Abstract
Objective
To characterize global structural features of large-scale biomedical terminologies using currently emerging statistical approaches.
Design
Given rapid growth of terminologies, this research was designed to address scalability. We selected 16 terminologies covering a variety of domains from the UMLS Metathesaurus, a collection of terminological systems. Each was modeled as a network in which nodes were atomic concepts and links were relationships asserted by the source vocabulary. For comparison against each terminology we created three random networks of equivalent size and density.
Measurements
Average node degree, node degree distribution, clustering coefficient, average path length.
Results
Eight of 16 terminologies exhibited the small-world characteristics of a short average path length and strong local clustering. An overlapping subset of nine exhibited a power law distribution in node degrees, indicative of a scale-free architecture. We attribute these features to specific design constraints. Constraints on node connectivity, common in more synthetic classification systems, localize the effects of changes and deletions. In contrast, small-world and scale-free features, common in comprehensive medical terminologies, promote flexible navigation and less restrictive organic-like growth.
Conclusion
While thought of as synthetic, grid-like structures, some controlled terminologies are structurally indistinguishable from natural language networks. This paradoxical result suggests that terminology structure is shaped not only by formal logic-based semantics, but by rules analogous to those that govern social networks and biological systems. Graph theoretic modeling shows early promise as a framework for describing terminology structure. Deeper understanding of these techniques may inform the development of scalable terminologies and ontologies.
Introduction
Terminologies serve as shared data standards for public health reporting, health care reimbursement, indexing the biomedical literature, and interoperability between systems. While many are simply lists of concepts, some include explicitly defined properties and relationships. It is widely acknowledged that local design constraints affect large-scale structure or topology. In this descriptive study we use standard graph theoretic measures of network structure to analyze 16 biomedical terminologies, and we consider how these emerging approaches may contribute to terminology development, maintenance, and auditing.
Background
Network Modeling Has Been Used to Study Many Phenomena
Network analysis methods have been used to model many phenomena, including biological networks, transportation networks, and the Internet. The methods draw upon the mathematical formalisms of graph theory and upon analytic methods refined over decades of social network research. Networks consist of nodes, which represent entities, and lines, or links, drawn between the nodes to indicate a connection between them.
Analysis of network topology has been used in a variety of domains. In the informatics community, early uses of network modeling approaches have occurred mainly in bioinformatics and computational biology. 1 Various biological systems such as protein-protein interaction and genetic regulatory networks have been studied, sometimes yielding new insights into cellular and molecular pathways and interdependencies. Despite the prominence of these methods in the computational biology community, they are only just beginning to be adopted in other domains of informatics.
Standard Measures Are Used to Characterize Terminology Structure
Several articles summarize the graph theoretic measures used in this research, including a review by Steyvers (2005). 2 Additional details can be found in our recent review 3 and in the glossary in Appendix A (available as a JAMIA online data supplement at www.jamia.org). To summarize, in graph-theoretic modeling, a graph is comprised of a set of nodes, also referred to as vertices, along with a set of links which connect pairs of nodes. 2 The number of nodes to which a given node is immediately connected is its degree. A link from a node to itself is a loop. A node with no links is an isolate.
Networks vary widely depending on topological features. Five measures used in this research are the average node degree, the node degree distribution, the average path length, the diameter, and the clustering coefficient. The average node degree, a measure of the density of a graph, is the average number of links per node. It is calculated by dividing the number of links by the number of nodes, and then multiplying by two. The degrees of all the nodes in a network can be characterized as a node degree distribution. In this research the distribution is represented as a scatterplot with node degree plotted logarithmically on the x-axis, and frequency logarithmically on the y-axis.
The average path length, sometimes called the “average shortest path,” refers to the average distance between any two nodes. A simple algorithm determines the minimum distance between any node and any other node. An average is then calculated based on all of these values. The diameter is the longest distance between any two nodes in the network.
Finally, the clustering coefficient refers to the level of clustering in a graph at the local level. It is calculated for a given node by counting the number of links between the node’s neighbors and then dividing by all their possible links. This results in a value between 0 and 1, which is then averaged over all nodes in a network. 4
Two important concepts in this research are small-world and scale-free characteristics. Several articles 2,5 offer concise descriptions of these features. In networks with small-world properties there are highly clustered neighborhoods and it is possible to move from one node to another in a relatively small number of steps. Scale-free networks have a power law distribution in average node degree. This is a distribution in which one variable is proportional to a power of the other. When plotted on a double logarithmic scale, individual points are distributed about a straight line. There are a small number of nodes (the hubs) which have many neighbors and a large number of nodes that have only a few neighbors.
In research on large-scale network structure it is customary to simplify networks in two ways: First, although networks of semantic entities often have links of several types, it is common to treat all types of links equally when measuring structural features. Second, the directionality of links is often disregarded. Because these simplifications facilitate comparison of large-scale structure, the networks studied in this research are simplified in both of these ways.
Because graphs vary topologically, two graphs with the same number of nodes and links can diverge with respect to average path length and clustering coefficient. Given this variability, networks are often compared with an equivalent randomly configured network. To confirm the presence of statistically significant differences between selected parameters, in this research we use three random networks per terminology network. Average path length and diameter measures are particularly stable and only one random network is needed (see Appenidx B online at www.jamia.org).
Network Modeling Falls within a Wide Spectrum of Terminology Research
If large network modeling is adopted by terminologists, the approach will find its place within a wide spectrum of research. One of the large bodies of research within this spectrum involves ontologies, which are formal models of the concepts in a given domain. In ontologies, entities are assigned properties and relations between entities are defined explicitly. By investigating combinations of logical rules, ontological researchers have sought to find an optimal balance between expressiveness and computational tractability.
