Abstract
The role of CD8+ T lymphocytes in controlling replication of live, attenuated simian immunodeficiency virus (SIV) was investigated as part of a vaccine study to examine the correlates of protection in the SIV/rhesus macaque model. Rhesus macaques immunized for >2 yr with nef-deleted SIV (SIVmac239Δnef) and protected from challenge with pathogenic SIVmac251 were treated with anti-CD8 antibody (OKT8F) to deplete CD8+ T cells in vivo. The effects of CD8 depletion on viral load were measured using a novel quantitative assay based on real-time polymerase chain reaction using molecular beacons. This assay allows simultaneous detection of both the vaccine strain and the challenge virus in the same sample, enabling direct quantification of changes in each viral population. Our results show that CD8+ T cells were depleted within 1 h after administration of OKT8F, and were reduced by as much as 99% in the peripheral blood. CD8+ T cell depletion was associated with a 1–2 log increase in SIVmac239Δnef plasma viremia. Control of SIVmac239Δnef replication was temporally associated with the recovery of CD8+ T cells between days 8 and 10. The challenge virus, SIVmac251, was not detectable in either the plasma or lymph nodes after depletion of CD8+ T cells. Overall, our results indicate that CD8+ T cells play an important role in controlling replication of live, attenuated SIV in vivo.
Keywords: simian immunodeficiency virus, live attenuated vaccine, OKT8F, CD8+ T lymphocytes, differential PCR
Introduction
A distinguishing feature of live, attenuated vaccines is the persistence of the vaccine strain in the immunized host. Rhesus macaques immunized with nef-deleted simian immunodeficiency virus (SIVmac239Δnef) have a peak of viremia (105–107 RNA copies/ml) in the first few weeks after infection, and virus often remains detectable at low levels for years in peripheral blood and LNs. In most cases, virus replication is well controlled and infected animals remain clinically healthy. However, in some instances, infection with live, attenuated SIV leads to high levels of viremia and disease progression in both infant and adult macaques 1 2 3 4, raising questions as to the safety of these candidate vaccines. Although vigorous immune responses are induced after immunization with live, attenuated SIV (for a review, see reference 5), the mechanisms involved in controlling virus replication in the host are unknown.
Despite persistence of the vaccine strain, rhesus macaques immunized with live, attenuated SIV are protected from pathogenic infection with wild-type SIV 3 6 7 8 9 10 11 12 13 14. Because of the robust protection provided by this vaccine model, immune responses induced as a result of immunization have been investigated as possible correlates of protection (for a review, see reference 5). Several studies have demonstrated the presence of antibodies against SIV proteins in immunized macaques 3 12 15 16 17 18 19. However, no correlation has been found between protection from pathogenic SIV infection and the ability of anti-SIV antibodies to neutralize the challenge strain 3 12 13 17 18, nor could protection be conferred by the transfer of immune serum from protected macaques 20. Induction of SIV-specific CTLs has also been documented in peripheral blood 21 22 and tissues 23 of rhesus macaques immunized with live, attenuated SIV, suggesting that cellular immune responses play a key role in protection.
Several lines of evidence support the concept that CTLs are critical in controlling virus replication. In vitro assessments of CTL activity have demonstrated a temporal relationship between the induction of CTLs and suppression of HIV-1 24, and SIV 25 26 replication during primary infection. Low viral loads in HIV-1–infected long-term nonprogressors 27 28 and in rhesus macaques infected with live, attenuated SIV 22 have been associated with high levels of virus-specific CTL activity. Recently, cellular immune responses have been shown to directly control replication of wild-type SIV 29 30 and simian/human immunodeficiency virus (SHIV) 31 in vivo during acute and chronic infection.
Whether CD8+ T lymphocytes are involved in controlling replication of SIVmac239Δnef in vivo and whether these cells play a direct role in mediating protection from pathogenic SIV challenge are not known. Studies aimed at in vivo depletion of CD8+ T cells are often complicated by difficulties in affecting complete and sustained depletion of cells from the periphery and LNs 32. Moreover, in the setting of vaccine studies, in vivo depletion of CD8+ T cells often necessitates the use of quantitative assays capable of discriminating between the vaccine and challenge strains.
In this study, we investigated the effect of in vivo depletion of CD8+ T cells on virus replication in rhesus macaques immunized with SIVmac239Δnef. A total of six adult macaques were studied: three infected with SIVmac239Δnef, and three immunized with SIVmac239Δnef and challenged with pathogenic SIVmac251. To distinguish between the vaccine strain (SIVmac239Δnef) and the challenge virus (SIVmac251), a novel quantitative assay was developed based on the use of real-time PCR and molecular beacons 33 34 35. Our results indicate that CD8+ T cells play an important role in controlling replication of SIVmac239Δnef in immunized macaques.
Materials and Methods
Immunization of Rhesus Macaques.
Six adult rhesus macaques (Macaca mulatta) were immunized with SIVmac239Δnef (provided by Dr. Ronald Desrosiers, New England Regional Primate Center, Harvard Medical School, Southborough, MA) by intravenous administration of 4 × 103 50% tissue culture infective doses (TCID50) as described previously 3 (Table ). All six macaques became infected on the basis of detection of SIVmac239Δnef RNA in plasma 3. Three of these animals were then intravenously challenged at 15 or 25 wk after immunization with 10 animal infectious doses (AID) of uncloned SIVmac251 (provided by Dr. Desrosiers). The immunized and challenged macaques were monitored for the presence of SIVmac239Δnef and SIVmac251 by nested DNA PCR for 2 yr. During this time, there was no evidence of the challenge virus in PBMC samples from the peripheral blood (Metzner, K.J., unpublished data), suggesting that the macaques were protected from challenge.
Table 1.
Baseline Characteristics of Rhesus Macaques before CD8+ T Cell Depletion
| Lymphocyte count | SIVmac239Δnef viral load | ||||||
|---|---|---|---|---|---|---|---|
| Macaque | Immunized | Challenged | CD4+ T cells | CD8+ T cells | Chiron bDNA plasma | Real-time PCR plasma | Real-time PCR PBMC |
| cells/mm3 | cells/mm3 | RNA copies/ml | RNA copies/ml | copies/106 PBMCs | |||
| 1496/AT41 | SIVmac239Δnef | SIVmac251 | 581 ± 193 | 1,317 ± 254 | <104 | 1.1 × 103 | <50 |
| 1502/AT44 | SIVmac239Δnef | SIVmac251 | 1,408 ± 218 | 895 ± 145 | <104 | 2.5 × 103 | <50 |
| 1506/AT45 | SIVmac239Δnef | SIVmac251 | 664 ± 141 | 519 ± 118 | <104 | 1.3 × 103 | <50 |
| 1518/AT49 | SIVmac239Δnef | – | 692 ± 199 | 738 ± 153 | <104 | 0.2 × 103 | <50 |
| 1520/AT50 | SIVmac239Δnef | – | 989 ± 97 | 1,709 ± 427 | <104 | 1.8 × 103 | <50 |
| 1522/AT51 | SIVmac239Δnef | – | 695 ± 98 | 1,288 ± 412 | <104 | 6.1 × 103 | <50 |
Depletion of CD8+ T Cells In Vivo.
CD8+ T cells were depleted in vivo by administration of the anti-CD8 antibody, OKT8F (provided by K. Demarest and T. Mercolino, The R.W. Johnson Pharmaceutical Research Institute, Raritan, NJ). Four macaques received OKT8F (1496, 1502, 1518, and 1520), whereas the remaining two macaques (1506 and 1522) were given an isotype-matched control antibody, P1.17 (TIB-10; American Type Culture Collection). All animal protocols were approved by the Institutional Animal Care and Use Committee at the Tulane Regional Primate Research Center.
Macaques were anesthetized with 10 mg/kg ketamine-HCl, then either OKT8F or P1.17 were given intravenously at 2 mg/kg/d for three consecutive days. 3–5 ml of peripheral blood were obtained frequently before and after antibody administration. Inguinal and axillary LN biopsies were obtained at baseline (day −1) and on days 7 and 14 after antibody administration. Each node was sectioned into triplicate samples and stored frozen at −80°C until processing.
