Abstract
The CD1 family of major histocompatibility complex (MHC)-like molecules specializes in presenting lipid and glycolipid antigens to α/β T lymphocytes, but little is known about the size of the CD1-restricted T cell population or the frequency of T lymphocytes specific for a given glycolipid antigen. Here, we report the generation and use of mouse CD1d1–glycolipid tetramers to visualize CD1d-restricted T cells. In contrast with previous BIAcore-based estimates of very short half-lives for CD1d–glycolipid complexes, we found that the dissociation rate of several different CD1d–glycolipid complexes was very slow. Fluorescent tetramers of mouse CD1d1 complexed with α-galactosylceramide (αGalCer), the antigen recognized by mouse Vα14-Jα281/Vβ8 and human Vα24-JαQ/Vβ11 natural killer T (NKT) cell T cell receptors (TCRs), allowed us for the first time to accurately describe, based on TCR specificity, the entire population of NKT cells in vivo and to identify a previously unrecognized population of NK1.1-negative “NKT” cells, which expressed a different pattern of integrins. In contrast, natural killer (NK) cells failed to bind the tetramers either empty or loaded with αGalCer, suggesting the absence of a CD1d-specific, antigen-nonspecific NK receptor. Mouse CD1d1–αGalCer tetramers also stained human NKT cells, indicating that they will be useful for probing a range of mouse and human conditions such as insulin-dependent diabetes mellitus, tumor rejection, and infectious diseases where NKT cells play an important role.
Keywords: T cell development, T cell receptor, Vα14 NKT cell, natural killer cell, antigen presentation
Introduction
CD1 is an MHC-like, but genetically unlinked, family of β2-microglobulin–associated molecules 1 that has recently been shown to present lipid antigens to α/β T cells 2. Human CD1b presents mycobacterial lipids such as mycolic acid, glucose monomycolate, and derivatives of lipoarabinomannan (LAM), whereas mouse and human CD1d bind glycosylceramides 3 4 5 and glycosylphosphatidylinositol (GPI) anchors 6. X-ray crystallography studies indicated that the groove of mouse CD1d is formed of two large hydrophobic pockets seemingly well adapted to bind fatty acid chains 7 and, in conjunction with functional experiments 8, suggested a model whereby the hydrophobic groove of CD1 serves to anchor the fatty acid chains of the lipid antigens, positioning the polar head groups for recognition by the TCR.
The size and diversity of the CD1-restricted T cell population is not fully characterized. Limited numbers of CD1-restricted CD8, CD4, or CD4−CD8− double negative (DN) T cell lines and clones expressing diverse TCRs have been generated in vitro, but the largest populations of CD1-restricted cells currently identified in vivo are the CD1d-restricted, IL-4 and IFN-γ–producing, CD4 and DN NK T (NKT) cells 9. These NKT cells, which represent up to 30% of all T cells in the liver or bone marrow, express semiinvariant Vα14-Jα281/Vβ8 (Vα24-JαQ/Vβ11 in humans) TCRs 10, and they share several general features with subsets of γ/δ T cells and B1 B cells, suggesting that they belong to the innate rather than the adaptive arm of the immune system. Their germline-encoded TCR-α specificity is autoreactive, leading to autoantigen-driven selection and accumulation 11. Similar to mouse VH11/Vκ9 and VH12/Vκ4 B1 B cells, which recognize the membrane lipid phosphatidylcholine, mouse and human NKT cells recognize a glycolipid, α-galactosylceramide (αGalCer). Originally purified from marine sponge extracts on the basis of its antitumor property 12, αGalCer appears to be presented by CD1d to activate both mouse and human NKT cells 3 4 5. Although it is unclear whether αGalCer represents the natural ligand of NKT cells or a cross-reactive antigen, its recognition by virtually all mouse Vα14-Jα281/Vβ8 and human Vα24-JαQ/Vβ11 NKT cells supports the idea that the semiinvariant NKT cell TCRs evolved to recognize a conserved family of glycolipids. NKT cells regulate both mouse and human insulin-dependent diabetes mellitus (IDDM) 13 14, exert protective functions against parasites such as Plasmodium 15 and Toxoplasma gondii 16, against bacteria such as Listeria 17, and against tumors 18. These effects appear to be mediated at least in part through their ability to explosively release cytokines such as IL-4 and IFN-γ 19 20 and to influence the Th1/Th2 outcome of adaptive T cell responses 21 22.
Unambiguous identification of NKT cells has proved difficult, confusing the interpretation of several important observations. Several mouse strains do not express the NKRP1-A allele recognized by the anti-NK1.1 mAb, PK136 23. Even in positive strains such as C57BL/6, NK1.1+ T cells that are neither CD1d-restricted nor Vα14 expressers have been found in various tissues 24 and conversely, CD1d-autoreactive Vα14-Jα281–expressing hybridomas have been derived from NK1.1− subsets 25. Furthermore, there exists a minor population of CD1d-autoreactive, NK1.1+ T cells that expresses non-Vα14 TCRs 10 25. In humans, a combination of anti-Vα24 and anti-Vβ11 mAbs is available 26, but it does not discriminate between JαQ+ (canonical) or JαQ− Vα24/Vβ11 TCRs and misses non–Vβ11-based canonical NKT cells.
The recent discovery that virtually all mouse and human canonical Vα14-Jα281 or Vα24-JαQ NKT cells recognized the glycolipid antigen αGalCer suggested that a similar approach to that recently developed for staining conventional T cells with tetramers of MHC–peptide complexes might be employed to identify the CD1d-restricted T cells 27. However, unlike peptides, which can be genetically linked to MHC molecules, lipid antigens must be bound to CD1d after the production and purification of recombinant CD1d molecules. Therefore, for this approach to be successful, CD1d–glycolipid complexes must be stable, i.e., their dissociation rate should be sufficiently slow.
Based on the ability of soluble recombinant CD1d1 molecules loaded with αGalCer and chased for various periods of time before exposure to IL-2–producing NKT cell hybridomas, we estimated that the half-life of CD1d1–αGalCer complexes was quite long, on the order of 1 d. This result is in sharp contrast with a recent BIAcore-based estimate of a very short half-life between 7 s and 3 min 28. Furthermore, we found that several other lipids bound tightly to CD1d1, suggesting that, like MHC molecules, CD1 molecules have the ability to trap antigens for extended periods. We exploited this property to show that mouse CD1d tetramers loaded with αGalCer could be used to stain both mouse and human cells expressing the canonical αGalCer-specific NKT cell TCRs. Tetramer staining also revealed a previously unrecognized subset of NK1.1− “NKT” cells with a different pattern of expression of α4 integrins. “Empty” or αGalCer-loaded CD1d tetramers failed to stain NK cells, suggesting the absence of a CD1d-specific, antigen-nonspecific NK receptor.
