B cell development is often portrayed as a series of decision points that expand an antigen-reactive cell to a clone producing a single antibody. This is hardly the case: B cell development is dependent on a series of error-prone, random rearrangement events that through ongoing diversification reach a compromise in which most cells are not autoreactive (except in disease) and the majority of clone members remain specific for the initial antigen. One familiar example of ongoing diversification is somatic mutation during clonal expansion 1. Another example, receptor editing, is the means by which immature bone marrow B cells become self-tolerant 2 3 4. Here rearrangements are induced by encounter with autoantigens to change specificity from self to non-self. Now, a third level of diversification, termed “receptor revision,” has been suggested to occur in mature B cells. Initial evidence for revision included recombination activating gene (RAG) expression in germinal centers along with attendant double-stranded breaks adjacent to recombination signal sequences (RSS) 5 6 7, but the strongest evidence comes from examples of cells that underwent revision after somatic mutation was initiated. The paper in this issue by Wilson et al. 8, along with two previous studies 9 10, identifies clones of B cells that include cells whose antibody genes have undergone concurrent mutation and revision.
These findings place receptor revision firmly into the environment of germinal centers. In addition to somatic mutation, this is where other important immunological processes happen, including H chain class switch and immune memory formation. The germinal center cell subset that expresses most RAG activity appears to be the noncycling, centrocyte cells 5 11. Unlike other peripheral B cells, these cells express many markers shared by bone marrow B cells, including surrogate L chain components, IL-7R, and in humans, terminal deoxynucleotidyl transferase 6 11 12. Furthermore, purified IgD+ splenic B cells express RAG upon exposure either to a combination of CD40 antibodies and IL-4 (agents that are thought to mimic T cell help), or to a combination of LPS and IL-4 7. More recent studies show that IL-7, rather than IL-4, may be the critical cytokine driving receptor revision in vivo since RAG expression is unperturbed in the germinal centers of immunized IL-4–deficient mice, but is blocked in anti–IL-7R–treated mice 12. Interestingly, IL-7 is also a key cytokine for immature B cell expansion. These parallels between the cells undergoing receptor revision and immature B cells supported the idea that germinal center B cells reinduce a gene expression program characteristic of less mature cells, a concept known as “neoteny” 5. Reprogramming might be initiated by a lethal mutation in VH or VL. Such a mutant might resemble a pro-B or pre-B cell, and other phenotypes such as RAG expression might be activated.
The similarities between RAG-expressing bone marrow and germinal center B cells raise the possibility that receptor editing is going on in immature B cells that have migrated to the periphery. Three recent papers examining RAG indicator mice 13 14 15 reinforce this concern. Nussenzweig and colleagues generated bac-transgenic mice expressing a green fluorescent protein (GFP) gene placed in the context of ∼100 kb of the RAG gene cis-acting elements 13. Here, the cells expressing GFP in the periphery had the phenotype of newly minted bone marrow B cells, not germinal center cells. Furthermore, stimuli that were thought to increase RAG expression in vitro or in vivo failed to demonstrate upregulation of GFP and may just have prolonged expression in immature cells that were initially GFP+ 13. A second mouse made by Alt and colleagues targeted the endogenous RAG-2 gene to generate a RAG-2–GFP fusion protein in the natural locus 14. This gene proved to be functional in the homozygous mice, which generated B and T cells. Because RAG-2 is in part regulated at the level of protein stability 16, these mice, unlike the bac-GFP mice, rapidly lose GFP protein with B cell maturation. Upon immunization to generate germinal centers, RAG expression was found, but appeared mainly in cells with little or no surface (s)Ig 14. It remains to be seen if these cells are typical germinal center cells. In a third study, Sakaguchi and colleagues 15 targeted GFP to the RAG-1 locus and studied its expression in B-1 cells, which had been reported previously to express RAG 17. As in the previous study, RAG was expressed in just 1% of peritoneal (CD5+) B-1 cells, but was found in a large subset of apparently newly formed B-2 cells. These studies say that few B cells reinitiate V(D)J recombination in the peripheral lymphoid system, and suggest that cells expressing RAG in the periphery are phenotypically immature. To reconcile these studies with those that demonstrate revision in cells undergoing hypermutation, one must assume either that immature B cells can participate in germinal center reactions or that germinal center B cells that revise are rare or difficult to detect.
