Abstract
Cancer staging provides a basis for planning clinical management, but also allows for meaningful analysis of cancer outcomes and evaluation of cancer care services. Despite this, stage data in cancer registries is often incomplete, inaccurate, or simply not collected. This article describes a prototype software system (Cancer Stage Interpretation System, CSIS) that automatically extracts cancer staging information from medical reports. The system uses text classification techniques to train support vector machines (SVMs) to extract elements of stage listed in cancer staging guidelines. When processing new reports, CSIS identifies sentences relevant to the staging decision, and subsequently assigns the most likely stage. The system was developed using a database of staging data and pathology reports for 710 lung cancer patients, then validated in an independent set of 179 patients against pathologic stage assigned by two independent pathologists. CSIS achieved overall accuracy of 74% for tumor (T) staging and 87% for node (N) staging, and errors were observed to mirror disagreements between human experts.
Introduction
Cancer stage categorizes the size and location of the primary tumor, the extent of lymph node involvement, and the presence or absence of metastatic spread to other body parts. The clinical management of most cancers according to evidence-based guidelines 1 is dependent on the stage of disease at diagnosis, and documentation of cancer stage at diagnosis is increasingly being recommended as a standard of care by national cancer bodies.
International standards for cancer staging have been developed, such as the TNM (Tumor Node Metastases) standard defined by the American Joint Committee on Cancer (AJCC) and International Union Against Cancer (UICC), summarized in ▶. 2
Table 1.
Table 1 Summary of the TNM Staging Protocol 2
| T: Primary Tumor | X | Primary tumor cannot be assessed |
| 0 | No evidence of primary tumor | |
| is | Carcinoma in situ | |
| 1,2,3,4 | Increasing size and/or local extent of the primary tumor | |
| N: Regional Lymph Nodes | X | Regional lymph nodes cannot be assessed |
| 0 | No regional lymph node metastasis | |
| 1,2,3 | Increasing involvement of regional lymph nodes | |
| M: Distant Metastasis | X | Distant metastasis cannot be assessed |
| 0 | No distant metastasis | |
| 1 | Distant metastasis |
Apart from the important role played by cancer staging in the clinical management of individual patients, there is increasing acknowledgement that outcomes analysis of cancer management or intervention programs on a population, governance, or facility level is meaningful only if interpreted in the light of this major prognostic factor. As the main population-based data repositories, cancer registries have moved to incorporate clinically relevant fields such as cancer stage to enable more accurate and useful outcomes analysis. Despite these changes, stage data in registries is still commonly absent or incomplete. After four years of mandated stage data collection for prostate cancer by the Maryland Cancer Registry, data were still missing in 13% of cases on average, and up to 20% in some regions. 3 A similar study in the Ottawa Regional Cancer Centre found missing staging information in 10% of lymphoma cases and 38% of breast cancer cases. 4 An earlier study at that center showed that mandated stage data collection across all cancer types led to complete stage data being available for 71% of cases on average. 5 Organized stage data collection as undertaken in these two North American centers is in contrast to many other regions. For instance, in 2005 the National Cancer Control Initiative reported that there was no ongoing population-based collection of staging information in any Australian state or territory. 9
Even when collected, there is evidence that stage data are often inaccurate. A study of demographic differences in prostate cancer staging in Connecticut found that 23% of cases in the registry were incorrectly coded, 6 because of either incomplete medical records or staging errors. A review of lung cancer stage data in the Maastricht Cancer Registry in the Netherlands found major discrepancies in 12% and minor discrepancies in 23% of cases. 7 Many of these were caused by incorrect application of staging guidelines, as well as data entry errors. Similarly, a review of stage data in Ottawa Regional Cancer Centre found staging errors occurred in 2% to 5% and data entry error in 3% to 6% of all cases. 5 There were differences between registry stage and stage as determined from available clinical information in 31% of lymphoma and 8% of breast cancer cases. 4
When not obtained directly from clinicians prospectively, it is possible to perform retrospective staging based on retrieved medical records. A Nottingham prostate cancer study that retrospectively assigned stage using case notes showed that stage information regarding the primary tumor (T stage) could be abstracted for 96% of cases; however, only limited information was available for staging lymph node and metastatic involvement (N and M stage). 8 The Western Australian Cancer Registry feasibility study of staging from medical records for 20 cancer types found that, under various assumptions, stage data could be collected using current data sources for seven cancer types, but were not a feasible or required system change for the others. 9 The same group subsequently undertook a project to retrospectively collect stage data for all colorectal cancer cases over a one-year period. 10 They were able to fully stage 76% of cases from available data sources (pathology reports, case notes, hospital registries, etc.), and a further 22% of cases if M stage was omitted. A study in which stage data were retrospectively sourced from medical reports was used to monitor cancer outcomes for indigenous Australians in the Northern Territory. 11
Therefore, although staging is a recognized component of providing quality cancer care, data on stage often are incomplete, inaccurate, or not recorded. Furthermore, although it is possible to retrospectively retrieve data from available medical reports, doing this manually can be time and labor intensive.
Motivated by these limitations, we developed CSIS (Cancer Stage Interpretation System), a prototype software system to assign cancer stage data by automatically extracting relevant information from free-text medical reports stored in clinical information systems. CSIS could be used by a cancer registry to support collection of staging information for those patients not formally staged by human experts, allowing more comprehensive population-level analysis of outcomes. Alternatively, if deployed at the point of reporting, it has the potential to improve the efficiency and consistency of staging by clinicians. Although the system was developed on lung cancer data available to us, it could in principle be applied to stage other cancer types. For an individual patient, input to the system consists of textual reports describing pathology tests. The objective is to estimate pathologic stage by applying machine learning text categorization techniques. 12 Because metastatic lung cancer is defined as involvement of other organs, it is not usually assessable from pathological studies of the lung; therefore, the current system does not attempt to determine the M stage.
