrangement, that was undetected by this analysis, as it expresses a TCR δ chain (data not shown). Only a single in-frame rearrangement was isolated from F1D.36 and F1D.91. The inability to detect more than a single rearrangement in these hybridomas could be due to the use of novel Vδ gene segments unable to be detected by the primer set used in this analysis. Alternatively, these hybridomas may have two rearrangements involving members of the same Vδ gene family that were not both detected upon nucleotide sequence analysis. Significantly, analyses of the 17 hybridomas with two defined V(D)Jδ joins revealed that 6 had two in-frame rearrangements, demonstrating that assembly of TCR δ variable region genes does not exhibit allelic exclusion (Table 2).
Discussion
To determine if assembly of TCR δ variable region genes is regulated in the context of allelic exclusion, we have analyzed a panel of 27 clonal hybridomas derived from mouse splenic γδ T cells. Of the 17 hybridomas with defined V(D)Jδ rearrangements on both alleles, 6 (35%) have two in-frame rearrangements. This demonstrates that TCR δ variable region gene assembly does not exhibit allelic exclusion. Although this percentage is higher than the 20% (see Materials and Methods for calculations), which would be expected in the absence of allelic exclusion, this difference is not statistically significant (P > 0.10). Two human γδ T cell clones with in-frame TCR δ rearrangements on both alleles have been described previously (33, 45). However, given the number of cells analyzed in these studies, it was not possible to determine whether these clones represented rare events or a general lack of TCR δ allelic exclusion. As TCR γ rearrangements do not exhibit allelic exclusion, failure of TCR δ allelic exclusion further increases the possibility that a single γδ T cell will express two or more distinct γδ TCRs (7).
It is possible that one of the TCR δ rearrangements in each of the six cells with two in-frame rearrangements encodes for a TCR δ chain that cannot be expressed on the surface of the cell and therefore would not signal a block of further TCR δ rearrangements. This may occur, for example, if the TCR δ chain were not able to pair with a TCR γ chain or a component of a γδ pre-TCR, if such a receptor exists. In this regard, it has recently been shown that 2–4% of peripheral B cells have two in-frame IgH rearrangements but that only one encodes for an IgH chain that is capable of forming a pre-B cell receptor (22). Our data is more consistent with the notion that assembly of TCR δ variable region genes exhibits properties of allelic inclusion as the percentage of γδ T cell hybridomas with two in-frame TCR δ rearrangements is in agreement with the percentage expected in the absence of allelic exclusion. Furthermore, this percentage is similar to that of αβ T cells with two in-frame rearrangements at the TCR α locus, which also exhibits allelic inclusion (3).
It has been proposed for the IgH locus (and by analogy for the TCR β locus) that the precise ordering of variable gene segment rearrangement during lymphocyte development may be important for effecting allelic exclusion (14). In both of these loci, D to J rearrangement occurs on both alleles before V to DJ rearrangement. Presumably V to DJ rearrangement proceeds initially on one allele, at which point the rearrangement is “tested.” If it encodes a protein that can be expressed, signals are generated that prevent further V to DJ rearrangements on the other allele. In accordance with this model, the expected number of B and T cells have V(D)J/DJ configured rearrangements of their IgH and TCR β alleles, respectively (3, 10).
Unlike the IgH and TCR β loci, assembly of TCR δ variable gene segments is not ordered during development, and we now show that the TCR δ locus is not regulated in the context of allelic exclusion. However, the finding that many γδ T cells have incomplete TCR δ rearrangements demonstrates that rearrangement is frequently terminated before completion. The events that lead to termination of TCR δ rearrangement are not known. Thymic γδ T cells do not express RAG-1 or RAG-2, and it is possible, as proposed for TCR α rearrangement, that down regulation of RAG expression leads to termination of TCR δ rearrangement (30, 46). Termination of TCR δ rearrangement, by whatever mechanism, may be part of a developmental program that is independent of TCR δ expression. Alternatively, rearrangement may cease upon TCR δ expression, and failure of allelic exclusion may be due to the unordered simultaneous rearrangement of TCR δ alleles.