Informed in part by ontological approaches, some have proposed desirable characteristics, 6–9 many of them structural properties, or quality control approaches. 10,11 Other research has focused on issues such as detecting specific structural problems in the Unified Medical Language System (UMLS), 12 for example redundant semantic type assignments, 13 cycles in is-a hierarchies, 14 and inconsistencies between the hierarchies of the UMLS Semantic Network and Metathesaurus. 15 Other research has focused on achieving semantic interoperability 16 via ontology merging and alignment. 17 Our effort is distinct from previous research in that we use a formal method to describe emergent large-scale structural properties in terminologies.
Application of social network methods to study terminologies is like zooming out to take a photograph. The results can help individual terminology developers to see where a given terminology fits structurally within the greater universe of terminologies. There has been some effort to establish a useful typology of terminological systems. 18,19 However, to our knowledge, there has not been research comparing large-scale topological structure of biomedical terminologies. Because these techniques have not yet been widely adopted in the terminology community, the implications of the methods for terminology science remain unclear. We thus propose a methodology that provides quantitative and qualitative evidence to support formal topological analysis of terminologies.
Methods
Sixteen UMLS source vocabularies were selected. We sought to form a balanced selection of larger terminologies covering a variety of domains. To boost the interpretability of the results we selected source vocabularies familiar to the terminological research community. For contrastive purposes we also included two sets of related terminologies (ICD9CM and ICD10; and SNOMEDCT, SNMI, and RCD). Further details are available in see Figure 5, Appendix C online at www.jamia.org.
To extract the selected terminologies from the Metathesaurus we used the MetamorphoSys program. 12 We used the RRF (Rich Release Format) of the 2007AA release. The RRF is the only UMLS format that allows for source transparency (the ability to see the terminologies in a format consistent with that obtainable from the terminology’s authority). 20
After importing selected tables into a relational database, we queried the MRREL table to select the links assigned by each terminology. Concepts existing in a source vocabulary but having no associated records in MRREL were excluded. (These nodes would be considered isolates in a network model; they were excluded because isolates do not contribute meaningful information to the statistical measures used in this study. The numbers of isolated nodes in each terminology network appear in ▶.) Each terminology was then modeled as a graph in which the concepts were nodes and a link was assigned between concept pairs appearing in MRREL.
Table 1.
Table 1 Summary of Topological Features of Selected UMLS 2007AA Source vocabularies, arranged by small-world and scale-free characteristics
| Vocabulary Network |
Random Control |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source Abbreviation 2007AA | Official Name | Nodes | Links | Isolates | Loops | Multiple Lines | Avg. Node Degree | Median Node Degree | Maximum Node Degree | Diameter | Avg. Path Length | Clustering Coefficient | Avg. Path Length | Clustering Coefficient |
| CPT† | Current procedural terminology, 4th ed., 2006 | 18622 | 18621 | 0 | 0 | 0 | 2.00 | 2 | 451 | 16 | 8.88 | 0 | 13.03 | 0.000035 |
| NCBI ∗ † | NCBI taxonomy, 2006 | 247151 | 246854 | 50409 | 0 | 0 | 2.00 | 1 | 4308 | 63 | 26.49 | 0 | 16.98 | 0.000005 |
| GO ∗ † | Gene ontology | 21234 | 30105 | 18503 | 0 | 0 | 2.84 | 2 | 300 | 22 | 10.51 | 0.001462 | 9.46 | 0.000135 |
| RCD ∗ † | Clinical terms version 3 (Read codes), 1999 | 320354 | 319620 | 27214 | 0 | 0 | 2.00 | 1 | 163 | 29 | 14.02 | 0.000278 | 17.36 | 0.000005 |
| SNOMEDCT ∗ † | SNOMED clinical terms, 2006 | 391279 | 1540680 | 647854 | 0 | 35878 | 7.88 | 4 | 67721 | 21 | 5.09 | 0.195960 | 6.48 | 0.000018 |
| SMNI ∗ † | Systematized nomenclature of human and veterinary medicine, 1998 | 144478 | 219201 | 19701 | 0 | 4 | 3.03 | 1 | 3336 | 19 | 9.18 | 0.002351 | 10.71 | 0.000022 |
| NCI ∗ † | NCI thesaurus, 2006 | 49056 | 208436 | 97468 | 1 | 15637 | 8.50 | 3 | 3168 | 20 | 6.56 | 0.170025 | 5.29 | 0.000165 |
| HL7V3.0 ∗ † | Health level seven vocabulary, 1998–2006 | 8063 | 8952 | 0 | 0 | 0 | 2.22 | 1 | 485 | 19 | 6.66 | 0.000882 | 10.76 | 0.000122 |
| MSH ∗ † | Medical subject headings, 2007 | 377540 | 446587 | 247717 | 1 | 0 | 2.37 | 1 | 3937 | 23 | 7.43 | 0.003792 | 14.50 | 0.000007 |
| LNC ∗ | Logical observation identifier names and codes, 2.17.2006 | 166843 | 539422 | 3246 | 0 | 0 | 6.47 | 2 | 40562 | 11 | 3.88 | 0.002902 | 6.65 | 0.000040 |
| NOC | Nursing outcomes classification, 1997 | 3007 | 3006 | 49 | 0 | 0 | 2.00 | 1 | 28 | 10 | 7.42 | 0 | 10.69 | 0.000830 |
| DSM4 | Diagnostic and statistical manual of mental disorders. 4th ed., 1994 | 490 | 489 | 0 | 0 | 0 | 2.00 | 1 | 36 | 8 | 5.62 | 0 | 7.60 | 0.003333 |
| ICD10 | International statistical classification of diseases and related health problems, 1998 | 12319 | 12318 | 1186 | 0 | 0 | 2.00 | 1 | 28 | 10 | 7.83 | 0 | 12.61 | 0.000112 |
| ICD9CM | International classification of diseases, 9th revision, clinical modification, 2007 | 20958 | 20957 | 2 | 0 | 0 | 2.00 | 1 | 25 | 12 | 9.20 | 0 | 13.38 | 0.000039 |
| NIC | Nursing interventions classifications, 2005 | 11256 | 12175 | 0 | 0 | 0 | 2.16 | 1 | 66 | 8 | 6.91 | 0 | 11.39 | 0.000071 |
| ICPC | International classification of primary care | 748 | 1432 | 305 | 0 | 0 | 3.83 | 2 | 364 | 4 | 3.03 | 0 | 5.09 | 0.005883 |
∗ Small-world networks have a short average path length and strong clustering at the neighborhood level.