Previous work investigated direct classification of the cancer stage using binary support vector machines (SVMs) operating on the concatenated reports of a given patient, 13,14 essentially posing the problem as document-level topic categorization. 12 Although results were promising, there was a need to improve system performance. Furthermore, the direct report-level stage classification meant it was not possible to detail reasons for the stage classification, which was desirable to interpret errors and build user trust. Traditional topic categorization models a document as a collection of words representing a number of topics. Although this is an appropriate model for tasks such as news report categorization, it does not well fit the current task. A better model of medical reports is a sequence of specific statements relating to different diagnostic factors. With this motivation, the system proposed in this article instead determines the stage indirectly, by first determining the presence or absence of specific staging factors using sentence-level classifiers. The staging protocol, such as shown in ▶, is then applied to assign the most advanced stage associated with a positive finding. As well as potentially improving the accuracy of the eventual stage assignment, decomposing the stage in this way declares reasons behind the decision, linked to the supporting sentences.
Background
This article presents a system to automatically extract cancer stage information using text categorization techniques. Other researchers have previously presented automatic cancer staging algorithms using high-level structured input data coding major diagnostic factors for cervical, ovarian, and prostate cancer. 26–31 Other than these automatic methods, software has been developed for staging with synoptic data entry forms, 32,33 as well as converting between different staging protocols. 33
In previous work, we reported a novel approach to automatic staging by direct report-level classification of the stage from free-text histology reports using SVMs and a bag-of-words representation. 13,14 By using available free-text reports rather than relying on expert coding, the approach allowed for broader applicability than previous staging software, particularly for retrospective data collection and when access to expert knowledge of staging is limited. A review of the literature on medical text categorization has been presented, 13 and is summarized here. Traditionally, text categorization is the task of determining whether a given document belongs to each of a predefined set of classes. 12 Most recent research has concentrated on machine learning approaches, which automatically build classifiers by learning the characteristics of each category from a set of preclassified documents. 12,15 These most commonly use a bag-of-words document representation and SVM classifiers, 16,17 although many other classifiers have been investigated. 12,18–20 Within the medical domain, a number of comparative studies 21,22,25 have demonstrated that SVMs outperform other classifier types across a range of medical text classification tasks.
The system proposed in this article builds on prior work 13,14 by determining the presence or absence of specific staging factors using a two-level sentence classification approach. For each factor from the staging guidelines, sentences are first classified for relevance and then as either a positive or negative finding. There has been little prior work in the text classification literature where the unit of classification is smaller than the entire document; however, related approaches have been proposed for extractive text summarization. Such systems generally include a step in which a subset of important sentences is classified from a document using various features, such as sentence length or location, as well as term frequencies. 34,35 A double classification methodology, in which sentences are first classified as containing relevant information or not, and then terms of interest are classified from within these relevant sentences, has been proposed. 37 In other related work, sentences from Medline abstracts were accurately categorized according to four types using a sentence-level bag-of-words SVM. 36
System Description
Architecture
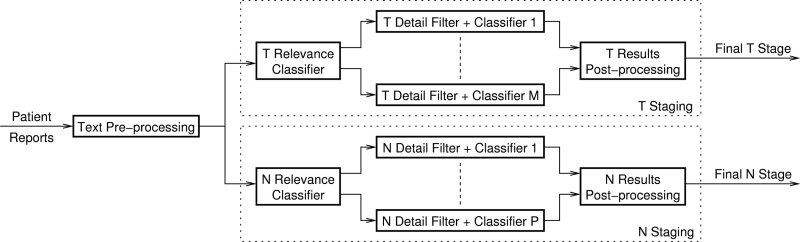
▶ shows the high-level architecture of the proposed system. The system components are described in the following subsections. CSIS is a prototype software system implementing these components in a command-line application, which inputs a list of patients with corresponding free-text report files, and outputs an XML file with the derived staging metadata. More specific implementation details, such as SVM training methods, follow under SVM Implementation in the section on System Development. The system uses a text preprocessing stage to standardize report texts, followed by SVM T and N relevance classifiers that assess the relevance of each report to staging tasks. Sentences from relevant reports are then each analyzed by a series of SVM-based and rule-based classifiers corresponding to specific contributing factors defined in the staging guidelines. Sentence-level classifier results are postprocessed to determine the final T and N stage.
Figure 1.
Proposed system high-level architecture.
Text Preprocessing
The purpose of text preprocessing is to standardize the report texts and to decrease variability by encoding common terms or phrases using a biomedical dictionary, the Unified Medical Language System (UMLS) SPECIALIST Lexicon. 38 The text preprocessing system in the current system is based on that reported by McCowan et al., 13 and consists of four steps: (1) normalization, (2) detection of negation phrases, (3) conversion to UMLS SPECIALIST term codes, and (4) negating relevant terms. The steps implementing normalization and conversion to UMLS SPECIALIST term codes are described in our previous work. 13 As in the prior work, the NegEx algorithm 23,24 was used to detect negation phrases. In the current system, the list of approximately 1,400 terms considered for negation in Step 4 comprised terms occurring in at least five reports in the development data set. Negation phrase codes inserted in Step 2 are removed after they have been applied to surrounding terms. ▶ shows example output of each step.
Table 2.
Table 2 Example Output of Each Text Preprocessing Step ∗
| Step 1 | There | is | no evidence of | lymph node | metastases |
| Step 2 | There | is | _PRENEG_ | lymph node | metastases |
| Step 3 | E0060550 | E0012152 | _PRENEG_ | E0038425 | E0039864 |
| Step 4 | E0060550 | E0012152 | NEG_E0038425 | NEG_E0039864 |
∗ In this case, the original eight-word sentence is mapped into a sequence of four input terms for subsequent classification.