Phenotypic Analysis of Cell Subsets.
Four-color flow cytometric analyses of whole blood samples were performed to monitor changes in T cell populations. In brief, 100 μl aliquots of whole blood were incubated with respective antibodies for 30 min at 4°C, then washed twice with PBS containing 2% FCS. The RBCs were lysed with FACS™ Lysing Solution (Becton Dickinson) and washed twice with PBS, followed by further washes and resuspension in 2% formaldehyde (Tousimis Research Corporation) before analysis on a FACSCalibur™ flow cytometer (Becton Dickinson). The antibodies used in the study were RhCD3-FITC (Immunotech), CD3-PE (BD PharMingen), CD4-allophycocyanin (APC) (Exalpha Biologicals, Inc.), CD8-peridinine chlorophyll protein (PerCP) (Becton Dickinson), CD16-FITC (Becton Dickinson), CD20-PE (Becton Dickinson), and control antibodies IgG1-FITC, IgG1-PE, IgG1-PerCP, and IgG1-APC (Caltag). The results were analyzed using CELLQuest™ software by first gating on small lymphocytes, then displaying the subset of cells on a two-dimensional density plot. The number of CD4+ and CD8+ T lymphocytes was determined using the Becton Dickinson TruCount™ method. The total number of T cells was determined (CD4 and CD8) and the percentage of lymphocyte subsets in the blood was assessed by staining PBMCs with directly conjugated antibodies, followed by FACS® analysis. The number of cells in each subset, e.g., CD16+ cells, was then calculated using the following formula: the number of CD16+ cells = (% CD16+ cells/% CD3+ cells) × the number of CD3+ cells.
Extraction of RNA and DNA from Plasma, PBMCs, and LNs.
Plasma samples collected from blood in EDTA anticoagulant were centrifuged at 2,000 g for 5 min to pellet cell debris. SIV RNA was purified from cell-free plasma (500 μl) using the QIAamp Viral RNA Mini kit (Qiagen) according to the manufacturer's instructions for large sample volumes. Genomic DNA was extracted from 1–2 × 106 PBMCs using the QIAamp DNA Mini kit (Qiagen) following the manufacturer's protocol with the exception that 60 μl of buffer was used to elute the DNA. To extract RNA from LNs, tissues were thawed, disrupted with a scalpel, and homogenized using a QIAshredder (Qiagen). RNA was isolated by using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions.
Differential Quantification of SIVmac239Δnef and SIVmac251 Viral and Proviral Load.
Reverse transcription (RT) with viral RNA from plasma or LN samples was performed in MicroAmp optical 96-well reaction plates (PE Biosystems). Viral RNA (10 μl) was added to 0.7 μM of a gene-specific primer recognizing either SIVmac239Δnef (5′-CTTCCAGTCCC-3′, nucleotide [nt] 9467; 36) or SIVmac251 (5′-CTCATCTATATCATCC-3′, nt 9352; 36), and incubated at 80°C for 5 min to dissolve secondary structures. The plate was then immediately placed on ice for at least 2 min. RT reactions consisted of PCR buffer II (PE Biosystems), 3 mM MgCl2, 1 mM dNTPs (GIBCO BRL), 10 mM dithiothreitol, 20 U RNase inhibitor (Roche Molecular Biochemicals), and 40 U SuperScript II RNase H− reverse transcriptase (GIBCO BRL) in a final volume of 30 μl. The reactions were incubated at 42°C for 50 min followed by inactivation at 70°C for 15 min.
Amplification of SIVmac239Δnef cDNA was performed using a hairpin-shaped PCR primer that hybridizes across the junction of the nef deletion (see Fig. 1 A). Use of this primer promotes independent amplification of SIVmac239Δnef regardless of the presence of SIVmac251 in the same plasma sample. A molecular beacon was used for detection of amplicons in real-time PCR. PCR reactions for amplification of SIVmac239Δnef consisted of 30 μl of cDNA products, 1× ROX-PCR buffer, 3.5 mM MgCl2, 0.4 μM upstream primer nef1 5′-GGGAGACTCTTAGGAGAGGTGGAAGATGG-3′ (nt 9161; 36), 0.4 μM hairpin-shaped downstream primer hs Δnef 5′-GGGACAGCCTTTTCTTTTATAAAATGAGACCTGTCCC-3′ (nt 9449; 36) (complementary arm sequences that form the stem are underlined), 0.4 μM molecular beacon mb Δnef FAM-5′-CACTC CCCAGGAGGATTAGACAAGGGAGTG-3′-DABCYL (nt 9204; 36), and 2.5 U AmpliTaq Gold DNA polymerase (PE Biosystems) in a final volume of 50 μl.
Figure 1.
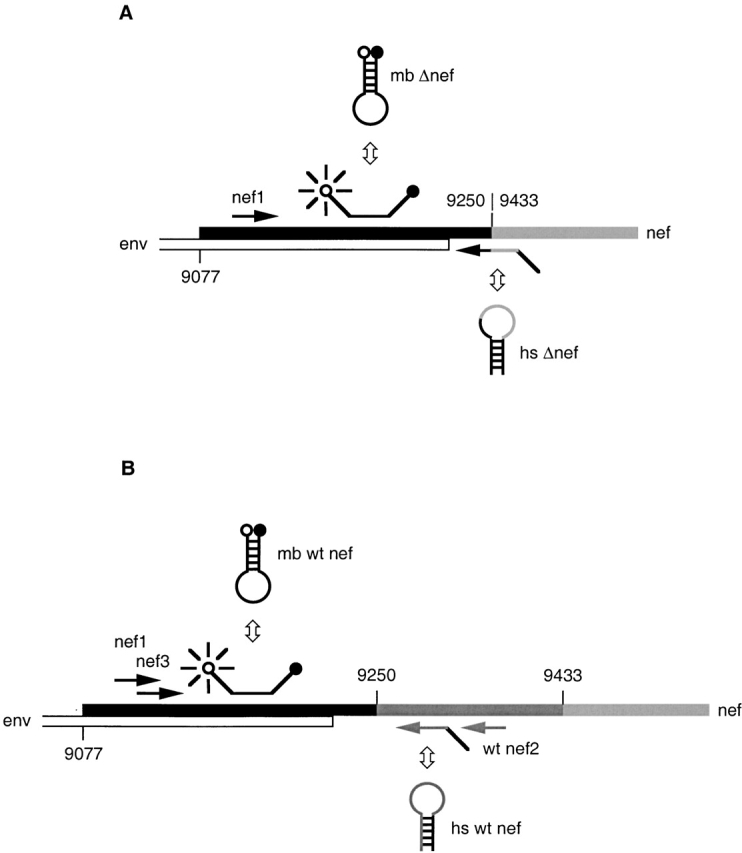
Scheme of differential amplification. (A) Amplification of SIVmac239Δnef. A linearized upstream primer (nef1) and hairpin-shaped primer (hs Δnef) were used to amplify a 106-bp amplicon. hs Δnef hybridizes across the junction of the deletion in nef, allowing amplification of SIVmac239Δnef in the presence of SIVmac251. A molecular beacon (mb Δnef) was used for detection of amplicons. (B) Amplification of SIVmac251. A linearized primer (nef1) and hairpin-shaped oligonucleotide (hs wt nef) were used to amplify a fragment of 142 bp. An additional PCR was performed using linearized oligonucleotides (nef3 and wt nef2), which amplify a fragment of 157 bp. Amplicons were detected using the molecular beacon (mb wt nef).
For amplification of SIVmac251 cDNA, a wild-type, nef-specific, hairpin-shaped downstream primer was used, which hybridizes to the deleted region of SIVmac239Δnef (Fig. 1 B) and allows amplification of SIVmac251 independently of SIVmac239Δnef in the same plasma sample. PCR reactions for amplification of SIVmac251 cDNA contained the reagents described above, with the substitution of 0.4 μM wild-type hairpin-shaped downstream primer hs wt nef 5′-CTCCATGGTCTTCAGCTGGGTTTCTCCATGGAG-3′ (nt 9303; 36), and 0.4 μM molecular beacon mb wt nef FAM-5′-CACTCCCTAGGAGGATTAGACAAG GGAGTG-3′-DABCYL (nt 9204; 36).