† Scale-free networks, characterized by a power law distribution in average node degree, have a small number of highly-connected hubs.
The vocabularies were modeled as networks in which concepts are nodes and terminology-asserted relationships constitute links between pairs of concepts. For each network we created three random controls of equivalent size and density. For these controls, clustering coefficient represents an average value of all three random networks.
To prepare for analysis, a series of preprocessing steps was performed. Each loop (relationship between a concept and itself) was removed. Sibling and allowed qualifier/qualified by relationships, which are superfluous and interfere with large network metrics, were also removed (see Figure 6, Appendix D online at www.jamia.org). Relationships with an inherent directionality, such as broader/narrower and parent/child, were replaced with undirected links. Multiple links connecting any given pair of nodes were replaced with single links. For each terminology network we also created three Erdös Rényi random graphs 1 with the same number of nodes and target average node degree (see Background section Standard Measures Are Used to Characterize Terminology; see also Appendix B online at www.jamia.org).
In determining the metrics to be included in the analysis we selected simple descriptive statistics and commonly-used measures of large network structure. For each terminology network and random network we measured the number of nodes, number of links, average node degree, and clustering coefficient. We also measured the average path length and diameter for all of the terminology graphs and for the first of the three random graphs created for each network (see Appendix B online at www.jamia.org). Finally, we represented the node degree distribution for each terminology using a scatterplot. We examined these plots to assess whether or not each terminology was scale-free. Analyses were conducted using Pajek, 21 NetDraw, 22 and the statistical program R. 23
Results
Summary Statistics for the Sixteen Terminology Networks and the Random Networks
The terminology networks ranged in size from 490 to 391,279 nodes (x̄ = 112,087, SD = 139,426) and from 489 to 1,540,680 links (x̄ = 226,803, SD = 378,958) (see Figure 7, Appendix E online at www.jamia.org). The numeric results of the research are summarized in ▶.
Node Degree Distribution and Scale-free Properties
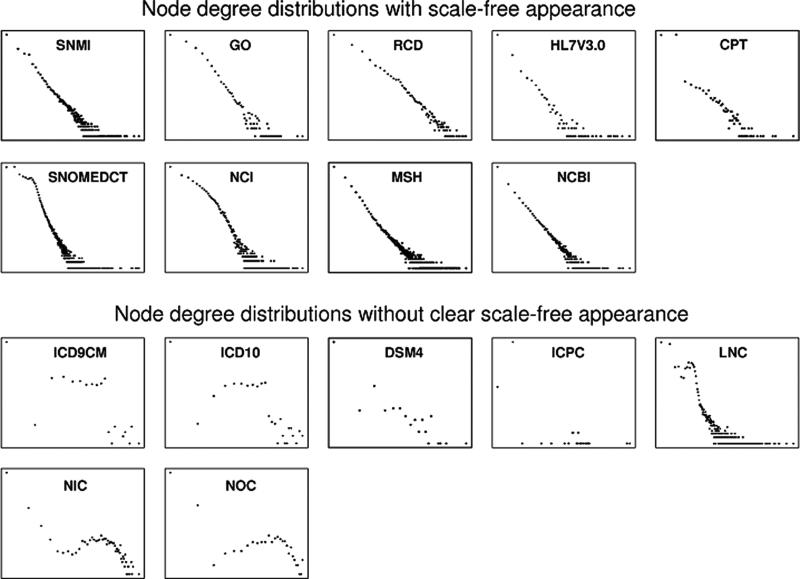
Nine of the networks (56.3%)—SNOMEDCT, SNMI, MSH, RCD, HL7V3.0, NCI, GO, CPT, and NCBI—exhibited a power law distribution in node degrees, indicative of a scale-free architecture (▶).
Figure 1.
Variations in node degree distributions among networks made from selected UMLS source vocabularies. Node degree is plotted logarithmically on the x-axis, and frequency logarithmically on the y-axis. The top nine networks have a clear power-law distribution and are thus scale-free; the bottom seven do not have clear scale-free features.
Average Path Length and Diameter
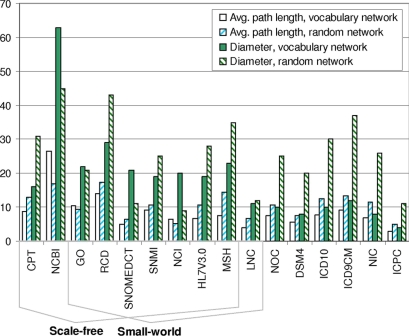
Average path lengths ranged from 3.03 to 26.49 (x̄ = 8.67, SD = 5.26). Diameters ranged from 4 to 63 (x̄ = 18, SD = 13.65). For the random networks, average path lengths ranged from 5.09 to 17.36 (x̄ = 10.34, SD = 3.76) and diameters ranged from 9 to 45 (x̄ = 18.44, SD = 13.26). Diameters and path lengths are graphed in ▶.
Figure 2.
Average path length and diameter for terminology network and corresponding random network, arranged by scale-free and small-world architecture. The diameters of the non scale-free terminologies were lower than those of their corresponding random networks.
Path lengths relative to the path lengths of the corresponding random networks ranged from 0.51 to 1.56 (x̄ = 0.79 SD = 0.27). As mentioned above, random networks have a short average path length. Since path lengths of all of the networks were similar to those of their corresponding random networks (up to 1.56 times higher, but still within an order of magnitude), it follows that all of the networks had a relatively “short” average path length. This is one of the hallmark features of a small-world network. (The second is strong local clustering.)