Report Relevance Classification
Pathology reports for lung cancer often contain insufficient macroscopic detail to enable T or N staging. Most reports on small lung biopsy samples, in which the emphasis is on microscopic findings, fall into this category. The purpose of relevance classification is to identify which of a patient’s reports are not useful for T or N staging so they are excluded in subsequent steps. If all reports for a patient are classified as irrelevant to T or N staging, then the patient is automatically assigned a stage of TX or NX, respectively. T and N report relevance classifiers are implemented using SVMs that classify a bag-of-words representation of each report.
Stage Detail Classifiers
The proposed classification strategy (see ▶) uses sentence-level classifiers corresponding to specific factors from the staging guidelines. Examples of factors influencing a T stage assignment are the maximum dimension of the tumor and whether it invades the main bronchus or chest wall. Factors that affect a particular N stage assignment are related to tumor involvement of particular anatomical lymph node groups (e.g., peribronchial, mediastinal, etc.).
The system starts with a default stage of T1/N0 (and thus assumes patients are known to have lung cancer) and upgrades this to the highest stage associated with any of the factors classified as positive across all sentences for that patient. Factors classified as negative are not explicitly taken into account when assigning the final stage.
▶ lists the sentence-level classifiers that were implemented, along with their type. All classifiers use keyword filtering as a first step to eliminate entirely unrelated sentences (e.g., a sentence must contain a dimension for it to be input to the tumor size classifier). Most sentence-level classifiers use a two-level SVM approach, in which a first-level SVM classifies a sentence as being either relevant (i.e., supports either a positive or a negative finding) or irrelevant to the factor in question. A relevant sentence is then classified as supporting a positive or a negative finding by the second-level SVM.
Table 3.
Table 3 List of Sentence-Level Classifiers Used in the Proposed System
| Classifier | Short Name | Stage Association | Classifier Type |
|---|---|---|---|
| Max. tumor dimension ≤ or >3 cm | TS | T2 | 2-level SVM |
| Visceral pleural invasion | VP | T2 | Invasion SVM ∗ |
| Main bronchus invasion | MB | T2 | Invasion SVM ∗ |
| Chest wall invasion | CW | T3 | Invasion SVM ∗ |
| Diaphragm invasion | DIA | T3 | Invasion SVM ∗ |
| Mediastinal pleural invasion | MEDP | T3 | Invasion SVM ∗ |
| Parietal pericardium invasion | PPER | T3 | Invasion SVM ∗ |
| Great vessel invasion | GV | T4 | Invasion SVM ∗ |
| Mediastinum/heart/trachea/esophagus/visceral pericardium invasion | T41 | T4 | Invasion SVM ∗ |
| Vertebral body/carina/vagus nerve invasion | T42 | T4 | Invasion SVM ∗ |
| Separate tumor nodules in same lobe | SEPN | T4 | Key-phrase |
| No nodal involvement | NONM | N0 | Key-phrase |
| Peribronchial lymph node involvement | PLN | N1 | 2-level SVM |
| Hilar lymph node involvement | HLN | N1 | 2-level SVM |
| Mediastinal lymph node involvement | MLN | N2 | 2-level SVM |
| Subcarinal lymph node involvement | SCLN | N2 | 2-level SVM |
∗ Common two-level SVM classifier with postprocessing to determine factor.
Individual two-level SVM classifiers were implemented for tumor size (TS) as well as for each type of lymph node involvement (PLN, HLN, MLN, SCLN; ▶ defines these classifier short names). For staging factors related to invasion of body sites by the primary tumor, a common “invasion” classifier was implemented. Each sentence is preprocessed to convert the UMLS SPECIALIST lexicon term representation of relevant body parts (e.g., visceral pleura, chest wall, etc.) to a common “_BODYPART_” term. A similar transformation is applied to tumor terms (e.g., mass, lesion → _TUMOUR_) and to terms/phrases implying invasion (e.g., involves, extends into → _INVADE_). Each transformed sentence is then input to a common two-level SVM classifier as described above. For sentences classified as positive by the two-level SVM, a rule-based postprocessing step examines the untransformed version of the sentence to discover the particular body part that is undergoing invasion by the primary tumor.
Because of lack of positive examples in development data set, the SEPN (defined in ▶) was implemented as a rule-based classifier that searches for phrases implying the existence of secondary tumor deposits in the same lobe. Similarly, the NONM searches for blanket statements commonly used by reporting pathologists to indicate that no lymph nodes are involved by the cancer. A positive finding from this classifier overrides the other N-stage factor decisions.
System Development
Development Corpus
To train and validate the system, a corpus of deidentified medical reports with corresponding pathological staging data was obtained after research ethics approval. The pathological staging data were obtained from a database 41 collected over the five-year period ending in December 2005. The corresponding medical reports were extracted from a pathology information system. A total of eight cases from the available data sources had pathological stages of T0, TX, Tis, or N3. Automatic classifiers for these stages were therefore not implemented, and cases with those stages were omitted from the corpus. The development corpus statistics are included in ▶.
Table 4.
Table 4 Key Statistics for the Development Data Set
| Data | Cases | Stage Breakdown | Reports | |||
|---|---|---|---|---|---|---|
| Pathology reports + pTNM | 710 | T1 | 204 | NX | 57 | 817 |
| T2 | 405 | N0 | 432 | |||
| T3 | 52 | N1 | 149 | |||
| T4 | 49 | N2 | 72 | |||
Training sets for the report relevance classifiers described above under Report Relevance Classification were derived from the development corpus by annotating each of the reports with a relevant/irrelevant label for both T and N staging.