Since peripheral B cells that express RAG seem to be a heterogeneous population including both immature bone marrow emigrants and germinal center–like cells, other properties (besides RAG expression) that distinguish mature and immature B cells must be considered to appreciate the role and significance of receptor revision. Several lines of evidence suggest that revision and editing, though similar in many ways, are distinct in much more than the anatomical location of the recombinationally active cells, particularly with respect to the consequences of antigen receptor signaling. First, when appropriately stimulated such as with LPS plus IL-4, mature but not immature B cells rapidly express RAGs and other germinal center markers shared with bone marrow B cells, including GL-7 and IL-7R 12 18 19. Importantly, addition of cognate antigens or B cell antigen receptor (BCR) antibodies to such cultures prevents RAG induction 11 20. On the other hand, simple BCR ligation induces editing in immature but not mature B cells, even when both types of cells are present together in the same microenvironment 21 22. These studies appear to rule out a direct role of receptor revision in immune tolerance.
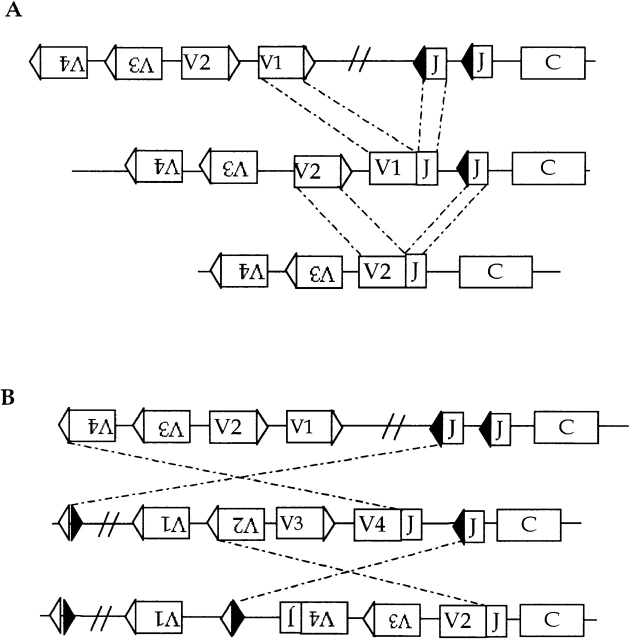
Another possible difference revealed in the Wilson et al. study 8 is V gene replacement at the H chain. Receptor editing was originally found at L chain loci 2 3 4. Secondary rearrangements at the κ locus replaced a VκJκ gene that contributed to the self-specificity of a BCR by another Vκ rearrangement to Jκ (genotypic editing, see Fig. 2) or formed a second, functional VJ allele that produced an L chain that could outcompete the first for association with the H chain (phenotypic editing; here the extent to which the edited B cell appears to be allelically excluded depends on the competitive advantage of the L chain for H chain). Two properties of L chain genes not shared with H chain loci seemed to favor editing of this subunit. The first is the grand organization of the κ locus: the asymmetry of the RSS of Vκ's and Jκ's allows secondary rearrangement of Vκ's upstream of and Jκ's downstream of the primary VκJκ rearrangement (Fig. 1 and Fig. 2 A). In theory, a κ allele could undergo up to five rearrangements (if one includes Vκ rearrangement to the Cκ deleting elements [κde]). Secondary rearrangements are sustained not just by multi-Vκ and Jκ loci, but also by the orientation of Vκ genes. Whereas most V genes, VH for example, are oriented vis a vis J so as to delete intervening DNA upon rearrangement 23 24, Vκ genes are oriented in both directions 25 26 27. Hence, about half of Vκ's invert intervening DNA, thereby conserving Vκ's that lie in the intervening DNA and converting deletion-oriented Vκ's to inversional Vκ's. This flip-flop potential optimizes the V repertoire for editing (Fig. 2 B). DNA deleting events also occur, leading to a directionality of rearrangement; hence a hallmark of editing in B (or T cells) is a bias toward downstream J genes and depletion of V genes.
Figure 2.