60 cycles of amplification (94°C for 15 s, 60°C for 30 s, and 72°C for 30 s) were performed in a 7700 Prism spectrofluorometric thermal cycler (PE Biosystems). In cases of undetectable SIVmac251, an additional real-time amplification was performed using 0.4 μM upstream primer nef3 5′-TAGGAGAGGTGGAAGATGGATMCTCGC-3′ (nt 9171; 36), 0.4 μM downstream primer wt nef2 5′-GCTAATTTTTCTCTCTCTTCAGCTGGG-3′ (nt 9318; 36), and 0.4 μM molecular beacon mb wt nef. Proviral SIVmac239Δnef and SIVmac251 DNA were measured by real-time PCR as described above using 5 μl genomic DNA extracted from PBMCs.
Duplicates of in vitro RNA transcripts or DNA quantified by OD260 measurements were used as standards for each experiment and were serially diluted from 107 to 20 or 10 copies per reaction, respectively. Viral RNA or genomic DNA samples were also tested in duplicate. As a control for potential DNA contamination of viral RNA, an additional reaction was tested in real-time PCR without the addition of reverse transcriptase. Copy numbers were calculated by interpolation of the experimentally determined threshold cycle for the test specimen onto the control standard regression curve 37. Both real-time RT-PCR assays have a detection limit of 50 SIV RNA copies/mL with a linear dynamic range of >5 logs. RNA copy numbers in LNs were normalized per 1 μg of total RNA.
Proviral DNA copy numbers were calculated per 106 genomic equivalents measured by real-time amplification of the single-copy CC chemokine receptor 5 (CCR5) gene (Kostrikis, L.G., and D.D. Ho, unpublished data). In brief, PCR reactions consisting of 5 μl genomic DNA, 1× ROX-PCR buffer, 3.5 mM MgCl2, 1 mM dNTPs (GIBCO BRL), 0.4 μM upstream primer rhesus (rh) CCR5 5′-GAGAAGGTCTTCATTACACCTGC-3′ (nt 750; sequence data available from EMBL/GenBank/DDBJ under accession no. MMU77672), 0.4 μM downstream primer rh CCR5 d 5′-GATTCCCGAGTAGCAGATGACC-3′ (nt 889), 0.4 μM molecular beacon mb rh CCR5 FAM-5′-CCGGTCTGAAAATTCTTCCAGAATTGATACTGACCGG-3′-DABCYL (nt 819), and 2.5 U AmpliTaq Gold DNA polymerase (PE Biosystems) were performed in a final volume of 50 μl.
Results
The six macaques described in this study were previously immunized with a live, attenuated strain of SIV (SIVmac239Δnef). Three of the six were subsequently challenged with pathogenic SIVmac251 (1496, 1502, and 1506), and all animals were monitored for SIV RNA in plasma and for changes in CD4+ and CD8+ T cell counts over a follow-up period of >2 yr. At the initiation of the current study, all macaques were clinically healthy, with CD4+ and CD8+ T cell counts in the normal range and undetectable plasma viremia as determined by the bDNA assay (<104 SIV RNA copies/ml of plasma; Chiron Corporation 38) (Table ).
To evaluate changes in viral load as a result of in vivo CD8+ T cell depletion, a novel assay was first developed to allow simultaneous quantification of both the vaccine strain (SIVmac239Δnef) and the challenge virus (SIVmac251) in the same sample. This assay is based on real-time PCR using molecular beacons and primers hybridizing within and flanking the 182-bp deleted region of nef, and allows for discrimination of nef-deleted and wild-type SIV nef genes (Fig. 1A and Fig. B). Initial evaluation of standard SIV RNA preparations defined a dynamic range of >5 logs for both nef-deleted and wild-type RNA when analyzed separately.
The discriminatory ability of the assays was tested in reciprocal mixing experiments by adding to a serial dilution (108 to 10 copies) of either SIVmac239Δnef or SIVmac251 RNA or DNA standard, 108 copies of noncomplimentary RNA or DNA. Amplifications were performed and results were compared with threshold cycles from PCR reactions performed without the addition of noncomplimentary viral RNA or DNA. Results from these experiments indicate that the assay for amplification of SIVmac239Δnef has a discriminatory ability of 1 copy of SIVmac239Δnef/106 copies of SIVmac251, whereas the amplification of SIVmac251 has a discriminatory ability of 1/107 (Fig. 2A and Fig. B).
Figure 2.
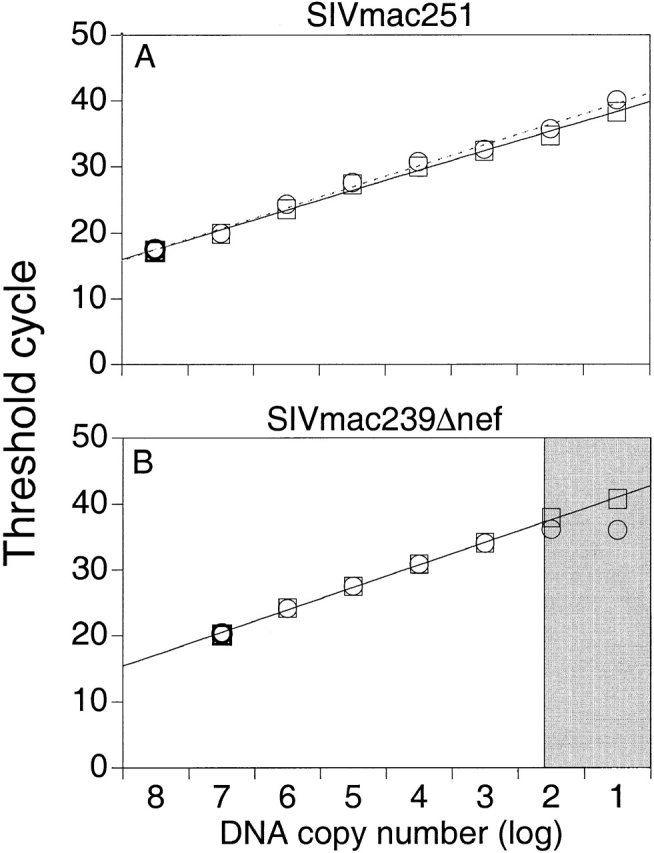
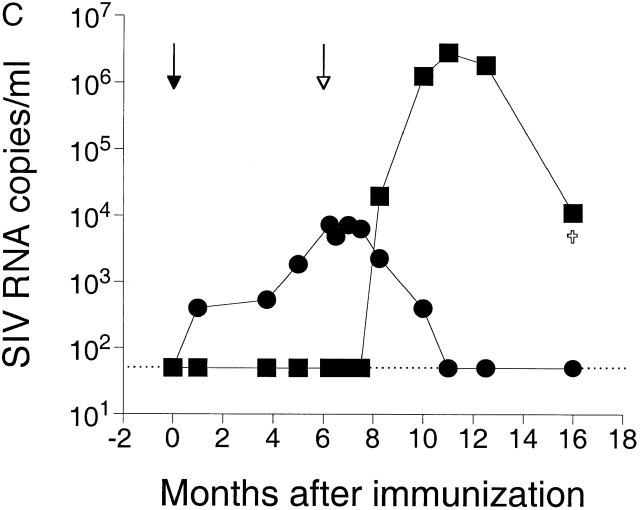
(A) Discriminatory ability of real-time PCR for differential amplification. Amplifications of SIVmac251 were performed using a serial dilution of SIVmac251 DNA standard with (○) and without (□) the addition of 108 copies of noncomplimentary SIVmac239Δnef DNA standard. Mean values of reactions performed in duplicate are shown. The dotted and solid lines indicate the linear standard curves as results of amplifications with and without the addition of noncomplimentary DNA, respectively. (B) Amplifications of SIVmac239Δnef were performed using a serial dilution of SIVmac239Δnef DNA standard with (○) and without (□) the addition of 108 copies of noncomplimentary SIVmac251 DNA standard. The linear standard curve for amplification of SIVmac239Δnef DNA alone is shown in black. The gray section indicates the area where the ratio of copies of SIVmac239Δnef to copies of SIVmac251 is ≥1/106. (C) Measurement of SIV RNA in plasma after immunization with SIVmac239Δnef and challenge with SIVmac251. Rhesus macaque 1510 was immunized with SIVmac239Δnef on day 0 (black arrow) and then challenged with SIVmac251 at 25 wk (white arrow). SIVmac239Δnef RNA (•) and SIVmac251 RNA (▪) in plasma were quantified using real-time PCR for differential amplification. The limit of sensitivity of the viral load assay is indicated by the dotted line.