A separate training set was derived from the development corpus for each of the sentence-level factor classifiers described above under Stage Detail Classifiers by splitting all reports into individual sentences, filtering out irrelevant sentences using the keyword filter for that classifier, and then annotating remaining sentences with one of three labels (irrelevant, −ve finding, +ve finding). Note that direct classification of NX was not done because this result was derived from the N report relevance classifier output.
SVM Implementation
The bag-of-words term weights used for text representation with all SVMs throughout the baseline and proposed systems were calculated according to the LTC weighting scheme. 39 The LTC weighting scheme is commonly used in state-of-the-art text categorization systems because it effectively de-emphasizes common terms (occurring often in many documents), produces normalized weights across different-length documents, and reduces the impact of large differences in frequency through use of the logarithm.
A common training strategy was used with all SVM-based classifiers. A cross-validation approach was used to optimize SVM training parameters and decision threshold and to obtain unbiased classifier output over the entire development training set. The SVMlight 40 toolkit was used for all SVM training and testing. The optimal parameters discovered through cross-validation were used to train a final classifier on all training data. Decision thresholds were selected by cross validation to equalize sensitivity and specificity. No attempt was made to adjust individual classifier decision thresholds to optimize the global T and N staging accuracy.
Development Results
Unbiased classifier outputs from the report relevance classifiers and the sentence-level staging factor classifiers described above were merged to obtain final T and N staging results on the 710-case development set. For T staging, 77.6% correct (95% confidence interval [CI] = 74.3 to 80.6) 1 was obtained for classifying the five T stages (TX, T1 to T4). For N staging, 81.8% correct (95% CI = 78.8 to 84.6) was obtained for classifying four N stages (NX, N0 to N2).
To compare the sentence-level classification with the previous direct report-level approach, a multiclass SVM system was used as a baseline. This approach directly classifies T and N stage from a concatenation of reports for each patient, with the multiclass classification implemented as a hierarchy of binary SVMs, and is fully described. 14 As TX and NX classes were not considered, 14 to allow direct comparison of results, the baseline system was augmented with the T and N report relevance classification stage from the current proposed system. On the same 710-case development set, baseline system performance was 62.8% (95% CI = 59.1 to 66.4) and 77.0% (95% CI = 73.7 to 80.1) correct for T and N staging respectively. This was used as the baseline system in the trial evaluation described in the following sections.
These development results indicate that accuracy has been improved by decomposition into sentence-level staging factor classifiers, as opposed to the more conventional document-level approach that directly classifies final T and N stages.
Status Report
To evaluate the reliability of the proposed system, a trial was conducted as described in the following sections. Some findings from this trial were presented in preliminary form. 42
Trial Objectives
1 To study the level of agreement in expert staging decisions. Subjectivity in the stage decision may arise from inconclusive examinations, varying interpretations of staging criteria, or ambiguity in the way the results are communicated. The first objective of the trial was to quantify the degree of variability between two independent human experts.
2 To evaluate the performance of automatic staging decisions. The second purpose of the trial was to evaluate the performance of the automatic cancer stage assignment, in comparison to a gold standard based on the same input information. For this purpose the gold standard consisted of stage independently assigned in perfect agreement between two human experts. For the few cases in which human experts disagreed, one expert’s decision was selected at random as the gold standard.
3 To evaluate the reliability of classifying key stage factors. Finally, in addition to overall T and N stage assignments, we evaluated how well the system classified specific factors based on key sentences in relation to the human experts.
Method
Input Data
The trial data set consisted of pathology reports for lung cancer cases that were not seen during the development phase, and was extracted from the same pathology information system as the development data set. The trial set consisted of reports for 116 cases that had been assigned a formal pathologic stage in the eight-month period subsequent to December 2005, along with 63 unstaged cases that had a report describing examination of a lung or lobe from a pneumonectomy or lobectomy procedure.
Output Data
Two expert pathologists competent in lung cancer staging were presented with the deidentified reports for the 179 patients. They then independently classified the TNM stage and specific factors (from ▶) for each patient and entered the data into an electronic form. Form validation required the pathologists to enter T and N stages; however, default values were set for all other data fields (M stage of MX, and negative for all other details). A text box was also provided on the form to allow any free-text comments to be entered.
To determine the gold standard TNM stage for system evaluation, after independent data collection from the pathologists, a meeting was convened to discuss cases in which the experts differed in the assigned TNM stage. In this meeting, a consensus TNM stage was assigned by the experts for as many cases as possible. If consensus was not reached for a case, due to ambiguity in the report language or staging guidelines, the two different TNM stages were retained.
System output consisted of the T and N stage, along with the output of the detail classifiers from ▶. To preclude bias, processing of the trial data was performed by technicians independent of the development team investigators so that investigators were blind to the trial data set.
Performance Measures
The following defines the measures used to evaluate results based on the total number of patients (N), along with counts of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) resulting from classification decisions. To evaluate the overall performance of the system for T and N staging, multiclass classification performance is measured using the accuracy,
Agreement between human experts was measured by the kappa statistic, which takes account of agreement occurring by chance
where P(A) is the observed agreement, and
is the agreement expected by chance, where N 1(c) is the number of times annotator 1 selected class c. Binary classification performance is measured using the sensitivity and specificity.
The confusion matrix is a two-dimensional tabulation of frequency counts according to assigned (test) class labels and actual (gold standard) class labels. By highlighting commonly occurring class confusions, the confusion matrix is a useful tool for analyzing multiclass classification systems.
Results
Expert Agreement
The interexpert agreement is shown in ▶ in terms of the kappa statistic and raw percentage agreement for T and N staging on the complete 179-case trial data set. The breakdown of cases by stage is shown in the confusion matrix in ▶.
Table 5.
Table 5 Interexpert Agreement for T and N Staging
| Stage | Kappa | % Agreement (95% CI) |
|---|---|---|
| T | 0.83 | 89.9 (84.3–93.8) |
| N | 0.96 | 97.8 (94.0–99.3) |
Table 6.