These results demonstrate that CD1d tetramers can be used to probe the glycolipid-specific CD1d-restricted T cell repertoire in vivo. Furthermore, they provide the first accurate identification of NKT and NKT-like CD1d-restricted T cells in vivo in mice and humans, allowing further dissection of their important regulatory functions in conditions such as IDDM, tumor rejection, and infectious diseases.
Materials and Methods
Mice.
C57BL/6 mice were obtained from Taconic Farms. CD1d-deficient mice were generated in our laboratory and used after 7–12 backcrosses to C57BL/6 29. Vα14-Jα281 TCR α chain transgenic mice were generated in our laboratory 10 on a C57BL/6 background and crossed to C57BL/6.TCR Cα2/− (The Jackson Laboratory). Mouse studies followed the Institutional Animal Care and Use Guidelines.
Construction, Expression, and Purification of Mouse Biotinylatable CD1d1 Molecules.
Recombinant soluble CD1d1 molecules were produced in a fly expression system as reported for other class I and class II MHC molecules 30 31. The cDNA coding for the mouse CD1d1 heavy chain 32 was interrupted at the third domain–transmembrane junction by PCR and cloned in frame between the EcoR1 and Bgl2 sites of the pRHMa3/6His inducible fly expression vector. The biotinylation sequence no. 45 of Schatz 33 was added by opening the Bgl2 site and ligating in the linker 5′-CTGGGTGGTATCTTCGAGGCTATGAAGATGGAGATGCGCGAT-3′. The 3′ end of the Bgl2 site was substituted by a GGA TCT sequence, to destroy the site and to encode G S instead of R S residues before the histidine tail. The mutagenesis was confirmed by sequencing. SC2 Drosophila melanogaster cells were transfected by calcium phosphate precipitation with the modified heavy chain and mouse β2-microglobulin cDNAs and a constitutive neomycin resistance gene at a ratio of 60:60:1. After 3 wk of G-418 selection, resistant cell lines were tested for expression after a 3-d induction with 0.7 mM CuSO4. Individual clones were subsequently derived by limiting dilution and selected for high expression. Large scale preparations were carried out in X-press serum-free medium (BioWhittaker) and expanded to 10–12 liters in roller bottles. Supernatants from induced cells were harvested by centrifugation and concentrated by tangential flow down to 400 ml. CD1d1 molecules were affinity purified by Ni-NTA-agarose chromatography from the concentrate. Further purification was carried out by anion exchange chromatography on a MonoQ 10/10 column (Amersham Pharmacia Biotech), and purity was checked by SDS-PAGE.
Biotinylation was done according to the manufacturer's instructions for 16–24 h at room temperature (Avidity). Biotinylation was then quantitated by using HABA colorimetric reagent (Pierce Chemical Co.) and immunoprecipitation with streptavidin-agarose beads (Pierce Chemical Co.). Efficiency was always >90%. Protein concentration was measured by microBCA (Pierce Chemical Co.).
Loading of CD1d1 with αGalCer and Generation of Fluorescent Tetramers of CD1d1–αGalCer for Flow Cytometry.
Soluble biotinylated recombinant CD1d1 molecules were incubated for 18 h with various concentrations of αGalCer diluted in PBS from a 220 μM stock solution in 0.5% Tween 20, and free αGalCer was removed by centrifugation dialysis in a Microcon YM-30 tube (Millipore). Tetramers were generated by mixing αGalCer-loaded monomers with fluorochrome-labeled streptavidin (streptavidin-PE or streptavidin-Cychrome, 5:1 ratio; BD PharMingen) without further separation by gel filtration. Staining was performed by incubating cells on ice for 3 h with tetramers at a concentration equivalent to 20 μg/ml of CD1d1. For staining with monomers, fluorochrome-conjugated streptavidin was added after the cells had been previously incubated for 3 h with 20 μg/ml of biotinylated αGalCer-loaded CD1d1.
Stimulation of Mouse T Cell Hybridomas by Preformed CD1d1/αGalCer Complexes.
The CD1d-restricted DN32.D3 and 431.G5 NKT cell hybridomas expressing canonical Vα14-Jα281/Vβ8 TCRs, and the CD1d-restricted hybridoma 431.A11 expressing Vα3.2-Jα8/Vβ8.2, were generated in our laboratory and described previously 10 34. RF33.70, a Kb/OVA-specific hybridoma was a gift of K. Rock (University of Massachusetts, Worcester, MA). For T cell hybridoma stimulation, purified recombinant mouse CD1d1 molecules were coated for 12–18 h at 2.5 μg/ml in PBS on 96-well plates. After washing with PBS, CD1d1-coated wells were incubated for 2 h with αGalCer (a gift from Dr. Y. Koezuka, Pharmaceutical Research Laboratory, Kirin Brewery Co., Ltd., Takasaki, Japan) diluted into PBS from a 220 μM stock solution made in 0.5% Tween 20 or from a 100 μM stock solution made in 50% DMSO. After extensive washes with PBS, 50,000 hybridoma cells were added in a 1:1 mixture of EHAA and RPMI (Biofluids) supplemented with 10% FCS, 5 × 10−5 M 2-ME, penicillin-streptomycin-gentamicin, glutamine. Supernatants were harvested after 18–24 h to measure IL-2 release using CTLL cells as described.
Lipid Binding to CD1d1.
To test whether other lipids bound CD1d1, we used a modification of the plate-bound CD1d1 hybridoma stimulation assay. Here, CD1d1 was preincubated for 6 h with LAM purified from Mycobacterium tuberculosis (a gift from J. Belisles, Colorado State University, Fort Collins, CO) or ganglioside GM1 (G-7641; Sigma-Aldrich) before washing and incubating for 2 h with αGalCer as described above. After washing, the αGalCer-responsive DN32D3 hybridoma cells were added to the microwells.
Human PBL Stimulation with αGalCer.
PBLs were centrifuged through Ficoll-Paque (Amersham Pharmacia Biotech) and stimulated with 0.1 μM αGalCer at 4 × 105 cells per microwell in RPMI with 10% FCS supplemented with glutamine and antibiotics. After 7 d in culture, blasts cells were recovered by centrifugation through Ficoll and processed for flow cytometry staining.
mAbs and Flow Cytometry.
FITC-conjugated anti–panTCR-β, Vβ8.1/8.2, Vβ7, Vβ2, Vβ6, Ly6C, CD44, CD49d, CD69, CD122, CD4, and CD8, CD5-Cychrome, CD5-allophycocyanin (APC), NK1.1-PE, and streptavidin-Cychrome were obtained from BD PharMingen. FITC- and PE-conjugated anti–human Vα24 and Vβ11 were obtained from Immunotech. FITC-conjugated Y3P anti–I-Ab was prepared in our laboratory. Samples were analyzed using a FACS Vantage™ (Becton Dickinson) equipped with argon and dye lasers or a four-color FACSort™ equipped with argon and 635-nm diode lasers (Becton Dickinson) and CELLQuest™ software.
Generation of A-LAK Cells.