Average path length values were associated with the types of links occurring in the networks. In terminologies that supplemented hierarchical links with lateral, associative relationships, average path length values tended towards lower values. Among the eight terminologies that were at least 95% hierarchical (ICPC, GO, NCBI, NIC, ICD10, ICD9CM, DSM4, and HL7V3.0), the mean value for average path length was 9.53. By contrast, the average path length for the terminologies with at least 5% lateral links was 7.81.
When the diameters of the terminology networks were compared with those of their corresponding random networks, the differences in value were associated with the presence or absence of scale-free features. Among the nine scale-free terminology networks, four had a diameter at least as high as the diameter of their corresponding random networks (average ratio for the eight scale-free terminology networks, 1.10:1) By contrast, all of the non-scale-free terminology networks had smaller diameters than those of their random networks (average ratio for the seven non-scale-free terminology networks, 0.49:1).
Clustering Coefficient and Small-world Properties
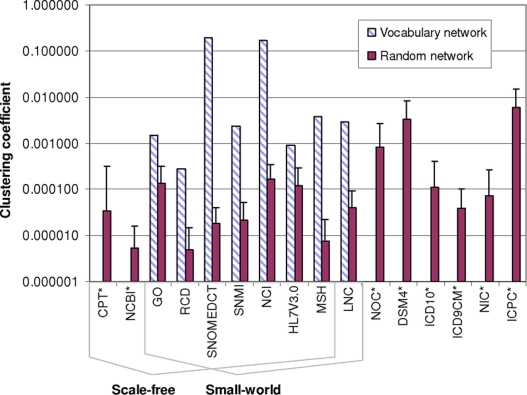
Eight (50%) of the terminology networks had a clustering coefficient of zero. When modeled using our approach, each of these networks (NCBI, CPT, ICD9CM, ICD10, NIC, ICPC, DSM4, and NOC) were trees or directed acyclic graphs (DAGs) with strictly hierarchical links. In these networks, even the links classified as other relationship or source asserted synonymy were hierarchical (see Appendix F online at www.jamia.org). In strict hierarchies, the only links between a node’s neighbors are via sibling relationships. Sibling relationships are not used in this research; as such, the clustering coefficient measure is zero for all nodes in the strictly hierarchical networks. Among the other eight terminology networks, clustering coefficients ranged from 0.00028 (for RCD) to 0.20 (for SNOMEDCT) (x̄ = 0.047, SD = 0.079). For all eight of these networks, clustering coefficient measures were higher than the average clustering coefficients for their three corresponding random networks (▶). Therefore, eight (50%) of the terminologies exhibited the small-world characteristics of a short average path length and strong local clustering.
Figure 3.
Clustering coefficient for terminology network and average clustering coefficient for corresponding random network, grouped by scale-free and small-world properties. Strong local clustering is one of the defining statistical properties of a small-world network; the other is a short average path length. Eight of the terminology networks had a clustering coefficient of zero. All eight nonzero clustering coefficients were significantly higher than the clustering coefficients of the corresponding three random networks.
Summary of Results
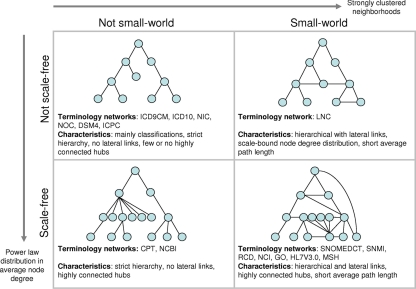
The results are summarized briefly in ▶. The last four columns indicate whether each terminology is a classification system, scale-free, at least 95 percent hierarchical, and small-world. This research yields an alternate grouping based on topological structure and shows that the terminologies fall into four groups based on scale-free and small-world properties (▶).
Table 2.
Table 2 Summary of Terminology Network Properties and Their Descriptions as Reported in the UMLS Documentation(24)
| Source Abbreviation, 2007 AA | Official Name | Category | Statistical Classification | Scale-Free | At least 95% Hierarchical | Small-World |
|---|---|---|---|---|---|---|
| GO | Gene Ontology, 2005 | Gene names | no | yes | yes | yes |
| HL7V3.0 | Health Level Seven Vocabulary, 1998–2006 | Miscellaneous | no | yes | yes | yes |
| NCBI | NCBI Taxonomy, 2006 | Gene names | no | yes | yes | no |
| CPT | Current Procedural Terminology, 4th ed., 2006 | Procedures only | no | yes | no | no |
| RCD | Clinical Terms Version 3 (Read Codes), 1999 | “Comprehensive” clinical vocabularies | no | yes | no | yes |
| SNOMEDCT | SNOMED Clinical Terms, 2006 | “Comprehensive” clinical vocabularies | no | yes | no | yes |
| SNMI | Systematized Nomenclature of Human and Veterinary Medicine, 1998 | “Comprehensive” clinical vocabularies | no | yes | no | yes |
| NCI | NCI Thesaurus, 2006 | Diseases | no | yes | no | yes |
| MSH | Medical Subject Headings, 2007 | Organisms ∗ | no | yes | no | yes |
| LNC | Logical Observation Identifier Names and Codes, 2.17,2006 | Thesaurus (used for indexing and retrieval of biomedical literature)† | no | no | no | yes |
| NOC | Nursing Outcomes Classification, 1997 | Nursing (currently used primarily for clinical documentation and research) | yes | no | no | no |
| DSM4 | Diagnostic and Statistical Manual of Mental Disorders. 4th ed., 1994 | Diagnoses/clinical problems/signs and symptoms | yes | no | yes | no |
| ICD10 | International Statistical Classification of Diseases and Related Health Problems, 1998 | Diagnoses only | yes | no | yes | no |
| ICD9CM | International Classification of Diseases, 9th Revision, Clinical Modification, 2007 | Diagnoses and procedures | yes | no | yes | no |
| NIC | Nursing Interventions Classification, 2005 | Nursing (currently used primarily for clinical documentation and research) | yes | no | yes | no |
| ICPC | International Classification of Primary Care, 1993 | Diagnoses/clinical problems/signs and symptoms | yes | no | yes | no |
∗ In UMLS documentation, 24 NCBI was listed under the category gene names.