Table 6 Confusion Matrices Comparing T and N Stage Assigned by Experts 1 and 2
| Expert 2 |
Expert 2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | NX | N0 | N1 | N2 | ||
| Expert 1 | Expert 1 | ||||||||
| T1 | 49 | 0 | 2 | 0 | NX | 16 | 1 | 0 | 0 |
| T2 | 1 | 94 | 3 | 2 | N0 | 0 | 107 | 1 | 2 |
| T3 | 0 | 3 | 7 | 2 | N1 | 0 | 0 | 35 | 0 |
| T4 | 0 | 5 | 0 | 11 | N2 | 0 | 0 | 0 | 17 |
The confusion matrices show there were 18 T-stage and four N-stage disagreements between the experts. In the subsequent meeting to determine gold standard stage data for system evaluation, as explained in the previous section, the experts were able to reach consensus for 10 of the 18 T-stage cases and all four of the N-stage cases. Two different T-stage assignments were retained for the remaining eight cases.
The interexpert agreement for each of the detailed staging factors is shown in ▶. A Kappa value of N/A (Not Applicable) indicates no instances found by experts (division by zero). Numbers for advanced T stage factors were small, so the significance of results for these factors is not clear.
Table 7.
Table 7 Interexpert Agreement for Detailed Staging Factors ∗
| Expert 1 vs. Expert 2 |
||||||
|---|---|---|---|---|---|---|
| Agree |
Disagree |
|||||
| Stage | Classifier | YY | NN | YN | NY | Kappa |
| T | TS | 96 | 80 | 1 | 2 | 0.97 |
| VP | 63 | 109 | 2 | 5 | 0.92 | |
| MB | 0 | 173 | 6 | 0 | 0.00 | |
| CW | 5 | 171 | 2 | 1 | 0.76 | |
| DIA | 0 | 179 | 0 | 0 | N/A | |
| MEDP | 0 | 176 | 3 | 0 | 0.00 | |
| PPER | 0 | 178 | 0 | 1 | 0.00 | |
| GV | 0 | 179 | 0 | 0 | N/A | |
| T41 | 2 | 170 | 1 | 6 | 0.35 | |
| T42 | 0 | 179 | 0 | 0 | N/A | |
| SEPN | 8 | 161 | 5 | 5 | 0.59 | |
| N | NONM | 76 | 71 | 19 | 13 | 0.64 |
| PLN | 27 | 146 | 6 | 0 | 0.88 | |
| HLN | 23 | 150 | 2 | 4 | 0.87 | |
| MLN | 12 | 165 | 2 | 0 | 0.92 | |
| SCLN | 6 | 172 | 1 | 0 | 0.92 | |
∗ See ▶ for classifier name definitions.
System Performance
Performance was evaluated against the gold standard of human expert stage assignments. As described above, the experts reached consensus on the T stage in only 171 cases. N stage consensus was attained for all 179 cases. For each of the remaining eight cases without T-stage consensus, one of the expert stage decisions was selected at random as the gold standard. Overall T and N stage accuracy with respect to the expert staging for a baseline system that classifies the stage directly from the concatenated reports for each patient, as described under Development Results in the System Description, as well as for the proposed CSIS is shown in ▶. The breakdown of cases by stage is demonstrated in the confusion matrix in ▶.
Table 8.
Table 8 Accuracy of System with Respect to Experts for T and N Stage
| Accuracy % (95% CI) |
|||
|---|---|---|---|
| Stage | Cases | Baseline | Proposed |
| T | 179 | 62.6 (55.0–69.6) | 74.3 (67.1–80.4) |
| N | 179 | 76.5 (69.5–82.4) | 86.6 (80.5–91.1) |
Table 9.
Table 9 Confusion Matrix Comparing T and N Stage Assigned by Experts and the Proposed System 2
| System |
System |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | NX | N0 | N1 | N2 | ||
| Experts | Experts | ||||||||
| T1 | 39 | 10 | 0 | 1 | NX | 10 | 6 | 1 | 0 |
| T2 | 6 | 80 | 2 | 13 | N0 | 2 | 105 | 0 | 1 |
| T3 | 0 | 7 | 2 | 1 | N1 | 0 | 8 | 27 | 1 |
| T4 | 1 | 5 | 0 | 12 | N2 | 0 | 3 | 2 | 13 |
2 Because there were no gold-standard cases, TX results are not reported; however, the system was successful in not inserting any false positive TX cases.
The performance of CSIS on cases with perfect expert agreement 3 for each of the detailed staging factors in terms of sensitivity, specificity, accuracy, and kappa statistic is shown in ▶. Again, the significance of results for advanced T factors is not clear due to low numbers of positive examples.
Table 10.