Longitudinal plasma samples obtained from a rhesus macaque immunized with SIVmac239Δnef but not protected from SIVmac251 challenge were then tested to determine the sensitivity and discriminatory ability of the assay under more biologically relevant conditions. The results demonstrate detection of both SIVmac239Δnef and SIVmac251 in the same plasma sample with a sensitivity of ≥50 SIV RNA copies/ml, and document a decline in SIVmac239Δnef replication that occurred temporally with an increase in replication of SIVmac251 (Fig. 2 C).
In further experiments, plasma samples from an unrelated rhesus macaque infected with wild-type SIV were also quantified in parallel using either real-time PCR or the Chiron bDNA assay, and the results were found to be highly similar (within threefold; data not shown), providing a basis for comparison with other quantitative SIV studies, including our previous report 3.
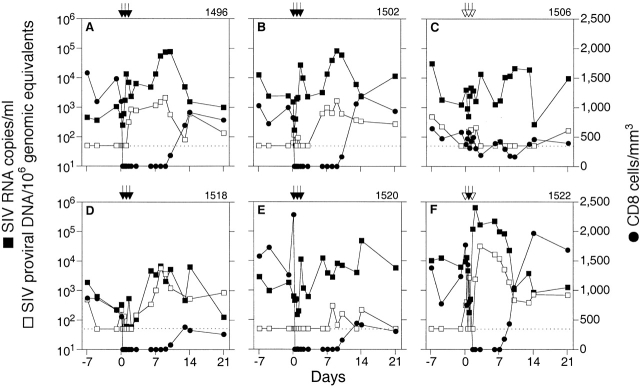
Using the real-time PCR assay to monitor changes in viral load, the anti-CD8 antibody OKT8F was administered to 4/6 rhesus macaques (1496, 1502, 1518, and 1520) at a concentration of 2 mg/kg/d for three consecutive days. The two remaining macaques (1506 and 1522) received an isotype-matched control antibody, P1.17. Blood samples taken at frequent intervals before and after administration of the antibodies were assayed to monitor changes in CD8+ T cells and viral load (Fig. 3). Within 1 h after administration of the first dose of antibody, CD8+ T cells decreased in the peripheral blood by an average of 99% (from a mean of 1,151 ± 428 to 0 cells/ml) in the four macaques receiving OKT8F, whereas the CD8+ T cell counts remained stable in the two macaques receiving the control antibody (1506 and 1522). In error, OKT8F was given at the second injection to one of the control macaques (1522), resulting in a similar, rapid depletion of CD8+ T cells (Fig. 3 F). To evaluate the effect of a single dose of OKT8F, the control antibody was then administered to macaque 1522 for the third injection. For the four macaques receiving three doses of OKT8F, the number of CD8+ T cells remained low for 8–9 d, and began to recover in all animals by day 10. A slightly more rapid recovery of CD8+ T cells, occurring on day 8, was observed for the macaque receiving one dose of OKT8F, suggesting a less sustained depletion.
Figure 3.
Changes in viral load after in vivo depletion of CD8+ T cells. SIV RNA in plasma and SIV proviral DNA in PBMCs were determined for each sample in duplicate by real-time PCR and differential amplification. SIV proviral DNA data were normalized per 106 genomic equivalents measured of the single-copy gene CCR5. Data are shown for CD8+ T cells (•), SIVmac239Δnef RNA (▪), and SIVmac239Δnef proviral DNA (□). SIVmac251 RNA or DNA were not detectable. Administrations of OKT8F (black arrows) or the control antibody P1.17 (white arrows) were performed on days 0, 1, and 2. The limit of sensitivity of the viral load assay is indicated by the dotted line.
Concomitant with the decrease in peripheral blood CD8+ T cells was a dramatic increase in the levels of SIVmac239Δnef in plasma, which occurred in 5/5 macaques receiving OKT8F antibody. Viral loads at baseline were low in all immunized macaques (0.22–6.1 × 103 SIV RNA copies/ml), but increased by a minimum of 1–2 logs after CD8+ T cell depletion. Viremia reached a mean peak value of 1.7 × 105 SIV RNA copies/ml (0.067–6.4 × 105 copies/ml) between days 1 and 8 after antibody administration. Moreover, the decrease in SIVmac239Δnef viremia was temporally associated with recovery of the CD8+ T cell population in all macaques. Although fluctuations in both CD8+ T cells and viral load occurred in the one macaque receiving three doses of the control antibody, there was no clear temporal association between these changes and administration of the antibody (Fig. 3 C), similar to what has been observed using the same control antibody in rhesus macaques chronically infected with SIV 29.
SIVmac239Δnef proviral DNA at baseline was low or undetectable (<50 copies/106 genomic equivalents) in rhesus macaques receiving OKT8F and increased by 0.6–2 logs after administration of OKT8F. Interestingly, the increase in SIVmac239Δnef proviral DNA occurred 1–3 d after the increase of viral RNA in plasma. Peak values for proviral DNA ranged from 3 × 102 to 3.1 × 104 copies/106 genomic equivalents. A decrease of proviral DNA was temporally associated with the increase of CD8+ T cells. Changes in SIVmac239Δnef proviral load in rhesus macaque 1506 were not associated with administration of the control antibody (Fig. 3 C).
Simultaneous quantification of SIVmac239Δnef and SIVmac251 was carried out using plasma and PBMC samples from the three macaques that were immunized and challenged (1496, 1502, and 1506; Table ). In an earlier study using conventional DNA PCR, we were unable to detect wild-type nef sequences in PBMCs from these macaques over a period of 2 yr after challenge (Connor, R., unpublished data). Before depletion of CD8+ T cells in vivo, baseline samples were assayed by real-time PCR and SIVmac251 RNA was not detected. After administration of either the anti-CD8 (1496, 1502) or control antibody (1506), SIVmac251 remained undetectable in plasma and PBMCs for the duration of CD8+ T cell depletion (data not shown).
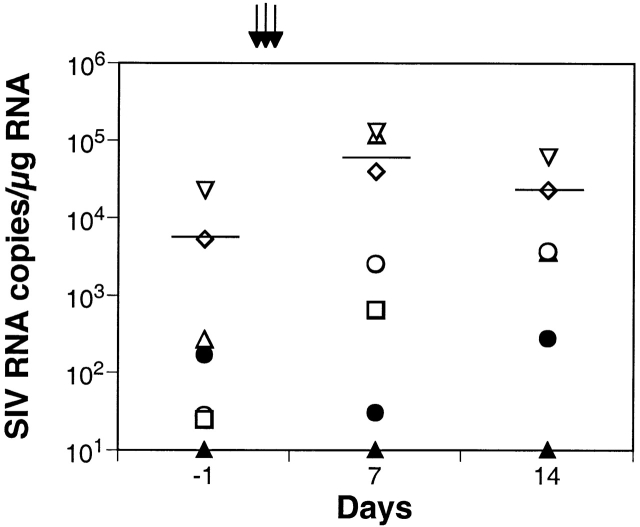
We could not rule out the possibility of low level replication of SIVmac251 in tissues such as LNs, which may have been below threshold levels of detection in the peripheral blood. To investigate this possibility, LN biopsies collected at baseline (day −1) and on days 7 and 14 after antibody administration were assayed for the presence of SIVmac239Δnef and SIVmac251 RNA. Our results indicate that SIVmac239Δnef RNA in the LNs at baseline ranged from 2.3 × 101 to 2.1 × 104 RNA copies/1 μg total RNA (mean 5.5 × 103 SIV RNA copies/1 μg total RNA) among the six immunized macaques (Fig. 4). On day 7 after OKT8F administration, SIVmac239Δnef increased by 0.6–3 logs (mean 5.6 × 104 RNA copies/1 μg total RNA). In three rhesus macaques, viral loads decreased at day 14 (mean 2.2 × 104 RNA copies/1 μg total RNA), but were still higher than baseline. Viral RNA values in LN samples from rhesus macaque 1518 were 2.5 × 103 at day 7 and 3.6 × 103 RNA copies/1 μg total RNA at day 14. The macaque receiving control antibody (1506) had no significant change in viral RNA in LNs (Fig. 4). RNA from the challenge virus, SIVmac251, was below the limit of detection (<10 RNA copies/1 μg total RNA) in all LN samples (Fig. 4).