Splenic A-LAK cells were generated after stimulation of nylon wool–passed C57BL/6 splenic cells with 1,000 U IL-2/ml for 5–7 d, as described by Chang et al. 35. Both day 3 adherent and day 3 nonadherent cells that subsequently readhered to plastic were examined.
Results
Long Life Span of CD1d1–Glycolipid Complexes.
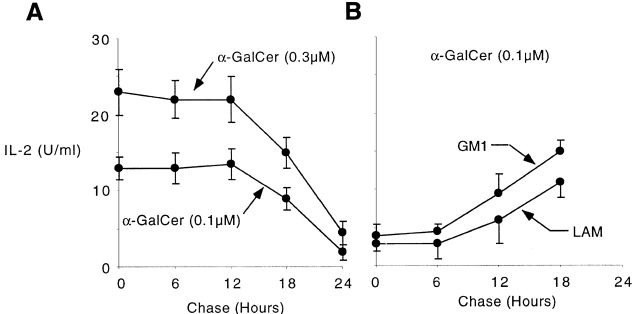
Stable binding of glycolipid antigens to CD1d1 is a prerequisite for tetramer staining of glycolipid-specific T cells. Fig. 1 A shows that plate-bound recombinant CD1d1 was able to present αGalCer to NKT cells (in line with a recent report 28), and that the half-life of CD1d1–αGalCer complexes was remarkably long. We performed chase experiments by washing the wells after loading plate-bound CD1d1 with αGalCer. As a readout assay, we used IL-2 production from the αGalCer-responsive T cell hybridoma DN32D3, which expresses the canonical Vα14-Jα281/Vβ8 TCR, and found that stimulation after loading with limiting concentrations of αGalCer was virtually unchanged 12 h after washing off αGalCer. Given that prolonged engagement of the TCR is needed for optimal IL-2 production by T cells in the readout assay, a half-life well in excess of 12 h can be estimated, a result that is more than two orders of magnitude higher than the previous estimates of 7 s to 3 min derived from the BIAcore studies 28. Experiments using CD1d1-transfected cells pulsed with αGalCer also supported a long life span of the CD1d1–αGalCer complexes, as these cells maintained their stimulatory properties for several days after chase (not shown). However, it is difficult to rule out the possibility of lipid uptake and gradual release from some cellular storage compartment in this system.
Figure 1.
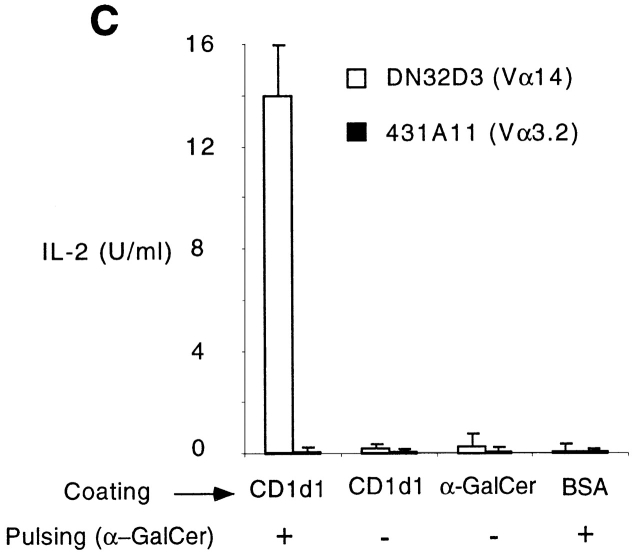
Long life span of CD1d1–glycolipid complexes. (A) Plate-bound CD1d1 (2.5 μg/ml) was loaded with 0.1 or 0.3 μM αGalCer for 2 h, washed, and chased for different periods of time before adding the Vα14-Jα281 NKT cell hybridoma DN32D3 and measuring IL-2 release in the supernatant (mean ± SD). (B) Plate-bound CD1d1 was first incubated with 30 μg/ml of LAM or GM1 for 6 h, washed, and chased for different periods of time. 0.1 μM αGalCer was added for 2 h and washed before incubation with DN32D3. (C) Microwells were coated overnight with 2.5 μg/ml CD1d1 or BSA or 0.1μM αGalCer, washed, then pulsed for 2 h with 0.1 μM αGalCer as indicated, and washed again before adding the CD1d-restricted Vα14 hybridomas DN32D3 or a control CD1d-restricted non-Vα14 hybridoma, 431.A11 (Vα3.2-Jα8/Vβ8), and measuring IL-2 release (mean ± SD). 431A11 and DN32D3 produced similar amounts of IL-2 when stimulated with CD1d1-expressing thymocytes.
The remarkable antigen-trapping property of CD1d1 was not limited to αGalCer. Soluble CD1d1 molecules preincubated with various lipids including a ceramide- and a phosphatidylinositol-based glycolipid (GM1 and LAM, respectively) were refractory to αGalCer binding as judged by inhibition of NKT cell stimulation. Loading of αGalCer was inhibited for extended periods of time (>6 h) after these lipids had been washed off the wells (Fig. 1 B). These competition experiments suggest that, in addition to αGalCer, other ceramide- or phosphatidyl-containing glycolipids also bind tightly, with a slow off rate, to CD1d1.
Fig. 1 C shows that CD1d1 alone did not stimulate DN32D3, suggesting that the natural ligand recognized by this CD1d1-autoreactive hybridoma was not present in the preparation made in Drosophila cells. CD1d1–αGalCer complexes did not stimulate 431.A11, a control CD1d1-restricted Vα3.2-Jα8/Vβ8 hybridoma also derived from thymic NKT cells. Presentation of αGalCer required CD1d1, as uncoated wells or wells coated with BSA were not stimulatory. In addition, several control lipids failed to stimulate NKT cells but nevertheless inhibited presentation of αGalCer to NKT cells (Fig. 1 B, and data not shown), suggesting that they competed with αGalCer for binding to CD1d1.
Staining with Monomers and Tetramers of CD1d1–αGalCer.
We exploited this long half-life of biotinylated CD1d1–αGalCer complexes to stain NKT cells expressing the canonical Vα14-Jα281 TCR α chain. Fig. 2 A shows that CD1d1 monomers that had been loaded overnight with 20 μM αGalCer significantly, albeit weakly, stained a Vα14-Jα281 NKT cell hybridoma. The αGalCer-loaded monomers retained their ability to stain NKT cells for extended periods of time after dialysis (not shown), confirming the very slow rate of dissociation previously measured with the T cell hybridoma assay. As expected, the same concentration of CD1d1–αGalCer complexes in tetrameric form showed much higher level of binding to NKT cells (Fig. 2 B). No staining was observed with empty tetramers, whereas examination of the binding curve of tetramers loaded over a range of concentration of αGalCer (measured via the fluorescence intensity of the T cell hybridoma, Fig. 2 C) revealed evident cooperative effects at ranges between 3 and 10 μM of αGalCer. The Hill coefficient of 4.5 measured in Fig. 2 C is in line with the need for occupancy of all four CD1d1 molecules of the tetramer by αGalCer to confer high avidity to the Vα14-Jα281 TCR.