Table 10 Performance of System for Classifying Detailed Staging Factors
| Experts vs. System |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Agree |
Disagree |
||||||||
| Stage | Classifier | YY | NN | YN | NY | Sensitivity | Specificity | Accuracy | Kappa |
| T | TS | 93 | 67 | 3 | 13 | 0.97 | 0.84 | 0.91 | 0.81 |
| VP | 55 | 96 | 8 | 13 | 0.87 | 0.88 | 0.88 | 0.74 | |
| MB | 0 | 171 | 0 | 2 | 1.00 | 0.99 | 0.99 | 0.00 | |
| CW | 3 | 170 | 2 | 1 | 0.60 | 0.99 | 0.98 | 0.66 | |
| DIA | 0 | 179 | 0 | 0 | 1.00 | 1.00 | 1.00 | N/A | |
| MEDP | 0 | 176 | 0 | 0 | 1.00 | 1.00 | 1.00 | N/A | |
| PPER | 0 | 178 | 0 | 0 | 1.00 | 1.00 | 1.00 | N/A | |
| GV | 0 | 178 | 0 | 1 | 1.00 | 0.99 | 0.99 | 0.00 | |
| T41 | 0 | 170 | 2 | 0 | 0.00 | 1.00 | 0.99 | 0.00 | |
| T42 | 0 | 179 | 0 | 0 | 1.00 | 1.00 | 1.00 | N/A | |
| SEPN | 5 | 148 | 3 | 13 | 0.62 | 0.92 | 0.91 | 0.34 | |
| N | NONM | 61 | 67 | 15 | 4 | 0.80 | 0.94 | 0.87 | 0.74 |
| PLN | 23 | 141 | 4 | 5 | 0.85 | 0.97 | 0.95 | 0.81 | |
| HLN | 20 | 143 | 3 | 7 | 0.87 | 0.95 | 0.94 | 0.77 | |
| MLN | 9 | 162 | 3 | 3 | 0.75 | 0.98 | 0.97 | 0.73 | |
| SCLN | 5 | 171 | 1 | 1 | 0.83 | 0.99 | 0.99 | 0.83 | |
A final point regarding system performance is the incurred processing time. For each report, on a single processor 3 GHz Pentium 4 PC, the report-level baseline system required 1.14 seconds, whereas the sentence-level proposed system required 1.20 seconds. In both cases, the major component was the text preprocessing stage, which required approximately 1 second.
Discussion
Trial Objectives
To Study the Level of Agreement in Expert Staging Decisions
The comparison between the stages assigned by the two experts shows that there is a degree of subjectivity in determining a patient’s T and N stage based purely on the available pathology reports, particularly for the T-stage decision. After initial coding, there were 18 disagreements between Experts 1 and 2 for T staging on the full 179 patient set, and four disagreements for N staging. The confusion matrices in ▶ show that the most common confusions were between T2 and T3, and T2 and T4. These findings broadly correspond with agreement levels found in reviews of registry data. 4–7 After discussion between the two experts, consensus was reached on 10 of these T stages, and all four N stages. The 10 original T-stage disagreements were attributed to six reports with ambiguous language and four interpretation errors. The four original N-stage disagreements were attributed to two data entry errors, one interpretation error, and one report with ambiguous language. The remaining eight T-stage decisions on which no consensus could be reached consisted of four cases in which the staging guidelines are ill-defined for distinguishing a single primary tumor from multifocal tumors (leading to T2M1 and T4M0 stage confusion), and four cases in which the report was imprecise regarding tumor extent (leading to T2/T3 confusion).
To Evaluate the Reliability of Automatic Staging Decisions
CSIS had T-stage accuracy of 74% and N-stage accuracy of 87% on the trial data. This represents an improvement of approximately 10% over the previous baseline system for both T and N staging. These results are similar to those observed on the development data set (see Development Results under System Description). In general, higher accuracy for N stage as compared with T stage mirrors the trend observed in the expert disagreements, and the CSIS confusions predominantly occurred between the same advanced T stages as for the human experts.
To Evaluate the Reliability of Classifying Key Stage Factors
The results in ▶ show that agreement between CSIS and the experts for individual key stage factors also follows the same patterns observed between human experts in ▶. The sentence-level factor classifier results in ▶ explain the reasons for CSIS stage errors. Confusion between T1 and T2 cases (observed in ▶) is due to both false-positive findings for the tumor size (TS) classifier and to the imperfect sensitivity and specificity of the visceral pleural invasion (VPI) classifier. Erroneous T3 and T4 stage classifications are mostly due to the chest wall invasion (CW) and the SEPN (separate tumor nodules in same lobe) classifiers. The lower performance for those factors is consistent with both their rarity and the subjectivity seen in the corresponding expert decisions, as shown in ▶.
Higher accuracy for N-stage sentence-level factor results are likely to reflect the higher prevalence of N-stage factors than T-stage factors in the reports; however, there is substantial agreement between the automatic classifiers and the experts for all N-stage factors. As seen in the confusion matrix in ▶, most of the system-level N-stage errors are false-positive findings of N0. These result from false-negative findings from the lymph node involvement classifiers (HLN, PLN, MLN, and SLN) coupled with false positives from the NONM classifier.
Other Considerations and Limitations
CSIS has been developed and evaluated for T and N staging of lung cancer based on reports from pathological studies of the lung, as proof-of-concept to determine the potential accuracy of an automatic system. There are several issues to be addressed for the system to be generalized to other cancers, or to process other input modalities before deployment in practice.
The current system was developed on a specific data set, and there is a risk that over-fitting may limit broader application. Using more complex natural language processing, richer medical terminologies (SNOMED CT, MetaMap), as well as larger and more varied training data sets may improve the generalization and portability of classifiers to new cancers or reporting modalities.
A practical consideration is the expert time required for training SVM classifiers during system development. This involves annotation of sentences for each staging factor, which was done manually by the development team in the current system. It is estimated that the present lung cancer system involved up to 40 hours of annotation work during development. Although this is not negligible, it must only be done once for each new cancer type, and is therefore not a major concern given eventual productivity gains from automatic stage data collection. Ongoing research is investigating methods for reducing annotation work in several ways, such as by identifying reusable classifiers across different cancers (e.g., tumor dimension, or the common invasion classifier in the present system), analyzing convergence with training set size, and using active learning.
Another practical consideration is the need to automatically discard irrelevant reports. The report relevance stage in the current system discards reports with no information for T or N staging, leading to TX or NX classifications. The system, however, assumes the input reports do relate to lung cancer. This has been achieved in the development and trial data sets by filtering on report metadata (e.g., disease codes, examination type) from the source databases; however, a practical system may require a more general report filtering stage.