V gene editing at a κ-like L chain locus. V gene replacement can occur by secondary VJ rearrangement since the V RSS and J RSS are asymmetric, i.e. fit the 1 turn/2 turn or 12/23 bp rule. V genes in the same transcriptional orientation as J such as V1 and V2 delete DNA upon rearrangement (A). V3 and V4 invert the DNA between V and J, thereby conserving V genes (B). Moreover, V genes in the deletionogenic orientation such as V1 and V2 are now turned around and will conserve DNA on subsequent rearrangement.
Figure 1.
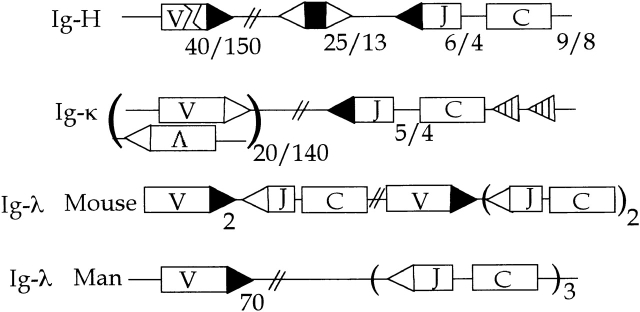
Organization of antibody genes. Gene segment numbers for humans and mice are indicated as n man/n mouse. Sequences involved in recombination are designated: ▸, 2 turn RSS; ▹, 1 turn RSS; striped ◃, κ deleting element (κde); >, VH central embedded heptamer (5′-CACAGTG-3′); and <, VH terminal embedded heptamer (5′-TACTGTG-3′).
The second relevant property of L chains is that they are usually encoded by two or more autonomously expressed loci: κ, λ1, λ2, etc. Multiple L chain loci (isotypes) also increase editing opportunities: if one isotype is terminally—but unsuccessfully—rearranged, then the other can take its place. Indeed, the editing potential of two isotypes is optimized by sequential rearrangement, i.e., κ before λ 28. In humans, this transition opens a large (∼70) Vλ gene library for further diversification and editing 29, but in the mouse the transition offers little variety. Yet mouse immunity is fine without λ, so the foreshortened mouse λ serves mainly to rescue κ-deleted B cells from oblivion. In the same sense that tolerance by editing influences Jκ usage, it drives the repertoire toward λ.
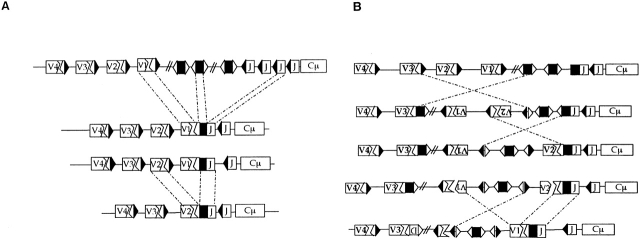
H chain genes do not have these features. V(D)J gene replacement by secondary VH to JH cannot work because the VH and JH RSS do not meet the 1 turn/2 turn requirement for recombination and because D segments, the guardians of this rule, are deleted by the primary V(D)J recombination (Fig. 3). But various types of recombination and H chain modification hinted that editing might be possible. Artificial recombination substrates have shown that the 1 turn/2 turn rule is relaxed and that heptamers alone can serve as recombination targets 30. B cell lymphomas constitutive for recombination provided in vivo evidence for VH gene replacement at a heptamer embedded in the primary V(D)J rearrangement 31 32. This phenomenon inspired surveys of VH genes for RSS-like sequences that showed remarkable conservation (and presumed significance) of the heptamer embedded at the end of most VH genes 33. Yet VH replacement as seen in VH transgenic mice suggested that VH replacement occurred early, at the pro-B stage, and could simply be a variation of primary V(D)J recombination played out on an inherited V(D)J substrate 34. Wilson et al. 8 now show that VH replacement is real, may happen often, and can work in strange ways.
Figure 3.
VH replacement. Secondary VH rearrangement to embedded heptamers can replace most of a V(D)J if at the VH terminal heptamer, or about half of the V(D)J if at the central heptamer (B). In the former case, DNA between the donor and recipient VHs is deleted since most VH genes are in the same transcriptional orientation as JH (A). But DNA can be inverted if the initial rearrangement is VH to the 3′ RSS of D (B). This inverts intervening DNA and would allow a VH1 replacement of a VH2 (D)J, as observed by Wilson et al. (reference 8).