Figure 2.
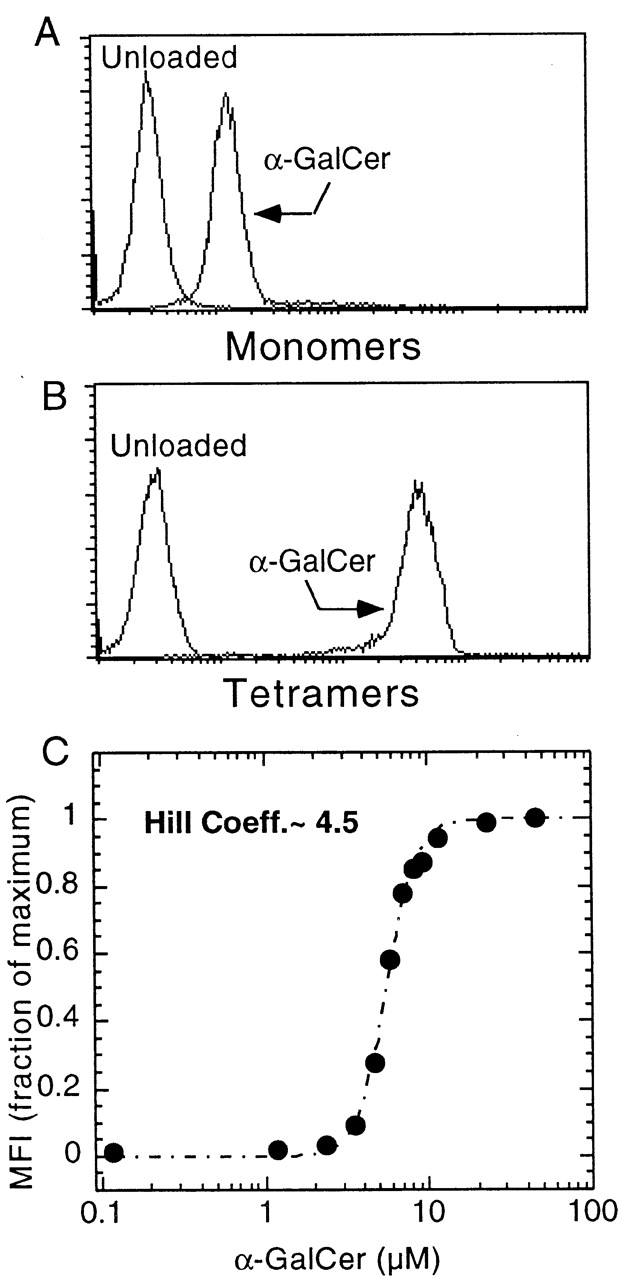
Binding of CD1d1–αGalCer complexes to a Vα14-Jα281/Vβ8 NKT cell hybridoma. (A) Biotinylated monomers, revealed with streptavidin-PE. (B) Streptavidin-PE tetramers. (C) Binding curve of CD1d1 tetramers loaded at different concentrations of αGalCer; binding was measured by flow cytometry analysis (mean fluorescence intensity, MFI). Hill Coeff., Hill coefficient (see Materials and Methods).
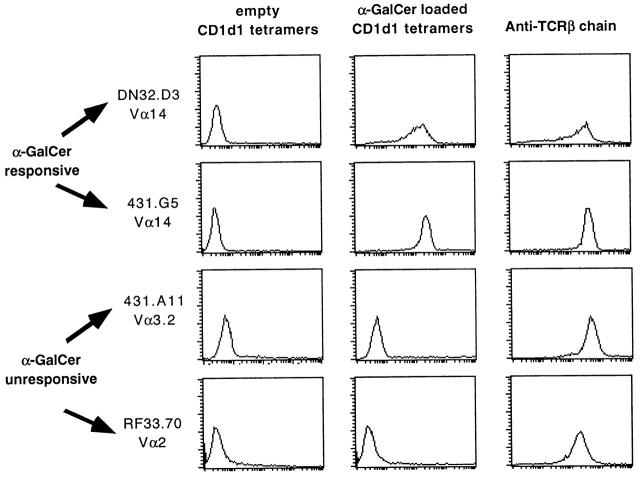
The specificity of CD1d1–αGalCer for canonical CD1d-restricted Vα14-Jα281 TCRs was investigated. Fig. 3 showed that the αGalCer-loaded but not the unloaded tetramers stained two different Vα14-Jα281 hybridomas (DN32D3 and 431G5, expressing different Vβ chains) with a similar profile as an anti–TCR-β antibody. In contrast, the tetramers did not stain a CD1d1-restricted Vα3.2-expressing hybridoma or a Kb-restricted Vα2-expressing hybridoma. Specificity for the TCRs was further demonstrated by blocking with anti-TCR antibodies (see Fig. 7) and by TCR gene transfer (see experiment with Vα14-Jα281 transgenic cells below).
Figure 3.
CD1d1–αGalCer tetramers specifically stained Vα14-Jα281 NKT cell hybridomas. “Empty” or αGalCer-loaded tetramers and anti–TCR-β mAb were used to stain two canonical Vα14-expressing, αGalCer-responsive hybridomas (DN32D3 and 431G5), one CD1d1-restricted αGalCer-unresponsive Vα3.2 hybridoma (431A11), and one Kb-restricted Vα2 hybridoma (RF33.70).
Figure 7.
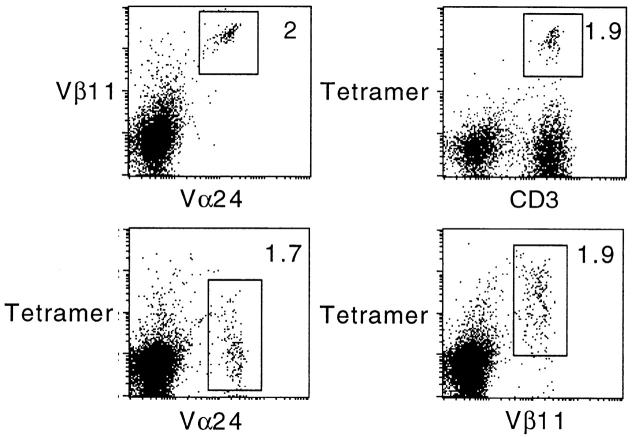
Mouse CD1d1–αGalCer tetramers stain human Vα24/Vβ11 NKT cells. Human PBLs were stimulated with 0.1 μM αGalCer in vitro and stained after 7 d in culture. In this experiment, CD1d1-mediated αGalCer presentation to NKT cells is provided by the PBLs themselves. After centrifugation through Ficoll, the cells were preincubated with anti-TCR antibodies for 1 h before adding tetramer-PE. Note that anti-Vα24 completely blocks tetramer binding.