Much analysis of cancer outcomes is based on the higher-level group stage, rather than the TNM stage. Because CSIS was developed on pathology reports and M staging is usually determined clinically or by medical imaging, M staging was omitted and CSIS therefore cannot output a proper group stage. Some indication of potential group stage accuracy can be given by assuming a known M stage. For all M0 cases with expert agreement on group stage from the trial, the present system attains an accuracy of 76.7% across Stages I–III (163 cases, Stage IV could not be assessed as it is defined as M1 with any T and N). Future work will investigate adaptability to using additional input sources, e.g., radiology or non-lung pathology reports, to determine M stage.
Conclusions
We developed a prototype software system to automatically determine a patient’s cancer stage from medical reports of lung cancer patients. The system uses SVM classification techniques to classify a range of detailed staging factors at the sentence level, and then combines these into a global stage decision. CSIS was compared against direct report-level classification and against staging by two independent pathology experts. The following conclusions can be made:
1 There is a significant level of disagreement in stage assigned by independent human experts based on pathology reports, particularly for T staging.
2 In comparison with human experts, CSIS achieved overall accuracy of 74% for T staging and 87% for N staging.
3 The two-level sentence classification approach improves on previous direct report-level stage classification by approximately 10% for both T and N staging.
4 The CSIS error pattern mirrors that observed between two independent experts.
The level of accuracy required for practical deployment of such a system would necessarily depend on the use case, and whether it involved a step of human validation. The results achieved do, however, lie within bounds of human staging accuracy observed in studies of registry data. 4–7 A productive avenue of research may be to improve the sensitivity of the N-stage detail classifiers through more sophisticated natural language processing techniques. The limitations with the T staging system mostly reflect uncertainty in the report language, as well as the fact that the stage protocols do not cater to every contingency for more advanced cancer cases, thus leading to subjective interpretations. As well as investigating new classification strategies to improve sensitivity of detail classifiers, ongoing work will focus on addressing these issues for practical deployment of the technology.
Acknowledgments
The authors acknowledge Hazel Harden, Shoni Colquist, and Steven Armstrong from QCCAT; Jaccalyne Brady and Donna Fry from the Queensland Integrated Lung Cancer Outcomes Project; and Wayne Watson from the AUSLAB Support Group.
Footnotes
The research in this article was done in partnership with the Queensland Cancer Control Analysis Team (QCCAT) within Queensland Health.
All 95% confidence intervals reported in this article are calculated using the Wilson procedure.
It was not feasible to resolve disagreements on detailed staging factors in the post-trial consensus meeting.
References
- 1.Australian Cancer Network Management of Lung Cancer Guidelines Working Party Clinical Practice Guidelines for the Prevention, Diagnosis and Management of Lung CancerSydney, Australia: The Cancer Council Australia; 2004.
- 2.Greene FL, Page DL, Fleming ID, et al. AJCC Cancer Staging Manual6th ed. Chicago, IL: Springer; 2002.
- 3.Klassen AC, Curriero F, Kulldorff M, Alberg AJ, Platz EA, Neloms ST. Missing stage and grade in Maryland prostate cancer surveillance data, 1992-1997 Am J Prev Med 2006;30:577-587. [DOI] [PubMed] [Google Scholar]
- 4.Yau J, Chan A, Eapen T, Oirourke K, Eapen L. Accuracy of the oncology patients information system in a regional cancer centre Oncol Rep 2002;9:167-169. [PubMed] [Google Scholar]
- 5.Evans WK, Crook J, Read D, Morriss J, Logan DM. Capturing tumour stage in a cancer information database Cancer Prev Control 1998;2:304-309. [PubMed] [Google Scholar]
- 6.Liu W-L, Kasl S, Flannery JT, Lindo A, Dubrow R. The accuracy of prostate cancer staging in a population-based tumor registry and its impact on the Black-White stage difference (Connecticut, US) Cancer Causes Control 1995;6:425-430. [DOI] [PubMed] [Google Scholar]
- 7.Schouten LJ, Langendijk JA, Jager JJ, van den Brandt PA. Validity of the stage of lung cancer in records of the Maastricht cancer registry, the Netherlands Lung Cancer 1997;17:115-122. [DOI] [PubMed] [Google Scholar]
- 8.Silcocks P, Needham P, Hemsley F. Audit of prostate cancer: validity and feasibility of registry-based staging Public Health 1999;113:157-160. [DOI] [PubMed] [Google Scholar]
- 9.Threlfall T, Wittorff J, Boutdara P, et al. Collection of Population-based Cancer Staging Information in Western Australia—A Feasibility Study Popul Health Metr 2005;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutard P, Platell C, Threlfall T. Model for collecting colorectal cancer staging information in Western Australia ANZ J Surg 2004;74:895-899. [DOI] [PubMed] [Google Scholar]
- 11.Condon JR, Barnes T, Armstrong BK, Selva-Nayagam S, Elwood JM. Stage at diagnosis and cancer survival for indigenous Australians in the Northern Territory Med J Aust 2005;182:277-280. [DOI] [PubMed] [Google Scholar]
- 12.Sebastiani F. Machine learning in automated text categorization ACM Computing Surveys 2002;34:1-47. [Google Scholar]
- 13.McCowan I, Moore D, Fry M-J. Classification of cancer stage from free-text histology reports Proceedings of the IEEE Engineering in Medicine and Biology Conference. New York, NY: IEEE Press; 2006. pp. 5153-5156. [DOI] [PubMed]
- 14.Nguyen A, Moore D, McCowan I, Courage M-J. Multi-class classification of cancer stages from free-text histology reports using support vector machines Proceedings of the IEEE Engineering in Medicine and Biology Conference. Lyon, France: IEEE Press; 2007. pp. 5140-5143. [DOI] [PubMed]
- 15.Aas K, Eikvil L. Text Categorisation: A SurveyTechnical Report. Norwegian Computing Center; 1999.