The functional VH replacements described so far have used an embedded heptamer at the end of framework region 3 (FR3) that is oriented in the same direction as those RSS to which VH ordinarily rearranges, for example, VH to (D)J. The example described by Wilson et al. 8 uses the heptamer located at the beginning of FR3 of VH, but this heptamer points in the opposite direction (Fig. 3). Wilson et al. reason that VH replacement can happen at this site through hybrid joint formation. This is one of the four possible products of the recombination intermediate that are detected in in vitro recombination systems and one that nicely explains the VH chimera 30. Similar chimeric VHs are seen in expanded B cell clones found in synovia of rheumatoid arthritis (RA) patients and here VH replacements at both heptamers are found (Chiorazzi, N., manuscript in preparation). Wilson et al. 8 argue that VH revision may be common but unrecognized. For example, VH replacement at the downstream heptamer is essentially “invisible,” since most of the recipient gene is erased. Another perhaps much more profound reason for underestimating the extent of VH editing is that VH lacks the backup equipment that L chain has. In effect, VH editing has one shot, and given the high likelihood of out-of-frame joins and inadvertent use of a pseudo V for replacement, successful VH replacement happens at best 20% of the time. In addition, VH genes include cryptic heptamers that have been shown to act as replacement targets but that lead to abnormal VH sequences 35. Taken together, these factors conspire against successful editing and the apparent preference for L chain editing is mainly due to the loss of B cells with two defunct VH alleles.
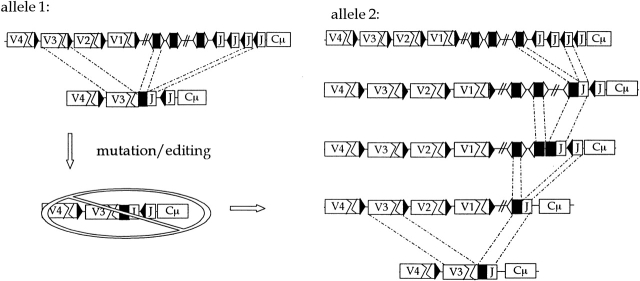
Of course, the key distinction between editing and revision is the setting in which revision appears to happen. The examples reported so far are in highly expanded clones as judged by the high frequency of mutations. This correlation suggests that revision may be a rare event and apparent only in special circumstances such as chronic antigen drive or autoimmunity. An example of the latter is found in the MRL mouse. Monestier and colleagues show that the unusual H chain junctions of autoantibodies resulting from DD fusion and D inversions (Fig. 4) are formed extensively in this strain 36. Since these unusual junctions are already found in preimmune B cells, they interpret this to mean that the D to J window of rearrangement is extended in autoimmunity. This idea helps to explain why autoantibodies are heavily biased in favor of JH4 (mouse) or JH5 and JH6 (humans). The bias is a puzzle because secondary V to J rearrangements as at the κ locus (Fig. 2) are not possible after the primary VHDHJH rearrangement (Fig. 3). But, as shown in Fig. 4, the bias can be understood by extended rearrangement at the D to J stage. During clonal expansion, lethal mutation or aberrant editing kills the primary V(D)J (allele 1, Fig. 4) but the cell can be rescued by rearranging allele 2. In the interim, this allele may have undergone several introductory DJ rearrangements that might have led (especially in MRL mice) to DJ4 or DDJ4 (Fig. 4). There is evidence for extended receptor editing in cells that overexpress cell survival proteins 37 38 39, and receptor revision has been seen in Fas-deficient (lpr/lpr) mice 9, lupus 40, and RA (Chiorazzi, N., manuscript in preparation).
Figure 4.
Rearrangement events at an H chain locus. Rearrangement occurs on both alleles, leading to a B cell with a V(D)J allele and a (D)J allele. If rearrangement can resume at both alleles, then allele 1 might be replaced and allele 2 might undergo further (D)J rearrangements. If allele 1 becomes nonfunctional during clonal expansion, then allele 2 can become functional by a V to (D)J event. Such V(D)Js should be enriched for downstream JHs.