In Vivo Staining of Fresh NKT Cells.
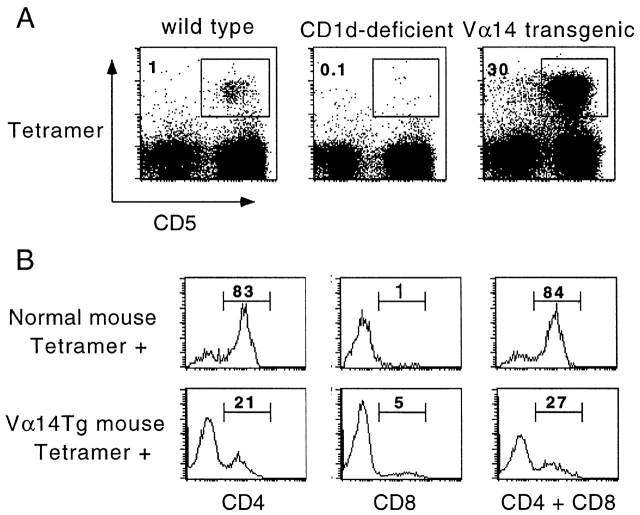
Fig. 4 A shows that CD1d1–αGalCer tetramers stained 1% of the MHC class II–negative population of splenocytes in C57BL/6 mice. These CD1d1–αGalCer tetramer-positive splenocytes corresponded to the CD1d-restricted cells expressing the Vα14-Jα281 canonical TCR α chain by the following criteria. They expressed the T cell marker CD5, they were absent in CD1d-deficient mice, and their frequency increased up to 30-fold in Vα14-Jα281 TCR α chain transgenic mice. In normal mice, most tetramer-positive spleen or liver cells exhibited a CD4+ phenotype and the remainder were DN, with very few CD8 cells. In the Vα14-Jα281 transgenic mice, the ratios were reversed: 73% cells were of the DN phenotype, with only 21% CD4+ and 5% CD8+ (Fig. 4 B).
Figure 4.
CD1d1–αGalCer tetramers stained CD1d1-restricted Vα14-Jα281 NKT cells in vivo. (A) Spleen cells from wild-type, CD1d-deficient, or Vα14-Jα281 TCR α chain transgenic mice in a C57BL/6 background were stained with CD1d1–αGalCer tetramers-PE, CD5-Cychrome, and anti–I-Ab–FITC. Dot plots were gated on MHC class II− splenocytes. (B) Splenocytes from normal mice and Vα14-Jα281 TCR α chain transgenics were stained with tetramer-Cychrome, CD5-APC, and anti-CD4 or CD8-FITC. Histograms display the frequency of CD4+ and/or CD8+ cells among gated CD5+ tetramer+ splenocytes.
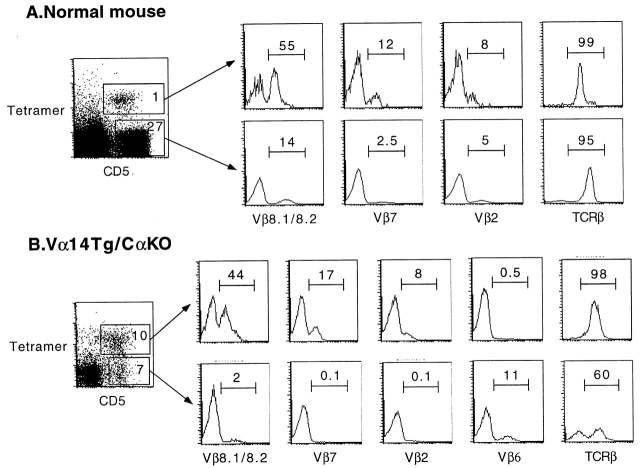
The TCR Vβ repertoire of these cells was next analyzed by flow cytometry. Fig. 5 A shows that, as reported for NKT cells, CD1d1–αGalCer tetramer-positive splenocytes from normal mice were considerably enriched in Vβ8-, Vβ7-, and Vβ2-expressing cells compared with the mainstream T cells that are MHC, rather than CD1d restricted. Moreover, tetramer-positive cells expressed intermediate levels of surface TCR (Fig. 5 A, TCR-β histograms).
Figure 5.
CD1d1–αGalCer tetramer+ cells exhibit intermediate levels of surface TCR and a bias in Vβ usage. Splenocytes from normal mice (top) and Vα14-Jα281 TCR α chain transgenics in a C57BL/6.TCR Cα2/− background (bottom) were stained with tetramer-Cychrome, CD5-APC, and anti-Vβ–FITC. Histograms display the frequency of Vβ1 cells among CD5+ tetramer+ and CD5+ tetramer− splenocytes gated as indicated.
In the Vα14-Jα281 transgenic mice, a subset of CD5+ cells did not bind the CD1d1–αGalCer tetramer (Fig. 4 A, right). As a proportion of these cells was likely composed of T cells expressing endogenous rather than transgenic TCR α chains, we examined Vα14-Jα281 TCR α chain transgenic cells from mice bred to a TCR Cα null background. Fig. 5 B shows that ∼40% of CD5+ cells that solely expressed the Vα14-Jα281 TCR α chain nevertheless remained unstained by the tetramers. Further, in contrast with the tetramer-positive subset, these cells were uniformly NK1.1− (not shown) and did not exhibit an increased frequency of Vβ8, Vβ7, or Vβ2 usage. In fact, they were almost completely depleted of these Vβs while expressing compensatory increases in other Vβs such as Vβ6 (Fig. 5 B). These results demonstrate that virtually all Vβ8, Vβ7, or Vβ2 TCR β chains combined with Vα14-Jα281 have specificity for αGalCer, suggesting that the TCR CDR3β region is permissive for but does not specifically contribute to recognition of CD1d1–αGalCer complexes.
Altogether, these striking results parallel previous observations on TCR-β and CD4/8 expression by natural as well as transgenic Vα14-Jα281 NKT cells, indicating that αGalCer is a very close mimic of the natural ligand of Vα14-Jα281 NKT cells and underscoring the value of CD1d1–αGalCer tetramers as probes for identifying all CD1d1-dependent Vα14-Jα281+ cells.
A Distinct Population of NK1.1− Vα14-Jα281+ NKT-like Cells.
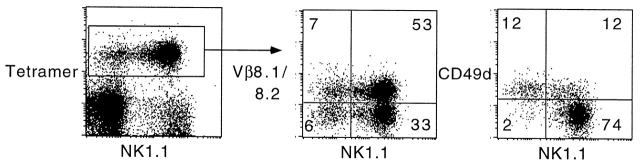
A subset of CD1d1–αGalCer tetramer-positive cells appeared to be NK1.1−, in normal as well as in Vα14-Jα281 transgenic mice. The frequency of these cells varied between 15 and 50% of tetramer-positive cells in different tissues and in individual mice. We studied their phenotype by multiparameter flow cytometry. Like NK1.1+ T cells, they exhibited other features of NKT cells, such as the biased expression of Vβ8 (Fig. 6), Vβ7, and Vβ2, and a CD44hiCD69+CD122+Ly6Chi phenotype (not shown), but they differed strikingly from NK1.1+ cells by their upregulated expression of CD49d (α4 integrins) (Fig. 6). Therefore, these data identify a novel subset of CD1d-restricted Vα14-Jα281+ cells that escaped previous recognition based on NK1.1 staining and may express a number of distinct properties, in particular with respect to migration patterns.