† In UMLS documentation, 24 MSH was listed under the category “Comprehensive” vocabularies.
Scale-free networks, which are characterized by a power law distribution in average node degree, have a small number of highly-connected hubs. Small-world networks have a short average path length and strong clustering at the neighborhood level.
Figure 4.
Selected UMLS source vocabularies arranged into four groups based on scale-free and small-world properties. The schematic diagrams portray the key differences between the terminology structures. Scale-free networks, which are characterized by a power law distribution in average node degree, have a small number of highly-connected hubs. Although a scale-free architecture is common in many general-purpose semantic networks, several terminologies in our sample (mainly the statistical classifications) did not exhibit a scale-free topology. This is thought to result from constraints introduced into the development process of such classifications. The other terminologies were scale-free. Small-world networks have a short average path length and relatively strong clustering at the neighborhood level. All eight terminologies with lateral (nonhierarchical) links were found to have a small-world architecture.
Discussion
At the core of any information system is its schema for representing data and conceptual knowledge. Many biomedical systems rely on rigid taxonomies that impose a single conceptual framework 25 on underlying semantic information. In the interdisciplinary world of biomedicine, there is no one ‘correct’ information model, and the heterogeneity of terminologies, even those dealing with identical subjects, cannot be controlled. 26 In hopes of using natural semantic networks to derive design principles for more flexible biomedical terminologies, one of the goals of this research is to understand the similarities and differences between the two.
As we have discussed in a recent review, 3 many networks made from natural language are both scale-free and small-world. These include networks made from corpora, 27 word association studies in which a human subject is shown a particular word and is asked to name a related word, 2 and an English-language thesaurus. 28 Unlike these natural language networks, terminologies are developed for the purpose of standardizing biomedical language for use in systems and applications. Our research demonstrates that while terminologies are regarded as synthetic, grid-like structures, some possess the large-scale structural features of natural language networks. This paradoxical result suggests that it is not solely formal logic-based semantics that governs terminology structure: Rules analogous to those governing social networks and biological systems also shape topology.
To summarize, six of the terminologies exhibited both the scale-free and small-world properties common in natural language networks, and 10 exhibited at least one of these properties. These features were more prevalent in larger terminologies. Among the eight smallest in terms of nodes, six had neither scale-free nor small-world features; among the eight largest, all were either scale-free, small-world, or both. Small-world and scale-free features were also associated with a relaxation of specific design constraints: Small-world terminologies were less restrictive as to the nature of relationships between entities, while scale-free terminologies placed no upper limit on connectivity (the number of nodes to which a given entity can be connected).
The Terminologies Fell into Four Groups Based on Their Structural Features
The terminologies fell into four groups (▶) based on their structural features. Half the terminologies exhibited the small-world characteristics of a short average path length and strong local clustering. These properties were associated with terminologies that were not restricted to subsumption (is-a and broader/narrower) relationships, but which also contained lateral links often signifying an associative relationship.
An overlapping subset of terminologies exhibited a power law distribution in node degrees, indicative of a scale-free architecture. This feature was found in comprehensive medical terminologies but not in statistical classifications with imposed constraints on node connectivity. Constraints in statistical classifications are intentional; to facilitate the study of the phenomenon being modeled, they have a limited number of categories. 29 As we will illustrate with examples, these design constraints result in scale-bound node degree distributions.
Large-scale Terminological Structure Reflects Small-scale Design Constraints
The most apparent difference between the terminology networks was between those having only hierarchical links and those that contained a mixture of hierarchical and lateral links. All the vocabularies in our sample had a relatively short path length when compared with their random networks, so they possessed the first qualification for small-world architecture. However, it was only those terminologies with lateral links that had strong local clustering, which is the second requirement. (Since the value of the clustering coefficient is interpreted in relation to each terminology’s random network, strong local clustering refers to networks with various degrees of stronger than random clustering.)
The relationship between lateral links and clustering coefficient can be illustrated by examining a pair of related terminologies. While the clustering coefficient for SNMI, the 1998 version of SNOMED, was 0.0024, that for SNOMEDCT was more than 80 times higher (0.20). This can be explained by examining the relationship type profiles (see Figure 8, Appendix F online at www.jamia.org). While 52 percent of the relationships in the SNMI network are strictly hierarchical parent/child or broader/narrower links, only 39 percent of the links in the SNOMEDCT network are strictly hierarchical. The average node degree of SNOMEDCT, which is more than double that of SNMI, also contributes to an increase in the clustering coefficient.
Although a scale-free architecture is common in many general-purpose semantic networks,2,3 several terminologies in our sample (mainly the statistical classifications) did not exhibit a scale-free topology; they had a distinctive scaling in average node degree. The scale-bound topology of these classifications is due in part to the fact that they are, by design, confined to a limited number of categories. 29 Each of the scale-bound terminologies had a constraint on growth that accounted for the deviation from the more organic scale-free connectivity.
The reason for scaling in the node degree distribution of the two ICD terminologies can be explored by comparing ICD10 with CPT. ICD10 and CPT were remarkably similar in most measures. They had similar numbers of nodes and links, and (with the exception of two anomalous source-asserted synonymy relations in CPT) were both comprised entirely of parent/child relationships. They both had an average path length near 9.0 and, as DAGs, a clustering coefficient of zero. The key difference between the two was in the distribution of node degrees. While the node degree distribution for CPT was scale-free, that for ICD10 was scale-bound.
To understand this difference it helps to consider the structure of the ICD framework. In both ICD networks, there is a disproportionately high number of nodes with exactly 10 or 11 neighbors. The structure of the identifier in ICD has long included a decimal point followed by one or more digits from 0 to 9. This has resulted in a strong disincentive, albeit unintentional, for concepts to have more than 9 or 10 children. This disincentive manifests as a spike in the distribution (▶); in ICD9CM, several thousand concepts have exactly 10 or 11 neighbors (9 or 10 children, plus one parent), while only a few hundred have 12 or 13. It has been suggested that biomedical terminology developers should avoid structural constraints that unintentionally limit the expansion of a terminology. 7 This research confirms the fact that constraints on numeric identifiers can affect the topology of a network.