Acknowledgments
We thank F. Livak and D. Schatz for providing us with probes and C.H. Bassing for critical review of the manuscript.
This work is supported by the Howard Hughes Medical Institute and by National Institutes of Health grants AI20047 (F.W. Alt) and AI01297-01 (B.P. Sleckman). B.P. Sleckman is a recipient of a Career Development Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Abbreviations used in this paper
- BW
BW-1100.129.237
- D
diversity
- DN
double negative
- DP
double positive
- H
heavy
- J
joining
- L
light
- RAG
recombinase activating gene
- V
variable
Footnotes
B.P. Sleckman's present address is Department of Pathology, Washington University School of Medicine, 660 S. Euclid Ave., St. Louis, MO 63110.
The first two authors contributed equally to this study.
References
- 1.Sleckman BP, Gorman JR, Alt FW. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 2.Okada A, Alt FW. Mechanisms that control antigen receptor variable region gene assembly. Sem Immunol. 1994;6:185–196. doi: 10.1006/smim.1994.1024. [DOI] [PubMed] [Google Scholar]
- 3.Malissen M, Trucy J, Jouvin-Marche E, Cazenave P-A, Scollay R, Malissen B. Regulation of TCR α and β gene allelic exclusion during T cell development. Immunol Today. 1992;13:315–322. doi: 10.1016/0167-5699(92)90044-8. [DOI] [PubMed] [Google Scholar]
- 4.Alt FW, Enea V, Bothwell AL, Baltimore D. Activity of multiple light chain genes in murine multiple myeloma cells producing a single, functional light chain. Cell. 1980;21:1–12. doi: 10.1016/0092-8674(80)90109-9. [DOI] [PubMed] [Google Scholar]
- 5.Casanova J-L, Romero P, Widmann C, Kourilsky P, Maryanski JL. T cell receptor genes in a series of class I major histocompatibility complex-restricted cytotoxic T lymphocyte clones specific for Plasmodium bergheinonapeptide: Implications for T cell allelic exclusion and antigen-specific repertoire. J Exp Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Expression of two T cell receptor α chains: dual receptor T cells. Science. 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 7.Davodeau F, Peyrat M-A, Houde I, Hallet M-M, De Libero G, Vie H, Bonneville M. Surface expression of two distinct functional antigen receptors on human γδ T cells. Science. 1993;260:1800–1802. doi: 10.1126/science.8390096. [DOI] [PubMed] [Google Scholar]
- 8.Coleclough C, Perry RP, Karjalainen K, Weigert M. Aberrant rearrangements contribute significantly to allelic exclusion of immunoglobulin gene expression. Nature. 1981;290:372–378. doi: 10.1038/290372a0. [DOI] [PubMed] [Google Scholar]
- 9.Lansford, R., A. Okada, J. Chen, E.M. Oltz, T.K. Blackwell, F.W. Alt, and G. Rathbun. Mechanism and control of immunoglobulin gene rearrangement. In Molecular Immunology-Frontiers in Molecular Biology. 2nd edition. D.B. Hames, and D.M. Glover, editors. IRL Press, New York. 1–100.