Figure 6.
A subset of CD1d1–αGalCer tetramer+ cells are NK1.1− and express upregulated levels of CD49d. Liver lymphocytes from normal C57BL/6 mice were stained with tetramer-Cychrome, NK1.1-PE, and anti-Vβ–FITC or CD49d-FITC. Note that most tetramer+ NK1.1+ cells were CD49d− whereas tetramer+ NK1.1− cells were CD49d+. Both subsets exhibited the same bias in Vβ8 (middle) and Vβ7 and Vβ2 (not shown). Similar results were found for splenocytes and in Vα14-Jα281 TCR α chain transgenic mice.
Human Vα24-JαQ/Vβ11 NKT Cells Bind to Mouse CD1d1–αGalCer Tetramers.
The well-known conservation of mouse and human CD1d and NKT TCRs was recently extended by reports that αGalCer could be recognized in both species, irrespective of the mouse or human CD1d1 molecule used as the antigen-presenting molecule 4 5. These observations suggested that mouse CD1d1–αGalCer tetramers might also be a useful tool to identify human Vα24-JαQ NKT cells. Fig. 7 shows that 2% of human PBLs expressed Vα24-JαQ/Vβ11 after expansion with one round of stimulation with human CD1d-expressing cells and αGalCer. These cells were brightly stained by mouse CD1d1–αGalCer tetramers, and this staining was specific for the Vα24/Vβ11 cells as shown by costaining with anti-Vβ11. Furthermore, preincubation of the cells with anti-Vα24 inhibited tetramer staining of Vβ11+ cells, formally demonstrating that the CD1d1–αGalCer tetramers bind the TCRs (Fig. 7, bottom). A similar inhibition of tetramer staining could be observed in the mouse system with some but not all anti-Vβ antibodies (not shown), probably depending on their TCR binding site.
Neither Empty Nor αGalCer-loaded CD1d1 Tetramers Stained NK Cells.
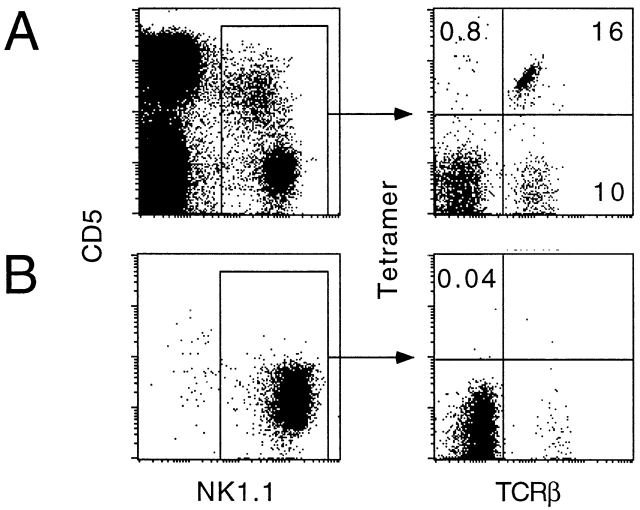
The NK receptors of NK cells can be stained with mono- or multimers of their MHC ligands, sometimes in a peptide-independent manner 36. As CD1d1 was reported to inhibit killing of RMA/s cells by spleen-derived NK cells 35, we examined the NK1.1+ splenic population for staining with CD1d1–αGalCer tetramers. Fig. 8 A shows that only NK1.1+ T cells (TCR-β1) were stained by the CD1d1–αGalCer tetramers, whereas fresh NK cells (TCR-β2) exhibited near background staining level (<1%). Unloaded “empty” tetramers also failed to stain fresh NK cells (not shown). Fig. 8 B shows that A-LAK cells generated from IL-2–activated splenic cultures also failed to bind the CD1d1 tetramers.
Figure 8.
CD1d1–αGalCer tetramers do not stain NK cells. Fresh splenocytes (A) or spleen-derived A-LAK cells (B) were stained with CD5-APC, NK1.1-PE, tetramer-Cychrome, and TCR-β–FITC. Less than 1% of NK cells (α/β sTCR−NK1.1+) were tetramer+.
Thus, these results suggest the absence of antigen-independent CD1d-specific NK receptor. It remains possible, as shown for HLA-E/Qa1–specific CD94/NKG2 receptors 37 38, that NK receptor binding to CD1d1 requires the presence of a particular antigen, or is influenced by the glycosylation of CD1d1, which is altered in molecules produced in insect cells.
Interestingly, Fig. 8 A also showed that a substantial fraction (∼20–40%) of splenic NK1.1+ TCR-β1 cells did not bind the CD1d1–αGalCer tetramers. These cells exhibited a predominantly DN phenotype with no bias in Vβ8 expression (not shown), and likely correspond to the CD1d-independent NKT cells described by others 24.
Discussion
This report demonstrates several important features relevant to CD1d-mediated antigen presentation and recognition by T cells. First, using soluble recombinant mouse CD1d1 to present lipid antigens to CD1d-restricted T cell hybridomas, we found that CD1d1 could tightly bind (i.e., with a slow dissociation rate) several lipids, including αGalCer, the marine sponge antigen recognized by mouse and human NKT cells. These results stand in marked contrast to those recently reported using plasmon resonance to measure the on and off rates of lipid binding to CD1d1. A very rapid off rate with a half-life of CD1d–αGalCer complexes on the order of 7 s to 3 min was measured in these experiments 28, thus underestimating by two to three orders of magnitude the actual life span of these complexes as read out by T cell hybridomas. Although it should be stressed that our studies are based on an indirect functional assay, the findings do indicate that CD1d–glycolipid complexes are stable and the results are more likely to be relevant to normal physiological situations than the markedly different results reported in the BIAcore studies. The discrepancy may be related to several technical issues associated with BIAcore technology and/or the use of a biotinylated form of αGalCer with potentially altered binding to CD1d1.
The remarkable antigen-trapping property of CD1d1 that we found is similar to that of MHC class I and II and consistent with the antigen-presenting function of CD1d1 in that it would enable the antigen-presenting cells to retain, for an extended period of time, the lipid antigens captured in infected tissues. We exploited it to generate monomers and tetramers of biotinylated CD1d1 stably complexed with αGalCer, and found that they specifically bound to NKT cells as revealed by multiparameter flow cytometry. The staining intensity of NKT cells by the preformed CD1d1–αGalCer complexes did not vary even after free αGalCer was removed by dialysis and complexes were chased for up to 6–12 h (not shown), further demonstrating the stability of αGalCer binding to CD1d1. As expected given the avidity gain associated with the tetrameric form of CD1d1, CD1d1–αGalCer tetramers stained NKT cells with ∼50-fold greater fluorescence intensity than the monomers.