Figure 4.
SIVmac239Δnef RNA in LNs. SIV copy number was determined for duplicate samples by real-time PCR and differential amplification, and the data were normalized per 1 μg of total RNA. The mean of duplicates as well as the geometric mean are shown for all monkeys receiving OKT8F (black arrows): 1496 (⋄), 1502 (▵), 1518 (○), 1520 (□), or one dose of OKT8F: 1522 (▿). Data from 1506 receiving three doses of the control antibody P1.17 are shown (•). SIVmac251 RNA was below the limit of detection in all LN samples (▴).
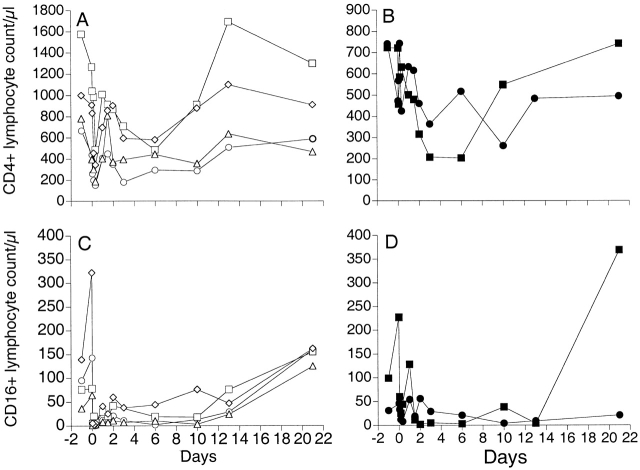
Our previous work has shown that the depletion of CD8+ T cells is accompanied by a reduction in CD4+ T cells in all animals receiving OKT8F antibody 29, suggesting that OKT8F may lead to nonspecific depletion of other cell subsets in the lymphoid system. In this study, in addition to confirming the previously observed depletion of CD4+ T cells (see Fig. 6A and Fig. B), we also examined changes in B cells expressing the CD20 marker and lymphocytes expressing the CD16 surface antigen in the SIVmac239Δnef-immunized macaques (Fig. 5). Our findings demonstrate no consistent changes in the CD20+ lymphocyte population as a result of administration of OKT8F antibody, although an increase in CD20+ cells occurred in four macaques after the first injection but was not sustained (data not shown).
Figure 6.
The reduction in total number of CD4+ T cells and NK cells after administration of OKT8F antibody. Variation in the number of CD4+ T cells (A and B) and CD16+ lymphocytes (C and D) over time in four macaques receiving OKT8F antibody (1496 [○], 1502 [□], 1518 [▵], and 1520 [⋄]) and the two macaques receiving the control antibody P1.17 (1506 [•] and 1522 [▪]). Only macaque 1506 received three doses of P1.17.
Figure 5.
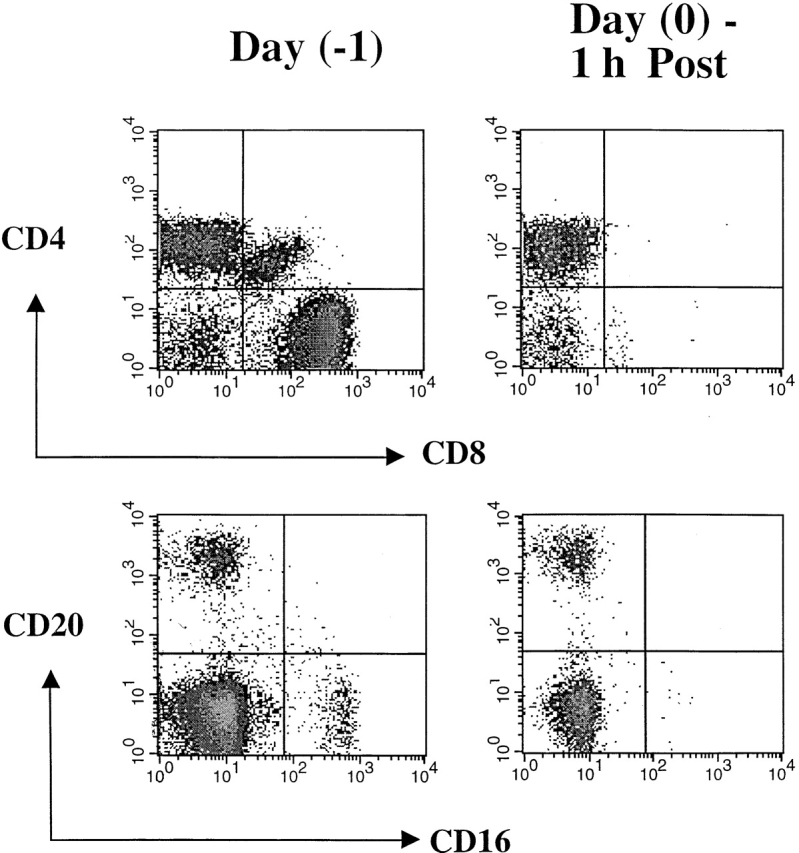
Phenotypic changes in lymphocyte subsets before and after administration of OKT8F. The impact of OKT8F antibody on various cell subsets was examined by FACS® as described in Materials and Methods. As early as 1 h after antibody injection, 99% of the CD8+ T cells were depleted and 89% of CD16+ NK cells were also depleted.
When changes in CD16+ cells were followed longitudinally in all macaques, we observed a partial depletion of cell numbers immediately after administration of OKT8F, with variable recovery of the cells over the next 2–21 d (Fig. 6C and Fig. D). Interestingly, similar decreases in CD16+ lymphocytes were also observed after administration of the control antibody, P1.17, to control macaque 1506 (Fig. 6 D), suggesting a nonspecific effect of the antibodies on CD16+ cells.
Evaluating more closely the rate of recovery of CD8+ T cells beginning 7–9 d after challenge, we found that the macaques segregate into two groups: those with a rapid recovery (group A) and those with a twofold slower recovery rate (group B) (Table ). We expressed the rate of CD8+ T cell recovery mathematically using the logistic growth law 39, which states that the rate of recovery depends on the T cell count and slows as the T cell count reaches some maximum, denoted T max. The equation for logistic growth is: dT/dt = gT (1 − T/T max), where T is the T cell count, g is the growth or recovery rate, and T max is the carrying capacity or maximum population size. Using nonlinear least squares to fit the CD8+ T cell recovery data to this equation, we found that the two challenged macaques, 1496 and 1502, as well as 1522 had growth rates g = 6 d−1, whereas the two other unchallenged macaques had growth rates of 3 d−1.
Table 2.
Kinetics of CD8+ T Cells and Viral Loads in the Rhesus Macaques Treated with Anti-CD8 Antibody
| Macaque | g | t dRNA | t 1/2 | t dDNA | Group | Challenged |
|---|---|---|---|---|---|---|
| d−1 | d | d | d | |||
| 1496 | 6.09 | 0.72 | 0.86 | 2.67 | A | Yes |
| 1502 | 5.98 | 0.64 | 0.95 | 2.67 | A | Yes |
| 1518 | 3.03 | 1.31 | 1.24 | 1.44 | B | No |
| 1520 | 2.98 | 3.47 | 2.31 | <8.66 | B | No |
| 1522 | 5.85 | 0.39 | 0.57 | 0.38 | A | No |
Analysis of t dRNA, the doubling time of SIVmac239Δnef in plasma in days, revealed that the group A macaques had a mean t dRNA of 0.58 d, whereas the group B macaques had a mean t dRNA of 2.4 d (Table ). t 1/2, the half-life of SIVmac239Δnef during the period of decline from the peak, also differed between the two groups. For group A, these values were 0.57, 0.86, and 0.95 d (mean = 0.79 d), whereas for group B the half-lives were 1.24 and 2.31 d (mean = 1.8 d). Linear regression was done to estimate the slope of SIV proviral DNA increase over the same period that the viral RNA increased (Table ). In the challenged macaques, SIV proviral DNA increased four times slower than plasma SIV RNA, whereas in the unchallenged macaques (1518 and 1522) the rates of RNA and DNA increase were essentially identical; in 1520, the rate appeared to be 2.5 times slower than the rate of RNA increase. However, the proviral DNA measurements for 1520 remained below the limit of detection for 4 d after anti-CD8 antibody administration and thus may have been increasing faster.