Another classification with a scale-bound node degree distribution is DSM4. The distribution is skewed in part as a result of the terminology’s association with ICD. Each version of the DSM is linked to a corresponding version of ICD; the fourth version has been coordinated with psychiatric diagnoses in ICD-10. 30 Many of the codes correspond with one another. DSM4 may thus have inherited its scaled node degree distribution from ICD.
The two nursing classifications, NIC and NOC, also had scale-bound node degree distributions. Again, the scaling is associated with constraints on structure. Both classifications are partitioned into domains. The structure of NOC will allow for its eventual expansion to 10 domains and 52 outcome classes, with 99 outcomes per class. 31
Among the terminologies that did not have a small-world architecture, some were scale-free (CPT and NCBI) while others (DSM4, ICD9CM, ICD10, ICPC, NIC, and NOC) were not. Both sets were either trees or DAGs. However, CPT and NCBI had a power-law distribution in average node degree. This implies that connectivity was allowed to develop organically; although the developers restricted the types of relationships to hierarchical (thereby ruling out small-world features) there was no upper restriction on the number of concepts to which any given concept could connect. This has allowed the CPT and NCBI trees to fan out freely. Recent changes in the Health Level Seven Vocabulary serve to illustrate the phenomenon of change in large-scale structure over time. Although the HL7 Vocabulary was not scale-free as represented in the 2005AA RRF version of the Metathesaurus, it is scale-free in the 2007AA RRF version. The nomenclature in the HL7 Vocabulary now includes entities of a more diverse range of semantic types, including medication types, terms pertaining to anatomy, and specific geographical areas.
The fact that some synthetic terminology networks are scale-free is a novel finding. Though initiated as tightly controlled systems, some terminologies have evolved in an organic-like manner. To our knowledge, this is the first study that uses graph theoretic approaches and statistics to establish the increasing relatedness between properties of large scale terminologies and natural language: Some biomedical terminologies, like many networks made from natural language, have scale-free and small-world features.
Emergent Structural Features Can Affect Terminology Usability
Terminologies are growing increasingly larger. While they were once mainly lists of terms with parent/child relationships, now more relationship types are being added and topological structure is becoming more complex. As the examples above demonstrate, defined growth constraints affect the layout of links between entities and therefore shape a terminology’s emergent large-scale topology. This topology, in turn, carries implications for the use of the terminology. Operations conducted in the course of terminology development and maintenance include finding, adding, editing, and deleting concepts. Features of network topology can help or hinder a user’s ability to perform these operations.
A small-world architecture implies short average path lengths and strong local clustering. These features may be useful when new terms are added. If related words are already arranged into highly-connected clusters, it should be easier to identify where a new term should be added. In terminologies, a short path length also implies an abundance of lateral links. As terminology developers are aware, these cross-links provide for efficient navigation. For example, when searching for the term hay fever in SNMI, which has other relationship lateral links, it is possible to navigate to the term via semantically related concepts such as nose, allergens, and pollen. However, in RCD, it is only possible to navigate to hay fever via one of its parents, allergic rhinitis, seasonal allergic rhinitis, or allergic reaction to substance, or via one of its children, such as hay fever with asthma, or other seasonal allergic rhinitis. Lateral links thus provide many options for navigating through the tree.
The existence of lateral links also carries implications when a term is changed. In SNOMEDCT, the concept diabetes mellitus is connected to more than 100 other concepts via other relationship links. By contrast, in ICD9CM, diabetes mellitus is only connected to a single parent (diseases of other endocrine glands) and to 10 children, such as diabetes with renal manifestations. A change to the term in ICD9CM, therefore, only has immediate effects on the meaning of these 11 neighbors, while any change to the term in SNOMEDCT subtly affects the meaning of all of these nearby concepts. This caveat contrasts with the flexibility conferred by inclusion of lateral links.
The unique structural properties of treelike terminologies have implications for maintenance. One is that the effects of changes are confined to the immediate neighborhood. The deletion of a highly-connected hub in a small-world, scale-free network can affect hundreds or thousands of nodes throughout the network. If a tree is scale-free, deletion of a concept can affect many nodes, but the nodes affected will all be parents, children, or siblings of deleted concept. If the tree is not scale-free, the deletion will typically only affect a small number of nearby concepts. Despite this fact, trees are not as robust as networks with lateral links. Removal of any non-leaf node (or any link) of a tree will split the tree into two or more subtrees (for an illustration, see Figure 9, Appendix G online at www.jamia.org). In addition, the constraints of a treelike structure restrict link assignment. If a new concept is added in a tree, it is impossible to assign links to concepts in other branches, or to concepts more than one level away. Therefore, the rigid constraints of a treelike structure confer advantages, but they result in restrictions on node connectivity and a less robust structure when nodes are deleted.
Scale-free structure also has implications for terminology maintenance. A scale-free architecture can help a user identify a starting place to look for a concept. Highly connected hubs, small in number, can serve effectively as landmarks. As a user delves deeper into the network, there are smaller hubs at every scale which can also be used for orientation.
Network Modeling Can Be a Tool for Terminology Developers
Software for visualization and analysis of large networks is becoming increasingly sophisticated. Some tools allow users to select nodes and links matching certain criteria and then to perform operations on the selected nodes. Users can view networks using a variety of layout algorithms, and zoom in or out, and display desired levels of detail about nodes and links. For example, the color of nodes can be assigned based on node degree, with high-degree nodes assigned a dark color and low-degree nodes assigned a light color. This technique highlights areas of high connectivity the network; areas of low connectivity can then be targeted for maintenance. 32 Figure 10 (Appendix H online at www.jamia.org) contains visualizations of the parent-child relationships in the 16 terminology networks examined in this research.