CD1d1–αGalCer tetramers exactly identified CD1d-restricted NKT cells expressing the canonical Vα14-Jα281 TCR α chain, including their intermediate level of surface TCR, Vβ bias and CD4/CD8 pattern of expression and, as expected, they also labeled the human NKT cell subset. In addition, they revealed a previously unrecognized NK1.1− “NKT” cell that closely resembled the NKT cell with regards to the expression of canonical TCRs with biased Vβs and of a large panel of phenotypic markers. One notable exception consisted in the upregulated levels of α4 integrins, which are not expressed on NK1.1+ NKT cells. The significance of this novel subset is unclear at present. It could reflect a different functional state of the mature Vα14-Jα281 T cell endowed with different functional or migratory properties. In that regard, experiments conducted in vitro showed that NK1.1 expression is downmodulated after TCR activation (39; and data not shown), strongly suggesting that the NK1.1− NKT cells are activated, migrating cells. An alternative, but not exclusive possibility, is that NK1.1− NKT cells represent a different developmental stage or product of Vα14-Jα281 T cell precursors. It will be interesting to further analyze potential functional differences between NK1.1+ and NK1.1− subsets since they could well be relevant in several conditions, such as IDDM, which is associated with generic defects in NKT cell number or function.
The remarkable identity between the populations defined by CD1d1–αGalCer tetramers and the NKT cell population suggests a high degree of similarity between αGalCer and the putative natural ligand(s) recognized by NKT cells, which extends to the human system. These data conflict somewhat with the recent suggestion 40 that GPI anchors, the prominent endogenous ligands associated with CD1d1, might be the actual antigens recognized by Vα14-Jα281 NKT cells, as GPI and αGalCer exhibit significant structural differences in both their fatty acid and carbohydrate components. Our own recent experiments do not lend support to this hypothesis, because we found that CD1d1-expressing GPI-deficient cell lines were as efficiently recognized by CD1d-autoreactive Vα14-Jα281 NKT cells as their GPI-sufficient counterparts (Bendelac, A., unpublished data). Thus, although the nature of the antigen recognized by Vα14-Jα281 NKT cells remains elusive and controversial, αGalCer certainly represents a near perfect mimic in terms of TCR recognition and thus function.
Our results demonstrate that stable CD1–glycolipid complexes can be generated and used to probe in vivo the glycolipid-specific CD1d-restricted T cell repertoire. The CD1d1–αGalCer tetramers will be useful in dissecting many aspects of the biology of mouse and human NKT cells, including their peculiar development and their important functions in IDDM, tumor rejection, and infectious diseases.
Acknowledgments
We thank Polly Matzinger for reviewing the manuscript, members of the Bendelac laboratory for discussions, and Lisa Antonucci for maintaining our mouse colonies. Dr. Yasuhiko Koezuka (Pharmaceutical Research Laboratory, Kirin Brewery Co., Ltd.) generously provided αGalCer.
This work was supported by grants from the National Institutes of Health (RO1 AI38339 to A. Bendelac and RO1 AI62267 to L. Teyton), The American Cancer Society (IM 788 to A. Bendelac), Juvenile Diabetes Foundation International (to A. Bendelac), and by The Leukemia and Lymphoma Society Fellowship (to K. Benlagha).
Footnotes
Abbreviations used in this paper: αGalCer, α-galactosylceramide; APC, allophycocyanin; DN, CD4−CD8− double negative; GPI, glycosylphosphatidylinositol; IDDM, insulin-dependent diabetes mellitus; LAM, lipoarabinomannan; NKT cell, natural killer T cell.
References
- Calabi F., Jarvis J.M., Martin L.H., Milstein C. Two classes of CD1 genes. Eur. J. Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- Porcelli S.A., Modlin R.L. The CD1 systemantigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu. Rev. Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- Kawano T., Cui J., Koezuka Y., Toura I., Kaneko Y., Motoki K., Ueno H., Nakagawa R., Sato H., Kondo E. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- Brossay L., Chioda M., Burdin N., Koezuka Y., Casorati G., Dellabona P., Kronenberg M. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J. Exp. Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada F.M., Koezuka Y., Porcelli S.A. CD1d-restricted recognition of synthetic glycolipid antigens by human natural killer T cells. J. Exp. Med. 1998;188:1529–1534. doi: 10.1084/jem.188.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce S., Woods A.S., Yewdell J.W., Bennink J.R., Silva A.D.D., Boesteanu A., Balk S.P., Cotter R.J., Brutkiewicz R.R. Natural ligand of mouse CD1d1cellular glycosylphosphatidylinositol. Science. 1998;279:1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- Zeng Z.H., Castano A.R., Segelke B., Stura E.A., Peterson P.A., Wilson I.A. The crystal structure of murine CD1an MHC-like fold with a large hydrophobic antigen binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- Moody D.B., Reinhold B.B., Guy M.R., Beckman E.M., Frederique D.E., Furlong S.T., Ye S., Reinhold V.N., Sieling P.A., Modlin R.L. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- Bendelac A., Lantz O., Quimby M.E., Yewdell J.W., Bennink J.R., Brutkiewicz R.R. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- Lantz O., Bendelac A. An invariant T cell receptor α chain is used by a unique subset of MHC class I–specific CD4+ and CD4−8− T cells in mice and humans. J. Exp. Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A., Rivera M.N., Park S.-H., Roark J.H. Mouse CD1-specific NK1 T cells. Development, specificity, and function. Annu. Rev. Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- Natori T., Morita M., Akimoto K., Koezuka Y. Agelasphins, novel antitumor and immunostimulatory cerebrosides from the sponge Agelas mauritianus . Tetrahedron. 1994;50:2771–2784. [Google Scholar]
- Lehuen A., Lantz O., Beaudoin L., Laloux V., Carnaud C., Bendelac A., Bach J.-F., Monteiro R.C. Overexpression of natural killer T cells protects Vα14-Jα281 transgenic nonobese diabetic mice against diabetes. J. Exp. Med. 1998;188:1831–1839. doi: 10.1084/jem.188.10.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.B., Kemt S.C., Patton K.T., Orban T., Jackson R.A., Exley M., Porcelli S., Schatz D.A., Atkinson M.A., Balk S.P. Extreme Th1 bias of invariant Vα24JαQ T cells in type I diabetes. Nature. 1998;391:177–181. doi: 10.1038/34419. [DOI] [PubMed] [Google Scholar]