Discussion
In this study, rhesus macaques immunized with a live, attenuated strain of SIV (SIVmac239Δnef) were treated with an anti-CD8 mAb (OKT8F) to evaluate the effects of CD8+ T cell depletion on viral replication in vivo. Of the five macaques receiving OKT8F antibody, 5/5 had a rapid and sustained decline in CD8+ T cells in the peripheral blood within 1 h after antibody administration. CD8+ T cells remained low for a period of 8–10 d during which time SIVmac239Δnef viremia increased by 1–2 logs in all animals. Control of SIVmac239Δnef replication was temporally associated with the reappearance of CD8+ T cells in the periphery. The challenge virus, SIVmac251, was not detected in either the peripheral blood or LNs after CD8+ T cell depletion, providing strong evidence that these immunized macaques were protected from pathogenic infection. These results provide the first direct evidence that CD8+ T cells play an important role in controlling replication of live, attenuated SIV in a vaccine setting and support the conclusions of several recent studies demonstrating the effects of CD8+ T cell depletion during chronic 29 and primary 30 SIV infection.
In an earlier study, we characterized SIVmac239Δnef replication during the acute and chronic phases of infection in 20 rhesus macaques 3. In all animals, plasma viremia increased in the first few weeks after infection, reaching peak values of 105–107 RNA copies/ml, and then declined to undetectable levels (<104 copies/ml by bDNA). In the majority of infected macaques, plasma viremia remained undetectable during a follow-up period of >2 yr. Here, we examined in detail the dynamics of SIVmac239Δnef replication in six of these animals after in vivo depletion of CD8+ T cells. We found that CD8+ T cell depletion results in a dramatic rise in SIVmac239Δnef plasma viremia to levels approaching those found during acute infection, suggesting that CD8+ T cells constitute a primary immune mechanism for control of SIVmac239Δnef replication in vivo.
Several possible mechanisms may contribute to the control of virus replication mediated by CD8+ T cells. These include direct lysis of virus-infected target cells by SIV-specific CTLs 5 and the production of soluble factors that suppress SIV infection 40 41. High levels of SIV-specific CTLs have been demonstrated in rhesus macaques chronically infected with live, attenuated SIV 22 42. In a recent study, CTLs against SIV Gag and Env antigens developed within 2–4 wk after infection with SIVmac239Δnef and persisted for up to 6 yr, even in macaques with relatively low viral loads 22. Many of the macaques in our study had low to undetectable viral loads in plasma and PBMCs, yet showed a dramatic rebound in viremia after CD8+ T cell depletion, suggesting that even minimal virus replication may be sufficient to provide antigenic stimulation and sustain potent antiviral cellular immune responses. Recent studies have also shown that SIV replication can be inhibited in vitro by soluble factors secreted by CD8+ T lymphocytes from rhesus macaques immunized with live, attenuated SIV 40 41. Depletion of CD8+ T cells in vivo might abrogate these suppressive effects and increase plasma viremia either by extending the life span of productively infected cells or, alternatively, by removing the source of inhibitory factors thereby allowing increased production of virions.
The role of CD8+ T cells in mediating protection against pathogenic SIV challenge in macaques immunized with live, attenuated SIV vaccines is less clear. Earlier studies failed to break protection after administering anti-CD8 antibodies in vivo 32; however, interpretation of these results is complicated by difficulties in achieving potent and sustained depletion of CD8+ T cells in the peripheral blood and tissues. In rhesus macaques immunized with SIVmac239Δnef and challenged with SIVmac251, we were unable to detect the challenge virus in peripheral blood or LNs upon CD8+ T cell depletion. It is unlikely that these results can be explained solely on the basis of incomplete CD8+ T cell depletion, as we were able to document a rapid and pronounced effect on replication of SIVmac239Δnef. Rather, failure to detect the challenge virus may reflect eradication from the host or, alternatively, the effects of other immune mechanism(s) that contribute to controlling replication of wild-type SIV. With respect to the latter, antibodies against SIV Gag and Env antigens have been documented after immunization with SIVmac239Δnef 3 10 and are maintained at high titers throughout infection. In earlier studies, we observed increasing titers of anti-SIV antibodies in macaques immunized with SIVmac239Δnef, but failed to find a correlation between protection from SIVmac251 challenge and the presence of antibodies capable of neutralizing the challenge virus in vitro 3. Whereas our current results do not preclude the possibility that anti-SIV antibodies play a role in mediating protection from pathogenic SIV challenge, they do indicate that CD8+ T cells are the primary mechanism of control of live, attenuated SIV replication. To determine whether CD8+ T cells are also involved in mediating protection from pathogenic SIV infection, it would be necessary to perform CD8+ T cell depletion immediately before challenge.
Similar to our previous study, we found that depletion of CD8+ T cells was accompanied by reduction in the CD4+ T cells and CD16+ cells in all animals receiving antibody injections 29. One explanation for this reduction is that OKT8F or P1.17 injection may act as a large dose of foreign antigen, activating numerous cell types including CD4+ T cells to later undergo activation-induced apoptosis. In agreement with our findings, previous studies have shown that treatment with OKT8F results in lowering of B cells and CD4+ T cells in rhesus macaques 43. Moreover, injection of OKT3 has also been shown to deplete non-T cells such as B cells and NK cells in humans 44. Although some NK cells express an α/α heterodimer of the CD8 molecule and may therefore be depleted by OKT8F, the extent of CD16+ lymphocyte depletion cannot be accounted for solely by this cross-reactivity. Nor does this explain the decline in CD16+ lymphocytes observed in macaques receiving the control antibody. These results indicate that some degree of nonspecific depletion occurred as a result of antibody injection, although the precise mechanism of this nonspecific depletion is unknown. It is interesting to note that not all anti-CD8 mAbs induce depletion of other cell populations. In a recent report by Schmitz et al. 30, intravenous administration of the CD8-specific mouse–human chimeric mAb, cM-T807, to rhesus macaques resulted in near total depletion of CD8-bearing lymphocytes while the CD4+ population remained unchanged 30. One notable difference between the OKT8F mAb used in this study and cM-T807 is the humanized portion of the antibody molecule. Because OKT8F is a murine mAb, nonspecific depletion of other cell subsets may be due to Fc-mediated interactions. This is supported by the observation that administration of an isotype-matched control antibody (P1.17) also resulted in nonspecific cell depletion. This effect may be minimized in rhesus macaques by the presence of a humanized Fc domain.
We were particularly interested in the difference in CD8+ T cell responses in the unchallenged and challenged macaques. The two challenged macaques, 1496 and 1502, and the unchallenged macaque 1522 (group A) showed a more rapid recovery of CD8+ T cells and a shorter half-life of virus during the period of decline from peak compared with the two unchallenged rhesus macaques, 1518 and 1520 (group B), suggesting that the group A macaques had a more vigorous CD8+ T cell response. Moreover, group B macaques did not experience a decline in SIVmac239Δnef until about day 14, even though the CD8+ T cells had recovered and reached close to their maximum value by day 14. Thus, the CD8+ T cells in the group B macaques appeared less effective at controlling viremia than those of the group A macaques. Consistent with this, we also observed that the loss of CD8+ T cells in group B did not lead to as rapid an increase in viremia as in group A macaques.