Limitations
The results of this research are subject to several limitations. The data used in this analysis were drawn from the UMLS Metathesaurus, a collection of terminological resources, and not directly from the terminologies themselves. Although the developers of the UMLS have attempted to represent each source vocabulary as accurately as possible, some subjective judgments are made when converting each source into the UMLS standardized database format. Specific limitations are discussed in detail in Appenidx C online at www.jamia.org.
Future Research
As we have shown in this research, terminology structures vary depending on design constraints. Although large-scale network modeling and analysis is not yet widely used in terminology science, the approaches show some early promise for visualizing terminology structure, demonstrating the effects of design constraints on structure, and for terminology development and maintenance. There are a number of compelling possibilities for future research.
First, although this research focused on a set of techniques that have been widely applied in the study of large networks, other topological measures and modeling techniques may be equally valuable. For example, inclusion of link directionality in models would allow for a more detailed analysis of local topological features such as motifs. Other techniques can be used to understand multiple structural patterns that may occur within any given terminology. One could assign weights or ordinal values to entities and links, create subnetworks of nodes and links of specific types, and then use cohesive subgroup detection algorithms to identify clusters of related entities. These techniques could be used to investigate unexpected results; for example, the fact that the SNMI network is scale-free even though, like ICD9CM, it uses a constrained scheme for its identifiers. One could also investigate whether specific types of lateral links are in fact strictly hierarchical.
Second, terminology metadata, such as age and number of participants, could be used to study the effects of social processes on structure. We suggest that mature terminologies with multiple contributors, having developed according to more organic processes, would be more likely to have scale-free and small-world features. These data could be combined to develop a fully automated classifier of a terminology as organic or synthetic.
As we have demonstrated, further research is needed to determine whether particular structural features are associated with flexibility. It is not yet clear how these features can benefit specific terminological functions such as billing support, coding for electronic health records, and computer-assisted decision support. Given the need for deterministic and reliable operations in biomedical information systems, it would be worthwhile to investigate the implications of highly-connected hubs, dense communities of nodes, and cycles, on computational tractability.
Conclusion
Although biomedical terminologies are initiated as tightly controlled systems, some have the scale-free and small-world structural features found in networks made from natural language. This paradoxical result suggests that it is not solely formal logic-based semantics that governs terminology structure: Rules analogous to those governing social networks and biological systems also shape topology. In biomedical terminologies, multiple link types and unrestricted node connectivity are associated with small-world and scale-free features, respectively. These features allow for efficient navigation and organic growth. Conversely, restrictions to hierarchical links and limits on node connectivity, which are common in statistical classifications, localize the effects of changes and deletions; however, these restrictions also result in synthetic structures that are less robust than organic networks. Since terminology networks with organic-like scale-free properties are particularly sensitive to the retirement of highly connected nodes, we propose that change management policies for scale-free terminologies should include additional procedures for altering highly-connected components of the network. Deeper understanding of terminology structure is a key next step in encouraging the development of increasingly flexible, scalable, and useful biomedical terminologies.
Note: References 33–46 are cited in the online data supplement to this article at jamia.org.
Footnotes
Support for this research was provided by NLM training grant 5T15LM07079. This work was partially supported by grants LM008308-01 and 1U54CA121852. The authors would like to thank Drs. Olivier Bodenreider, James Cimino, William Hole, and Adam Rothschild, who provided invaluable guidance during the preparation of this manuscript. We also thank Drs. Patrick Mary and David Auber for support with Tulip software. In addition, this manuscript has benefited greatly from the insightful comments of two anonymous reviewers. We thank them for their diligence.
References
- 1.Newman MEJ. The structure and function of complex networks Siam Rev. 2003;45(2):167-256. [Google Scholar]
- 2.Steyvers M, Tenenbaum JB. The large-scale structure of semantic networks: Statistical analyses and a model of semantic growth Cognitive Sci. 2005;29(1):41-78. [DOI] [PubMed] [Google Scholar]
- 3.Bales ME, Johnson SB. Graph theoretic modeling of large-scale semantic networks J Biomed Inform 2006;39(4):451-464. [DOI] [PubMed] [Google Scholar]
- 4.Sporns O, Chialvo DR, Kaiser M, Hilgetag CC. Organization, development and function of complex brain networks Trends Cogn Sci 2004;8(9):418-425. [DOI] [PubMed] [Google Scholar]
- 5.Motter AE, de Moura APS, Lai YC, Dasgupta P. Topology of the conceptual network of language Phys Rev E. 2002;65(6). [DOI] [PubMed]
- 6.Chute CG, Cohn SP, Campbell JR. A framework for comprehensive terminology systems in the United States: development guidelines, criteria for selection, and public policy implications J Am Med Inform Assoc 1998;5(6):503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century Method Inform Med 1998;37(4-5):394-403. [PMC free article] [PubMed] [Google Scholar]
- 8.Cimino JJ. In defense of the desiderata J Biomed Inform 2005;39(3):299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rector AL, Nowian WA, Kay S. Conceptual knowledge: the core of medical information systemsIn: Lun KC, Deguolet P, Piemme TE, Rienhoff O, editors. MEDINFO 92, Proceedings of the Seventh World Congress on Medical Informatics; 1992 Sep 6–10; Geneva, Switzerland. Amsterdam: North Holland. 1992. pp. 1420-1426.