- 10.ten Boekel E, Melchers F, Rolink A. The status of Ig rearrangements in single cells from different stages of B cell development. Int Immunol. 1995;7:1013–1019. doi: 10.1093/intimm/7.6.1013. [DOI] [PubMed] [Google Scholar]
- 11.Wabl M, Steinberg C. A theory of allelic exclusion for the immunoglobulin genes. Proc Natl Acad Sci USA. 1982;79:6976–6978. doi: 10.1073/pnas.79.22.6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonoda E, Pewzner-Jung Y, Schwers S, Taki S, Jung S, Eilat D, Rajewsky K. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–234. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 13.Alt FA, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MuLV-transformed cells: Rearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- 14.Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region genes. EMBO (Eur Mol Biol Organ) 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rusconi S, Kohler G. Transmission and expression of a specific pair of rearranged immunoglobulin μ and κ genes in a transgenic mouse line. Nature. 1985;314:330–334. doi: 10.1038/314330a0. [DOI] [PubMed] [Google Scholar]
- 16.Weaver D, Costantini F, Imanishi-Kari T, Baltimore D. A transgenic immunoglobulin mu gene prevents rearrangement of endogenous genes. Cell. 1985;42:117–127. doi: 10.1016/s0092-8674(85)80107-0. [DOI] [PubMed] [Google Scholar]
- 17.Nussenzweig MC, Shaw AC, Sinn E, Danner DB, Holmes KL, Morse HC, III, Leder P. Allelic exclusion in mice that express the membrane form of immunoglobulin μ. Science. 1987;236:816–819. doi: 10.1126/science.3107126. [DOI] [PubMed] [Google Scholar]
- 18.Storb U, Pinkert C, Arp B, Engler P, Gollahon K, Manz J, Brady W, Brinster R. Transgenic mice with μ and κ encoding anti-phosphorylcholine antibodies. J Exp Med. 1986;164:627–638. doi: 10.1084/jem.164.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uematsu Y, Ryser S, Dembic Z, Borgulya P, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. In transgenic mice the introduced functional T cell receptor β gene prevents expression of endogenous β genes. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- 20.Loffert D, Ehlich A, Muller W, Rajewsky K. Surrogate light chain expression is required to establish heavy chain allelic exclusion during B cell development. Immunity. 1996;4:133–144. doi: 10.1016/s1074-7613(00)80678-0. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura D, Rajewsky K. Targeted disruption of μ chain membrane exon causes loss of heavy-chain allelic exclusion. Nature. 1992;356:154–156. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- 22.ten Boekel E, Melchers F, Rolink AG. Precursor B cells showing H chain allelic inclusion display allelic exclusion at the level of pre-B cell receptor surface expression. Immunity. 1998;8:199–208. doi: 10.1016/s1074-7613(00)80472-0. [DOI] [PubMed] [Google Scholar]
- 23.Aifantis I, Buer J, von Boehmer H, Azogui O. Essential role of the pre-T cell receptor in allelic exclusion. Immunity. 1997;7:601–608. doi: 10.1016/s1074-7613(00)80381-7. [DOI] [PubMed] [Google Scholar]
- 24.Chien Y-H, Iwashima M, Kaplan KB, Elliot JF, Davis MM. A new T-cell receptor gene located within the α locus and expressed early in T cell differentiation. Nature. 1987;327:677–682. doi: 10.1038/327677a0. [DOI] [PubMed] [Google Scholar]
- 25.Satyanarayana K, Hata S, Devlin P, Roncarolo MC, De Vries JE, Spits H, Strominger JL, Krangel MS. Genomic organization of the human T-cell antigen-receptor α/δ locus. Proc Natl Acad Sci USA. 1988;85:8166–8170. doi: 10.1073/pnas.85.21.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehling HJ, von Boehmer H. Early αβ T cell development in the thymus of normal and genetically altered mice. Curr Opin Immunol. 1997;9:263–275. doi: 10.1016/s0952-7915(97)80146-x. [DOI] [PubMed] [Google Scholar]
- 27.Willerford DM, Swat W, Alt FW. Developmental regulation of V(D)J recombination and lymphocyte differentiation. Curr Opin Gen Dev. 1996;6:603–609. doi: 10.1016/s0959-437x(96)80090-6. [DOI] [PubMed] [Google Scholar]