Possible reasons for these differences might be that the challenge with SIVmac251 boosted the SIV-specific CD8+ T cell response in macaques 1496 and 1502. Therefore, CD8+ T cell depletion in these macaques resulted in a more decisive loss of inhibitory mechanisms responsible for controlling of virus replication and vice versa during the recovery of CD8+ T cells. The higher viral load at baseline in the unchallenged animal 1522 might also have stimulated a more vigorous anti-SIV CD8+ T cell response.
Concerns over safety have long been a focal point of discussions on live, attenuated SIV vaccines. Failure to control replication of highly attenuated strains of SIV in apparently healthy adult and neonatal macaques has been documented in several studies, including our own 1 2 3, raising doubts as to the feasibility of pursuing this approach in humans. While somewhat anecdotal in nature, these examples suggest an inability of the immune system to effectively contain ongoing viral replication. Our results indicate that CD8+ T cells provide a primary means of control over replication of SIVmac239Δnef in vivo, and raise the possibility that pathogenic infection with live, attenuated SIV may be associated with the inability to mount an effective cellular immune response against the virus or, alternatively, may represent escape from CTL control due to mutations in immunodominant epitopes 21 45 46 47. These findings underscore the importance of cellular immunity in the control of SIV replication in vivo and emphasize the need to induce a strong antiviral cellular response as a component of any candidate HIV-1 vaccine strategy.
Acknowledgments
We thank D. Nixon and L. Chakrabarti for helpful discussions; J. Blanchard, R. Bohm, M. Ratterree, and R. Rockar for animal care; W. Chen for graphics; K. Demarest, T. Mercolino (both from The R.W. Johnson Pharmaceutical Research Institute), and J. Safrit for assistance in obtaining OKT8F; and P. Dailey and J. Booth (from Bayer Reference Testing Laboratory) for SIV bDNA quantification.
M. Di Mascio is a Fellow from the Istituto di Ricerche Farmacologiche Mario Negri, Milan, Italy. This study was supported by National Institutes of Health grants RR06555 and AI43868, by the Deutsche Forschungsgemeinschaft, and by the Irene Diamond Fund.
Footnotes
Abbreviations used in this paper: AID, animal infectious dose; CCR5, CC chemokine receptor 5; nt, nucleotide(s); RT, reverse transcription; SIV, simian immunodeficiency virus; SIVmac239Δnef, nef-deleted infectious clone of SIV; TCID50, 50% tissue culture infective dose.
References
- Baba T.W., Jeong Y.S., Pennick D., Bronson R., Greene M.F., Ruprecht R.M. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- Baba T.W., Liska V., Khimani A.H., Ray N.B., Dailey P.J., Penninck D., Bronson R., Greene M.F., McClure H.M., Martin L.N. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat. Med. 1999;5:1–10. doi: 10.1038/5557. [DOI] [PubMed] [Google Scholar]
- Connor R.I., Montefiori D.C., Binley J.M., Moore J.P., Bonhoeffer S., Gettie A., Fenamore E.A., Sheridan K.E., Ho D.D., Daileyt P.J. Temporal analysis of virus replication, immune responses and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Virol. 1998;72:7501–7509. doi: 10.1128/jvi.72.9.7501-7509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M.G., Yalley-Ogunro J., Greenhouse J.J., Brennan T.P., Jiang J.B., VanCott T.C., Lu Y., Eddy G.A., Birx D.L. Limited protection from a pathogenic chimeric simian-human immunodeficiency virus challenge following immunization with attenuated simian immunodeficiency virus. J. Virol. 1999;73:1262–1270. doi: 10.1128/jvi.73.2.1262-1270.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.P., Desrosiers R.C. Protective immunity induced by live, attenuated simian immunodeficiency virus. Curr. Opin. Immunol. 1998;10:436–443. doi: 10.1016/s0952-7915(98)80118-0. [DOI] [PubMed] [Google Scholar]
- Daniel M.D., Kirchhoff F., Czajak S.C., Sehgal P.K., Desrosiers R.C. Protective effects of a live attenuated vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- Marthas M.L., Sutjipto S., Miller C.J., Higgins J., Torten J., Unger R.E., Marx P.A., Pedersen N.C. Efficacy of live-attenuated and whole-inactivated SIV vaccines against intravenous and vaginal challenge. In: Brown F., Chanock R.M., Ginsberg H.S., Lerner R.A., editors. Vaccines. Vol. 92. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1992. pp. 117–121. [Google Scholar]
- Lohman B.L., McChesney M.B., Miller C.J., McGowan E., Joye S.M., Van Romapy K.K.A., Reay E., Antipa L., Pedersen N.C., Marthas M.L. A partially attenuated simian immunodeficiency virus induces host immunity that correlates with resistance to pathogenic virus challenge. J. Virol. 1994;68:7021–7029. doi: 10.1128/jvi.68.11.7021-7029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J.E., Montelaro R.C., Zink M.C., Amedee A.M., Miller S., Trichel A.M., Jagerski B., Hauer D., Martin L.N., Bohm R.P. Cross-protective immune responses induced in rhesus macaques by immunization with attenuated macrophage-tropic simian immunodeficiency virus. J. Virol. 1995;69:2737–2744. doi: 10.1128/jvi.69.5.2737-2744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyand M.S., Manson K.H., Garcia-Moll M., Montefiori D., Desrosiers R.C. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl-Hennig C., Dittmer U., Nisslein T., Petry H., Jurkiewicz E., Fuchs D., Wachter H., Matz-Rensing K., Kuhn E.M., Kaup F.J. Rapid development of vaccine protection in macaques by live-attenuated simian immunodeficiency virus. J. Gen. Virol. 1996;77:2969–2981. doi: 10.1099/0022-1317-77-12-2969. [DOI] [PubMed] [Google Scholar]
- Norley S., Beer B., Binninger-Schinzel D., Cosma C., Kurth R. Protection from pathogenic SIVmac challenge following short-term infection with a nef-deficient attenuated virus. Virology. 1996;219:195–205. doi: 10.1006/viro.1996.0237. [DOI] [PubMed] [Google Scholar]
- Shibata R., Siemon C., Czajak S.C., Desrosiers R.C., Martin M.A. Live, attenuated simian immunodeficiency virus vaccines elicit potent resistance against a challenge with a human immunodeficiency virus type 1 chimeric virus. J. Virol. 1997;71:8141–8148. doi: 10.1128/jvi.71.11.8141-8148.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyand M.S., Manson K., Montefiori D.C., Lifson J.D., Johnson R.P., Desrosiers R.C. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J. Virol. 1999;73:8356–8363. doi: 10.1128/jvi.73.10.8356-8363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori D.C., Baba T.W., Li A., Bilska M., Ruprecht R.M. Neutralizing and infection-enhancing antibody responses do not correlate with the differential pathogenicity of SIVmac239Δ3 in adult and infant rhesus monkeys. J. Immunol. 1996;157:5528–5535. [PubMed] [Google Scholar]
- Cole K.S., Rowles J.L., Murphey-Corb M., Clements J.E., Robinson J., Montelaro R.C. A model for the maturation of protective antibody responses to SIV envelope proteins in experimentally immunized monkeys. J. Med. Primatol. 1997;26:51–58. doi: 10.1111/j.1600-0684.1997.tb00319.x. [DOI] [PubMed] [Google Scholar]
- Gundlach B.R., Reiprich S., Sopper S., Means R.E., Dittmer U., Matz-Rensing K., Stahl-Hennig C., Uberla K. Env-independent protection induced by live, attenuated simian immunodeficiency virus vaccines. J. Virol. 1998;72:7846–7851. doi: 10.1128/jvi.72.10.7846-7851.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson C., Mäkitalo B., Thorstensson R., Norley S., Binninger-Schinzel D., Cranage M., Rud E., Biberfeld G., Putkonen P. Live, attenuated simian immunodeficiency virus (SIV)mac in macaques can induce protection against mucosal infection with SIVsm. AIDS. 1998;12:2261–2270. doi: 10.1097/00002030-199817000-00006. [DOI] [PubMed] [Google Scholar]