The differential regulation of revision and editing by the antigen receptor predicts far-reaching effects on immune tolerance. Editing minimizes autoreactivity in immature, preimmune cells by specifically replacing autoreactive receptors, whereas revision occurs during antigen-driven immune responses and is suppressed, rather than induced, by sIg cross-linking. Therefore, revision, unlike editing, should complicate immune tolerance by generating new, often autoreactive receptors in activated, mature B cells. As a consequence, revision may be associated with autoimmunity for two reasons: extended clonal expansion (as in disease) may be necessary to realize significant frequencies of revision and when revision occurs, virgin repertoires that include autoantibodies are generated. Furthermore, because of their differential sensitivity to BCR signaling, revision and editing also differ in their predicted impact upon “allelic and isotypic (κ or λ) exclusion,” i.e., the propensity of cells to express at any given time a single pair of antibody H and L chains. Receptor editing that is stimulated by an autoreactive receptor is geared to promote continued secondary Ig L gene rearrangements until the offending receptor is eliminated or altered. This automatically diminishes double producers, at least in terms of cell surface expression. But because receptor revision in antigen-activated cells is suppressed by sIg binding 11 20, revision should allow multiple receptor production and that could lead to gratuitous autoantibody expression.
How can the phenomena of receptor editing and revision, which (along with somatic hypermutation) may be lumped under the rubric of “receptor selection,” be reconciled with the concept of “clonal selection”? Rather than being viewed as mutually exclusive pathways, these mechanisms complement each other by regulating independently the survival and propagation of cells and their receptors. In promoting lymphocyte cell death or proliferation, clonal selection reduces diversity, whereas receptor selection mechanisms enhance diversity. When receptors are autoreactive, receptor selection can destroy them, while often sparing the cell. If an antigen-reactive cell has made a useful improvement in specificity, then that specificity can be fixed, facilitating clonal expansion. On the other hand, if antigen reactivity is weak, receptor selection allows specificity to drift, sometimes generating saltatory improvements in antigen binding affinity, albeit rarely, but at other times generating self-reactive cells, which may in turn need to be controlled by clonal mechanisms. Working together, receptor selection and clonal selection account for the astonishing rapidity of the somatic evolution of immune specificity.
References
- McKean D., Huppi K., Bell M., Staudt L., Gerhard W., Weigert M. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA. 1984;81:3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D., Saunders T., Camper S., Weigert M. Receptor editingan approach by autoreactive B cells to escape tolerance. J. Exp. Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radic M.Z., Erikson J., Litwin S., Weigert M. B lymphocytes may escape tolerance by revising their antigen receptors. J. Exp. Med. 1993;177:1165–1173. doi: 10.1084/jem.177.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiegs S.L., Russell D.M., Nemazee D. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Zheng B., Schatz D.G., Spanopoulou E., Kelsoe G. Neoteny in lymphocytesRag1 and Rag2 expression in germinal center B cells. Science. 1996;274:2094–2097. doi: 10.1126/science.274.5295.2094. [DOI] [PubMed] [Google Scholar]
- Han S., Dillon S.R., Zheng B., Shimoda M., Schlissel M.S., Kelsoe G. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 1997;278:301–305. doi: 10.1126/science.278.5336.301. [DOI] [PubMed] [Google Scholar]