- 16.Vapnik VN. The Nature of Statistical Learning TheoryNew York, NY: Springer; 1995.
- 17.Joachims T. Text categorization with support vector machines: learning with many relevant features Proceedings of the 10th European Conference on Machine Learning. Heidelberg, Germany: Springer-Verlag; 1998. pp. 137-142.
- 18.Dumais ST, Platt J, Heckerman D, Sahami M. Inductive learning algorithms and representations for text categorization Proceedings of the ACM-CIKM98. New York, NY: ACM Press; 1998. pp. 148-155.
- 19.Lewis DD, Ringuette M. A comparison of two learning algorithms for text categorization Proceedings of the 3rd Annual Symposium on Document Analysis and Information Retrieval. 1994. pp. 81-93Las Vegas, Nevada.
- 20.Yang Y, Liu X. A re-examination of text categorization methods Proceedings of the 22nd International ACM SIGIR Conference on Research and Development in Information Retrieval. New York, NY: ACM Press; 1999. pp. 42-49.
- 21.Aphinyanaphongs Y, Tsamardinos I, Statnikov A, Hardin D, Aliferis CF. Text categorization models for high-quality article retrieval in internal medicine J Am Med Inform Assoc 2005;12:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Bruijn B, Cranney A, O’Donnell S, Martin JD, Forster AJ. Identifying wrist fracture patients with high accuracy by automatic categorization of x-ray reports J Am Med Inform Assoc 2006;13:696-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries J Biomed Inform 2001;34:301-310. [DOI] [PubMed] [Google Scholar]
- 24.Chapman WW, Bridewell W, Hanbury P, Cooper G, Buchanan BG. Evaluation of negation phrases in narrative clinical reports Proceedings of American Medical Informatics Association Symposium. Philadelphia, PA: Hanley & Belfus; 2001. pp. 105-109. [PMC free article] [PubMed]
- 25.Sordo M, Zeng Q. On sample size and classification accuracy: a performance comparison Lecture Notes in Computer Science 2005;3745:193-201. [Google Scholar]
- 26.Phinjaroenphan P, Bevinakoppa S. Automated prognostic tool for cervical cancer patient database Proceedings of International Conference on Intelligent Sensing and Information Processing. Chennai, India: IEEE Press; 2004. pp. 63-66.
- 27.Mitra P, Mitra S, Pal SK. Staging of cervical cancer with soft computing IEEE Trans Biomed Eng 2000;47:934-940. [DOI] [PubMed] [Google Scholar]
- 28.Renz C, Rajapakse JC, Razvi K, Liang SKC. Ovarian cancer classification with missing data Proceedings of the 9th International Conference on Neural Information Processing. Singapore, Singapore: IEEE Press; 2002.
- 29.Tewari A, Narayan P. Novel staging tool for localized prostate cancer: a pilot study using genetic adaptive neural networks J Urol 1998;160:430-436. [DOI] [PubMed] [Google Scholar]
- 30.Han M, Snow PB, Brandt JM, Partin AW. Evaluation of artificial neural networks for the prediction of pathologic stage in prostate carcinoma Cancer 2001;91:1661-1666. [DOI] [PubMed] [Google Scholar]
- 31.Gamito EJ, Stone NN, Batuello JT, Crawford ED. Use of artificial neural networks in the clinical staging of prostate cancer: implications for prostate brachytherapy Techn Urol 2000;6:60-63. [PubMed] [Google Scholar]
- 32.mTuitive 2007. Available at: http://www.mtuitive.com/. Accessed September 24.
- 33.Collaborative Staging 2007. Available at: http://www.cancerstaging.org/cstage/index.html. Accessed September 24.
- 34.Kupiec J, Pedersen J, Chen F. A trainable document summarizer Proceedings of 18th ACM SIGIR Conference. New York, NY: ACM Press; 1995. pp. 68-73.
- 35.Teufel S, Moens M. Sentence extraction as a classification task Proceedings of the ACL’97/EACL’97 Workshop on Intelligent Scalable Text Summarization. 1997. pp. 58-68Madrid, Spain.
- 36.McKnight L, Srinivasan P. Categorization of sentence types in medical abstracts Proceedings of the American Medical Informatics Association Symposium. Philadelphia, PA: Hanley & Belfus; 2003. pp. 440-444. [PMC free article] [PubMed]
- 37.De Sitter A, Daelemans W. Information extraction via double classification Proceedings of the International Workshop on Adaptive Text Extraction and Mining. 2003. pp. 66-73September, Cavtat-Dubrovnik, Croatia.
- 38.NIH Unified Medical Language System (UMLS) 2006. Available at: http://www.nlm.nih.gov/research/umls/. Accessed September 24, 2007.
- 39.Buckley C, Salton G, Allan J, Singhal A. Automatic query expansion using SMART: TREC 3 Proceedings of the 3rd Text Retrieval Conference. Gaithersburg, MD: National Institute of Standards and Technology; 1995. pp. 69-80.
- 40.Joachims T. Making large-scale SVM learning practicalIn: Scholkopf B, Burges C, Smola A, editors. Advances in Kernel Methods—Support Vector Learning. Cambridge, MA: MIT Press; 1999.
- 41.Fong KM, Bowman R, Fielding D, Abraham R, Windsor M, Pratt G. Queensland integrated lung cancer outcomes project (QILCOP): initial accrual and preliminary data from the first 30 months (abstr). Presented at The Thoracic Society of Australia and New Zealand Annual Scientific Meeting, Adelaide, Australia, April 4–9, 2003.
- 42.Moore D, McCowan I, Nguyen A, Fry M-J. Trial evaluation of automatic lung cancer staging from pathology reports Proceedings of 12th International Health (Medical) Informatics Congress (Medinfo). Amsterdam, Netherlands: IOS Press; 2007. pp. 365.