- Robinson J.E., Cole K.S., Elliott D.H., Lam H., Amedee A.M., Means R., Desrosiers R.C., Clements J., Montelaro R.C., Murphey-Corb M. Production and characterization of SIV envelope-specific rhesus monoclonal antibodies from a macaque asymptomatically infected with a live SIV vaccine. AIDS Res. Hum. Retroviruses. 1998;14:1253–1262. doi: 10.1089/aid.1998.14.1253. [DOI] [PubMed] [Google Scholar]
- Almond N., Rose J., Sangster R., Silvera P., Stebbings R., Walker B., Stott E.J. Mechanisms of protection induced by attenuated simian immunodeficiency virus I. Protection cannot be transferred with immune serum. J. Gen. Virol. 1997;78:1919–1922. doi: 10.1099/0022-1317-78-8-1919. [DOI] [PubMed] [Google Scholar]
- Dittmer U., Niblein T., Bodemer W., Petry H., Sauermann U., Stahl-Hennig C., Hunsmann G. Cellular immune response of rhesus monkeys infected with a partially attenuated nef deletion mutant of the simian immunodeficiency virus. Virology. 1995;212:392–397. doi: 10.1006/viro.1995.1496. [DOI] [PubMed] [Google Scholar]
- Johnson R.P., Glickman R.L., Yang J.Q., Kaur A., Dion J.T., Mulligan M.J., Desrosiers R.C. Induction of vigorous cytotoxic T-lymphocyte responses by live attenuated simian immunodeficiency virus. J. Virol. 1997;71:7711–7718. doi: 10.1128/jvi.71.10.7711-7718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranage M.P., Whatmore A.M., Sharpe S.A., Cook N., Polyanskaya N., Leech S., Smith J.D., Rud E.W., Dennis M.J., Hall G.A. Macaques infected with live attenuated SIVmac are protected against superinfection via the rectal mucosa. Virology. 1997;229:143–154. doi: 10.1006/viro.1996.8419. [DOI] [PubMed] [Google Scholar]
- Koup R.A., Safrit J.T., Cao Y., Andrews C.A., McLeod G., Borkowsky W., Farthing C., Ho D.D. Temporal association of cellular immune responses with the initial control of viremia in primary HIV-1 syndrome. J. Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann K.A., Tenner-Racz K., Racz P., Montefiori D.C., Yasutomi Y., Ransil B.J., Letvin N.L. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J. Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutomi Y., Reimann K.A., Lord C.I., Miller M.D., Letvin N.L. Simian immunodeficiency virus-specific CD8+ lymphocyte response in acutely infected rhesus monkeys. J. Virol. 1993;67:1707–1711. doi: 10.1128/jvi.67.3.1707-1711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Qin L., Zhang L., Safrit J., Ho D.D. Virological and immunological characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- Dyer W.B., Ogg G.S., Demoitie M.A., Jin X., Geczy A.F., Rowland-Jones S.L., McMichael A.J., Nixon D.F., Sullivan J.S. Strong human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte activity in Sydney Blood Bank cohort patients infected with nef-defective HIV type 1. J. Virol. 1999;73:436–443. doi: 10.1128/jvi.73.1.436-443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Bauer D.E., Tuttleton S.E., Lewin S., Gettie A., Blanchard J., Irwin C.E., Safrit J.T., Mittler J., Weinberger L. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus–infected macaques. J. Exp. Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J.E., Kuroda M.J., Santra S., Sasseville V.G., Simon M.A., Lifton M.A., Racz P., Tenner-Racz K., Dalesandro M.M., Scallon B.J. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Matano T., Shibata R., Siemon C., Connors M., Lane H.C., Martin M.A. Administration of anti-CD8 monoclonal antibody interferes with clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 1998;72:164–169. doi: 10.1128/jvi.72.1.164-169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbings R., Stott J., Almond N., Hull R., Lines J., Silvera P., Sangster R., Corcoran T., Rose J., Cobbold S. Mechanisms of protection induced by attenuated simian immunodeficiency virus. II. Lymphocyte depletion does not abrogate protection. AIDS Res. Hum. Retroviruses. 1998;14:1187–1198. doi: 10.1089/aid.1998.14.1187. [DOI] [PubMed] [Google Scholar]
- Kostrikis L.G., Tyagi S., Mhlanga M.M., Ho D.D., Kramer F.R. Spectral genotyping of human alleles. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- Tyagi S., Kramer F.R. Molecular beaconsprobes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Tyagi S., Bratu D.P., Kramer F.R. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- Regier D.A., Desrosiers R.C. The complete nucleotide sequence of a pathogenic clone of simian immunodeficiency virus. AIDS Res. Hum. Retroviruses. 1990;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- Suryanarayana K., Wiltrout T.A., Vasquez G.M., Hirsch V.M., Lifson J.D. Plasma SIV RNA viral load by determination by real-time quantification of product generation in reverse-transcriptase-polymerase chain reaction. AIDS Res. Hum. Retroviruses. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- Dailey P.J., Zamroud M., Kelso R., Kolberg J., Urdea M. Quantitation of simian immunodeficiency virus (SIV) RNA in plasma of acute and chronically infected macaques using a branched DNA (bDNA) signal amplification assay. J. Med. Primatol. 1995;24:209. [Google Scholar]
- Sachsenberg N., Perelson A.S., Yerly S., Schockmel G.A., Leduc D., Hirschel B., Perrin L. Turnover of CD4+ and CD8+ T lymphocytes in HIV-1 infection as measured by Ki-67 antigen. J. Exp. Med. 1998;187:1295–1303. doi: 10.1084/jem.187.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauduin M.C., Glickman R.L., Means R., Johnson R.P. Inhibition of simian immunodeficiency virus (SIV) replication by CD8+ T lymphocytes from macaques immunized with live attenuated SIV. J. Virol. 1998;72:6315–6324. doi: 10.1128/jvi.72.8.6315-6324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R.K., Nilsson C., Wang Y., Lehner T., Biberfeld G., Thorstensson R. Beta-chemokine production in macaques vaccinated with live, attenuated virus correlates with protection against simian immunodeficiency virus (SIVsm) challenge. J. Gen. Virol. 1999;80:1569–1574. doi: 10.1099/0022-1317-80-7-1569. [DOI] [PubMed] [Google Scholar]
- Nixon D.F., Donahoe S.M., Kakimoto W.M., Samuel R.V., Metzner K.J., Gettie A., Hanke T., Marx P.A., Connor R.I. Simian immunodeficiency virus-specific cytotoxic T lymphocytes and protection against challenge in rhesus macaques immunized with a live attenuated simian immunodeficiency virus vaccine. Virology. 2000;266:203–210. doi: 10.1006/viro.1999.0078. [DOI] [PubMed] [Google Scholar]
- Jonker M., Nooij F.J., van Suylichem P., Neuhaus P., Goldstein G. The influence of OKT8F treatment on allograft survival in rhesus monkeys. Transplantation. 1986;41:431–435. doi: 10.1097/00007890-198604000-00004. [DOI] [PubMed] [Google Scholar]
- Zlabinger G.J., Stuhlmeier K.M., Eher R., Schmaldienst S., Klauser R., Vychytil A., Watschinger B., Traindl O., Kovarik J., Pohanka E. Cytokine release and dynamics of leukocyte populations after CD3/TCR monoclonal antibody treatment. J. Clin. Immunol. 1992;12:170–177. doi: 10.1007/BF00918085. [DOI] [PubMed] [Google Scholar]
- Evans D.T., O'Connor D.H., Jing P., Dzuris J.L., Sidney J., da Silva J., Allen T.M., Horton H., Venham J.E., Rudersdorf R.A. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- Gallimore A., Cranage M., Cook N., Almond N., Bootman J., Rud E., Silvera P., Dennis M., Corcoran T., Stott J. Early suppression of SIV replication by CD8+ nef-specific cytotoxic T cells in vaccinated macaques. Nat. Med. 1995;1:1167–1173. doi: 10.1038/nm1195-1167. [DOI] [PubMed] [Google Scholar]
- Sharpe S.A., Whatmore A.M., Hall G.A., Cranage M.P. Macaques infected with attenuated simian immunodeficiency virus resist superinfection with virulence-revertant virus. J. Gen. Virol. 1997;78:1923–1927. doi: 10.1099/0022-1317-78-8-1923. [DOI] [PubMed] [Google Scholar]