- Hikida M., Mori M., Takai T., Tomochika K., Hamatani K., Ohmori H. Reexpression of RAG-1 and RAG-2 genes in activated mature mouse B cells. Science. 1996;274:2092–2094. doi: 10.1126/science.274.5295.2092. [DOI] [PubMed] [Google Scholar]
- Wilson P.C., Wilson K., Liu Y.-J., Banchereau J., Pascual V., Capra J.D. Receptor revision of immunoglobulin heavy chain variable region genes in normal human B lymphocytes. J. Exp. Med. 2000;191:1881–1894. doi: 10.1084/jem.191.11.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brard F., Shannon M., Prak E.L., Litwin S., Weigert M. Somatic mutation and light chain rearrangement generate autoimmunity in anti-single-stranded DNA transgenic MRL/lpr mice. J. Exp. Med. 1999;190:691–704. doi: 10.1084/jem.190.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wildt R., Hoet R.M.A., van Venrooij W.J., Tomlinson I.M., Winter G. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J. Mol. Biol. 1999;285:895–901. doi: 10.1006/jmbi.1998.2396. [DOI] [PubMed] [Google Scholar]
- Meffre E., Papavasiliou F., Cohen P., de Bouteiller O., Bell D., Karasuyama H., Schiff C., Banchereau J., Liu Y.J., Nussenzweig M.C. Antigen receptor engagement turns off the V(D)J recombination machinery in human tonsil B cells. J. Exp. Med. 1998;188:765–772. doi: 10.1084/jem.188.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikida M., Nakayama Y., Yamashita Y., Kumazawa Y., Nishikawa S.I., Ohmori H. Expression of recombination activating genes in germinal center B cellsinvolvement of interleukin 7 (IL-7) and the IL-7 receptor. J. Exp. Med. 1998;188:365–372. doi: 10.1084/jem.188.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Nagaoka H., Jankovic M., Misulovin Z., Suh H., Rolink A., Melchers F., Meffre E., Nussenzweig M.C. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature. 1999;400:682–687. doi: 10.1038/23287. [DOI] [PubMed] [Google Scholar]
- Monroe R.J., Chen F., Ferrini R., Davidson L., Alt F.W. RAG2 is regulated differentially in B and T cells by elements 5′ of the promoter. Proc. Natl. Acad. Sci. USA. 1999;96:12713–12718. doi: 10.1073/pnas.96.22.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata N., Igarashi H., Ohmura T., Aizawa S., Sakaguchi N. Cutting edgeabsence of expression of RAG1 in peritoneal B-1 cells detected by knocking into RAG1 locus with green fluorescent protein gene. J. Immunol. 1999;163:6355–6359. [PubMed] [Google Scholar]
- Lin W.C., Desiderio S. V(D)J recombination and the cell cycle. Immunol. Today. 1995;16:279–289. doi: 10.1016/0167-5699(95)80182-0. [DOI] [PubMed] [Google Scholar]
- Qin X.F., Schwers S., Yu W., Papavasiliou F., Suh H.Y., Nussenzweig A., Rajewsky K., Nussenzweig M.C. Secondary V(D)J recombination in B-1 cells. Nature. 1999;397:355–359. doi: 10.1038/16933. [DOI] [PubMed] [Google Scholar]
- Han S., Zheng B., Takahashi Y., Kelsoe G. Distinctive characteristics of germinal center B cells. Semin. Immunol. 1999;9:255–260. doi: 10.1006/smim.1997.0081. [DOI] [PubMed] [Google Scholar]
- Laszlo G., Hathcock K.S., Dickler H.B., Hodes R.J. Characterization of a novel cell-surface molecule expressed on subpopulations of activated T and B cells. J. Immunol. 1999;150:5252–5262. [PubMed] [Google Scholar]
- Hertz M., Kouskoff V., Nakamura T., Nemazee D. V(D)J recombinase induction in splenic B lymphocytes is inhibited by antigen-receptor signalling. Nature. 1998;394:292–295. doi: 10.1038/28419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz M., Nemazee D. BCR ligation induces receptor editing in IgM+IgD− bone marrow B cells in vitro . Immunity. 1997;6:429–436. doi: 10.1016/s1074-7613(00)80286-1. [DOI] [PubMed] [Google Scholar]
- Melamed D., Benschop R.J., Cambier J.C., Nemazee D. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor selection. Cell. 1998;92:173–182. doi: 10.1016/s0092-8674(00)80912-5. [DOI] [PubMed] [Google Scholar]