- 28.Seboun E, Joshi N, Hauser S. Haplotypic origin of beta-chain genes expressed by human T-cell clones. Immunogenetics. 1992;36:363–368. doi: 10.1007/BF00218043. [DOI] [PubMed] [Google Scholar]
- 29.Marolleau J-P, Fondell JD, Malissen M, Trucy J, Barbier E, Marcu KB, Cazenave P-A, Primi D. The joining of germ-line Vα to Jα genes replaces the preexisting Vα-Jα complexes in a T cell receptor α,β positive T cell line. Cell. 1988;55:291–300. doi: 10.1016/0092-8674(88)90052-9. [DOI] [PubMed] [Google Scholar]
- 30.Borgulya P, Kishi H, Uematsu Y, von Boehmer H. Exclusion and inclusion of α and β T cell receptor alleles. Cell. 1992;69:529–537. doi: 10.1016/0092-8674(92)90453-j. [DOI] [PubMed] [Google Scholar]
- 31.Kang J, Raulet DH. Events that regulate differentiation of αβ TCR+ and γδ TCR+ T cells from a common precursor. Semin Immunol. 1997;9:171–179. doi: 10.1006/smim.1997.0069. [DOI] [PubMed] [Google Scholar]
- 32.Chien Y-H, Iwashima M, Wettstein DA, Kaplan KB, Elliot JF, Born W, Davis MM. T-cell receptor δ gene rearrangements in early thymocytes. Nature. 1987;330:722–727. doi: 10.1038/330722a0. [DOI] [PubMed] [Google Scholar]
- 33.Migone N, Padovan S, Zappador C, Glachino C, Bottaro M, Matullo G, Carbonara C, De Libero G, Casorati G. Restriction of the T-cell receptor V delta gene repertoire is due to preferential rearrangement and is independent of antigen selection. Immunogenetics. 1995;42:323. doi: 10.1007/BF00179393. [DOI] [PubMed] [Google Scholar]
- 34.Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW. Function of the TCRα enhancer in αβ and γδ T cells. Immunity. 1997;7:505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- 35.White J, Blackman M, Bill J, Kappler J, Marrack P, Gold D, Born W. Two better cell lines for making hybridomas expressing specific T cell receptors. J Immunol. 1989;143:1822–1825. [PubMed] [Google Scholar]
- 36.Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc Natl Acad Sci USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yancopoulos GD, Blackwell TK, Suh H, Hood L, Alt FW. Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell. 1986;44:251–259. doi: 10.1016/0092-8674(86)90759-2. [DOI] [PubMed] [Google Scholar]
- 39.Korman AJ, Maruyama J, Raulet D. Rearrangement by inversion of a T-cell receptor δ variable region gene located 3′ of the δ constant region gene. Proc Natl Acad Sci USA. 1989;86:267–271. doi: 10.1073/pnas.86.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winoto A, Baltimore D. Separate lineages of T cells expressing the αβ and γδ receptors. Nature. 1989;338:430–432. doi: 10.1038/338430a0. [DOI] [PubMed] [Google Scholar]
- 41.Livak F, Petrie HT, Crispe IN, Schatz DG. In-frame TCR δ gene rearrangements play a critical role in the αβ/γδ T cell lineage decision. Immunity. 1995;2:617–627. doi: 10.1016/1074-7613(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 42.Elliott JF, Rock EP, Patten PA, Davis MM, Chen Y-H. The adult T-cell receptor δ-chain is diverse and distinct from that of fetal thymocytes. Nature. 1988;331:627–631. doi: 10.1038/331627a0. [DOI] [PubMed] [Google Scholar]
- 43.Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 44.Sleckman, B.P., C.H. Bassing, C.G. Bardon, A. Okada, B. Khor, J.-C. Bories, R. Monroe, and F.W. Alt. 1998. Accessibility control of variable region gene assembly during T cell development. Immunol. Rev. In press. [DOI] [PubMed]
- 45.Krangel MS, Yssel H, Brocklehurst C, Spits H. A distinct wave of human T cell receptor γ/δ lymphocytes in the early fetal thymus: evidence for controlled gene rearrangement and cytokine production. J Exp Med. 1990;172:847–859. doi: 10.1084/jem.172.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livak F, Wilson A, MacDonald HR, Schatz D. αβ lineage-committed thymocytes can be rescued by the γδ T cell receptor (TCR) in the absence of TCR β chain. Eur J Immunol. 1997;27:2948–2958. doi: 10.1002/eji.1830271130. [DOI] [PubMed] [Google Scholar]