- 10.Kohler J, Munn K, Ruegg A, Skusa A, Smith B. Quality control for terms and definitions in ontologies and taxonomies BMC Bioinformatics 2006;7:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers JE. Quality assurance of medical ontologies Method Inform Med. 2006;45(3):267-274. [PubMed] [Google Scholar]
- 12.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology Nucleic Acids Res 2004;32(Database issue):D267-D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng Y, Halper MH, Perl Y, Geller J. Auditing the UMLS for redundant classifications Proc Am Med Inform Assoc Annu Symp 2002:612-616San Antonio, TX. [PMC free article] [PubMed]
- 14.Bodenreider O. Circular hierarchical relationships in the UMLS: etiology, diagnosis, treatment, complications and prevention Proc Am Med Inform Assoc Annu Symp 2001:57-61Washington, DC. [PMC free article] [PubMed]
- 15.Cimino JJ, Min H, Perl Y. Consistency across the hierarchies of the UMLS Semantic Network and Metathesaurus J Biomed Inform 2003;36(6):450-461. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, Geller J. Semantic enrichment for medical ontologies J Biomed Inform 2006;39:209-226. [DOI] [PubMed] [Google Scholar]
- 17.Yu AC. Methods in biomedical ontology J Biomed Inform 2005(39):252-266. [DOI] [PubMed]
- 18.de Keizer NF, Abu-Hanna A. Understanding terminological systemsII: Experience with conceptual and formal representation of structure. Method Inform Med 2000;39(1):22-29. [PubMed] [Google Scholar]
- 19.de Keizer NF, Abu-Hanna A, Zwetsloot-Schonk JH. Understanding terminological systemsI: Terminology and typology. Method Inform Med 2000;39(1):16-21. [PubMed] [Google Scholar]
- 20.Hole WT, Carlsen BA, Tuttle MS, Srinivasan S, Lipow SS, Olson NE, et al. Achieving “source transparency” in the UMLS MetathesaurusIn: Fieschi M, Coiera E, Li YCJ, editors. MEDINFO 2004, Proc 11th World Congress Med Inform. 2004. pp. 57-61San Francisco, CA.
- 21.Batagelj V, Mrvar A. Pajek—analysis and visualization of large networksIn: Juenger M, Mutzel P, editors. Graph Drawing Software. Berlin: Springer; 2003. pp. 77-103.
- 22.Borgatti SP. NetDraw: Graph Visualization Software. 1.46 edHarvard: Analytic Technologies; 2002.
- 23.R Development Core Team R: A Language and Environment for Statistical Computing 2007. Vienna, Austria.
- 24.National Library of Medicine (US) Grouping UMLS Vocabularies by Category as of Jan. 2005 2006. Available at: http://www.nlm.nih.gov/research/umls/sources_by_categories.html. Accessed on: July 16, 2007.
- 25.Rose E. Life after go-live, Part 3: Struggling with structure J Healthc Inf Manag 2003;17(3):25-27. [PubMed] [Google Scholar]
- 26.National Center for Biomedical Ontology NCBO Dissemination Event 2006. Available at: http://www.bisti.nih.gov/ahm2006/ncbo_dissemination.htm. Accessed on: Jul 16, 2007.
- 27.Ferrer i Cancho R, Sole RV. The small world of human language P Roy Soc Lond B Bio 2001;268(1482):2261-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sigman M, Cecchi GA. Global organization of the Wordnet lexicon P Natl Acad Sci USA 2002;99(3):1742-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization (Switzerland) Manual of the International Statistical Classification of Diseases, Injuries, and Causes of DeathGeneva: The Organization; 1957.
- 30.American Psychiatric Association Committee on Nomenclature and Statistics Diagnostic and Statistical Manual of Mental DisordersFourth ed. Washington DC: The Association; 1994.
- 31.Medical-Billing-Coding.org History of outcomes classification 2007. Available at: http://www.medical-billing-coding.org/NewsArticleDetail1184.htm. Accessed on: July 16.
- 32.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches J Biomed Inform 2003;36(6):414-432Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: Simple building blocks of complex networks Science 2002;298(5594):824-827. [DOI] [PubMed] [Google Scholar]
- 34.University of Oklahoma College of Geosciences An abbreviated glossary of system terminology 2006. Available at: http://www.esse.ou.edu/glossary_st.html. Accessed on: July 16, 2007.
- 35.Barabasi AL, Albert R. Emergence of scaling in random networks Science 1999;286(5439):509-512. [DOI] [PubMed] [Google Scholar]
- 36.Gutwenger C, Mutzel P. Planar polyline drawings with good angular resolution Proc 6th Int Symp Graph Drawing; Lecture Notes Comp Sci 1998;1547:167-182. [Google Scholar]
- 37.Auber D. Tulip: A huge graph visualisation frameworkIn: Mutzel P, Jünger M, editors. Graph Drawing Software, Mathematics, and Visualization. Springer-Verlag; 2003. pp. 105-126.
- 38.National Center for Health Statistics (US) International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) 2006. Available at: http://www.cdc.gov/nchs/about/otheract/icd9/abticd9.htm. Accessed on: July 16, 2007.
- 39.Chute CG, Cohn SP, Campbell KE, Oliver DE, Campbell JR. The content coverage of clinical classificationsFor The Computer-Based Patient Record Institute’s Work Group on Codes & Structures. J Am Med Inform Assoc 1996;3(3):224-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stausberg J, Lang H, Obertacke U, Rauhut F. Classifications in Routine Use: Lessons from ICD-9 and ICPM in Surgical Practice J Am Med Inform Assoc 2001;8(1):92-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.International Health Terminology Standards Development Organization Historical perspectives: history of creating a world-class terminology 2007. Available at: http://www.snomed.org/about/history_summary.html. Accessed on: July 16, 2007.
- 42.McCray AT, Ide NC, Loane RR, Tse T. Strategies for supporting consumer health information seeking. MEDINFO 2004. Proc 11th World Congress Med Inform; San Francisco, CA, USA. p. 1152–6 (pt. 2). [PubMed]
- 43.Zeng QT, Crowell J, Plovnick RM, Kim E, Ngo L, Dibble E. Assisting consumer health information retrieval with query recommendations J Am Med Inform Assoc 2006;13(1):80-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text J Biomed Inform 2003;36(6):462-477. [DOI] [PubMed] [Google Scholar]
- 45.Cokol M, Iossifov I, Weinreb C, Rzhetsky A. Emergent behavior of growing knowledge about molecular interactions Nat Biotechnol. 2005;23(10):1243-1247. [DOI] [PubMed] [Google Scholar]
- 46.Koslow SH, Subramaniam S. Databasing the Brain: From Data to Knowledge (Neuroinformatics)Hoboken, NJ: John Wiley & Sons, Inc; 2005.