- Brodeur P.H., Osman G.E., Mackle J.J., Lalor T.M. The organization of the mouse Igh-V locus. Dispersion, interspersion, and the evolution of VH gene family clusters. J. Exp. Med. 1988;168:2261–2278. doi: 10.1084/jem.168.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F., Ishii K., Bourvagnet P., Kuma K., Hayashida H., Miyata T., Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J. Exp. Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S., Rosenberg N., Alt F., Baltimore D. Continuing kappa-gene rearrangement in a cell line transformed by Abelson murine leukemia virus. Cell. 1982;30:807–816. doi: 10.1016/0092-8674(82)90285-9. [DOI] [PubMed] [Google Scholar]
- Van Ness B.G., Coleclough C., Perry R.P., Weigert M. DNA between variable and joining gene segments of immunoglobulin kappa light chain is frequently retained in cells that rearrange the kappa locus. Proc. Natl. Acad. Sci. USA. 1982;79:262–266. doi: 10.1073/pnas.79.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachau H.G. The human immunoglobulin κ genes. In: Honjo T., Alt F., editors. Immunoglobulin Genes. Academic Press Limited; London: 1995. pp. 174–191. [Google Scholar]
- Mehr R., Shannon M., Litwin S. Models for antigen receptor gene rearrangement. I. Biased receptor editing in B cellsimplications for allelic exclusion. J. Immunol. 1999;163:1793–1798. [PubMed] [Google Scholar]
- Kawasaki K., Minoshima S., Nakato E., Shibuya K., Shintani A., Schmeits J.L., Wang J., Shimizu N. One-megabase sequence analysis of the human immunoglobulin lambda gene locus. Genome Res. 1997;7:250–261. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- Gellert M. Recent advances in understanding V(D)J recombination. Adv. Immunol. 1999;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- Kleinfield R., Hardy R.R., Tarlinton D., Dangl J., Herzenberg L.A., Weigert M. Recombination between an expressed immunoglobulin heavy-chain gene and a germline variable gene segment in a Ly 1+ B-cell lymphoma. Nature. 1986;322:843–846. doi: 10.1038/322843a0. [DOI] [PubMed] [Google Scholar]
- Reth M., Gehrmann P., Petrac E., Wiese P. A novel VH to VHDJH joining mechanism in heavy-chain-negative (null) pre-B cells results in heavy-chain production. Nature. 1986;322:840–842. doi: 10.1038/322840a0. [DOI] [PubMed] [Google Scholar]
- Kleinfield R.W., Weigert M.G. Analysis of VH gene replacement events in a B cell lymphoma. J. Immunol. 1989;142:4475–4482. [PubMed] [Google Scholar]
- Chen C., Nagy Z., Prak E.L., Weigert M. Immunoglobulin heavy chain gene replacementa mechanism of receptor editing. Immunity. 1995;3:747–755. doi: 10.1016/1074-7613(95)90064-0. [DOI] [PubMed] [Google Scholar]
- Taki S., Meiering M., Rajewsky K. Targeted insertion of a variable region gene into the immunoglobulin heavy chain locus. Science. 1993;262:1268–1271. doi: 10.1126/science.8235657. [DOI] [PubMed] [Google Scholar]
- Klonowski K.D., Primiano L.L., Monestier M. Atypical VH-D-JH rearrangements in newborn autoimmune MRL mice. J. Immunol. 1999;162:1566–1572. [PubMed] [Google Scholar]
- Rolink A., Grawunder U., Haasner D., Strasser A., Melchers F. Immature surface Ig+ B cells can continue to rearrange kappa and lambda L chain gene loci. J. Exp. Med. 1993;178:1263–1270. doi: 10.1084/jem.178.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J., Arnold B., Hammerling G., Harris A.W., Korsmeyer S., Russell D., Strasser A., Nemazee D. Enforced Bcl-2 expression inhibits antigen-mediated clonal elimination of peripheral B cells in an antigen dose-dependent manner and promotes receptor editing in autoreactive, immature B cells. J. Exp. Med. 1997;186:1513–1522. doi: 10.1084/jem.186.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W., Mueller D.L., Pennell C.A., Rivard J.J., Li Y.S., Hardy R.R., Schlissel M.S., Behrens T.W. Frequent aberrant immunoglobulin gene rearrangements in pro-B cells revealed by a bcl-xL transgene. Immunity. 1996;4:291–299. doi: 10.1016/s1074-7613(00)80437-9. [DOI] [PubMed] [Google Scholar]
- Dorner T., Foster S.J., Farner N.L., Lipsky P.E. Immunoglobulin kappa chain receptor editing in systemic lupus erythematosus. J. Clin. Invest. 1998;102:688–694. doi: 10.1172/JCI3113. [DOI] [PMC free article] [PubMed] [Google Scholar]