- Pied S., Roland J., Louise A., Voegtle D., Soulard V., Mazier D., Cazenave P.A. Liver CD4−CD8− NK1.1+ TCRαβ intermediate cells increase during experimental malaria infection and are able to exhibit inhibitory activity against the parasite liver stage in vitro. J. Immunol. 2000;164:1463–1469. doi: 10.4049/jimmunol.164.3.1463. [DOI] [PubMed] [Google Scholar]
- Denkers E.Y., Scharton-Kersten T., Barbieri S., Caspar P., Sher A. A role for CD4+ NK1.1+ T lymphocytes as major histocompatibility complex class II–independent helper cells in the generation of CD8+ effector function against intracellular infection. J. Exp. Med. 1996;184:131–139. doi: 10.1084/jem.184.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flesch I.E., Wandersee A., Kaufmann S.H. IL-4 secretion by CD4+ NK1+ T cells induces monocyte chemoattractant protein-1 in early listeriosis. J. Immunol. 1997;159:7–10. [PubMed] [Google Scholar]
- Kawano T., Cui J., Koezuka Y., Toura I., Kaneko Y., Sato H., Kondo E., Harada M., Koseki H., Nakayama T. Natural killer-like nonspecific tumor cell lysis mediated by specific ligand-activated Valpha14 NKT cells. Proc. Natl. Acad. Sci. USA. 1998;95:5690–5693. doi: 10.1073/pnas.95.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto T., Paul W.E. CD4pos, NK1.1pos T cells promptly produce IL-4 in response to in vivo challenge with anti-CD3. J. Exp. Med. 1994;179:1285–1295. doi: 10.1084/jem.179.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A., Hunziker R.D., Lantz O. Increased interleukin 4 and immunoglobulin E production in transgenic mice overexpressing NK1 T cells. J. Exp. Med. 1996;184:1285–1293. doi: 10.1084/jem.184.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Hong S., Scherer D.C., Serizawa I., Burdin N., Kronenberg M., Koezuka Y., Van Kaer L. Cutting edgeactivation of NK T cells by CD1d and alpha-galactosylceramide directs conventional T cells to the acquisition of a Th2 phenotype. J. Immunol. 1999;163:2373–2377. [PubMed] [Google Scholar]
- Cui J., Watanabe N., Kawano T., Yamashita M., Kamata T., Shimizu C., Kimura M., Shimizu E., Koike J., Koseki H. Inhibition of T helper cell type 2 cell differentiation and immunoglobulin E response by ligand-activated Vα14 natural killer T cells. J. Exp. Med. 1999;190:783–792. doi: 10.1084/jem.190.6.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama W.M., Seaman W.E. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cellsthe NK gene complex. Annu. Rev. Immunol. 1993;11:613–635. doi: 10.1146/annurev.iy.11.040193.003145. [DOI] [PubMed] [Google Scholar]
- Eberl G., Lees R., Smiley S.T., Taniguchi M., Grusby M.J., MacDonald H.R. Tissue-specific segregation of CD1d-dependent and CD1d-independent NK T cells. J. Immunol. 1999;162:6410–6419. [PubMed] [Google Scholar]
- Chiu Y.H., Jayawardena J., Weiss A., Lee D., Park S.H., Dautry-Varsat A., Bendelac A. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J. Exp. Med. 1999;189:103–110. doi: 10.1084/jem.189.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prussin C., Foster B. TCR Vα24 and Vβ11 coexpression defines a human NK1 T cell analog containing a unique Th0 subpopulation. J. Immunol. 1997;159:5862–5870. [PubMed] [Google Scholar]
- Altman J.D., Moss P.A.H., Goulder P.J.R., Barouch D.H., McHeyzer-Williams M.G., Bell J.I., McMichael A.J., Davis M.M. Phenotypic analysis of antigen-specific T lymphocytes Science. 274 1996. 94 96[published erratum at 280:1821] [DOI] [PubMed] [Google Scholar]
- Naidenko O.V., Maher J.K., Ernst W.A., Sakai T., Modlin R.L., Kronenberg M. Binding and antigen presentation of ceramide-containing glycolipids by soluble mouse and human CD1d molecules. J. Exp. Med. 1999;190:1069–1080. doi: 10.1084/jem.190.8.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Guy-Grand D., Lemonnier F.A., Wang C.R., Bendelac A., Jabri B. Selection and expansion of CD8α/α+ T cell receptor α/β+ intestinal intraepithelial lymphocytes in the absence of both classical major histocompatibility complex class I and nonclassical CD1 molecules. J. Exp. Med. 1999;190:885–890. doi: 10.1084/jem.190.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.A., Garcia K.C., Carbone F.R., Wilson I.A., Teyton L. Role of chain pairing for the production of functional soluble IA major histocompatibility complex class II molecules. J. Exp. Med. 1996;183:2087–2095. doi: 10.1084/jem.183.5.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K.C., Tallquist M.D., Pease L.R., Brunmark A., Scott C.A., Degano M., Stura E.A., Peterson P.A., Wilson I.A., Teyton L. Alphabeta T cell receptor interactions with syngeneic and allogeneic ligandsaffinity measurements and crystallization. Proc. Natl. Acad. Sci. USA. 1997;94:13838–13843. doi: 10.1073/pnas.94.25.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-H., Roark J.H., Bendelac A. Tissue specific recognition of mouse CD1 molecules. J. Immunol. 1998;160:3128–3134. [PubMed] [Google Scholar]
- Schatz P.J. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzymea 13 residue consensus peptide specifies biotinylation in Escherichia coli . Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.S., Brossay L., Kronenberg M., Kane K.P. The murine nonclassical class I major histocompatibility complex–like CD1.1 molecule protects target cells from lymphokine-activated killer cell cytolysis. J. Exp. Med. 1999;189:483–491. doi: 10.1084/jem.189.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K., Boyd L.F., Schuck P., Yokoyama W.M., Eliat D., Margulies D.H. Interaction of the NK cell inhibitory receptor Ly49A with H-2Ddidentification of a site distinct from the TCR site. Immunity. 1999;11:591–601. doi: 10.1016/s1074-7613(00)80134-x. [DOI] [PubMed] [Google Scholar]
- Braud V.M., Allan D.S., O'Callaghan C.A., Soderstrom K., D'Andrea A., Ogg G.S., Lazetic S., Young N.T., Bell J.I., Phillips J.H. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- Vance R.E., Kraft J.R., Altman J.D., Jensen P.E., Raulet D.H. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1b . J. Exp. Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang H., Paul W.E. NK1.1+ CD41 T cells lose NK1.1 expression upon in vitro activation. J. Immunol. 1997;158:5112–5119. [PubMed] [Google Scholar]
- Schofield L., McConville M.J., Hansen D., Campbell A.S., Fraser-Reid B., Grusby M.J., Tachado S.D. CD1d-restricted immunoglobulin G formation to GPI-anchored antigens mediated by NKT cells. Science. 1999;283:225–229. doi: 10.1126/science.283.5399.225. [DOI] [PubMed] [Google Scholar]