Abstract
T-shape radial spokes regulate flagellar beating. However, the precise function and molecular mechanism of these spokes remain unclear. Interestingly, Chlamydomonas reinhardtii flagella lacking a dimeric heat shock protein (HSP) 40 at the spokehead–spokestalk juncture appear normal in length and composition but twitch actively while cells jiggle without procession, resembling a central pair (CP) mutant. HSP40− cells begin swimming upon electroporation with recombinant HSP40. Surprisingly, the rescue doesn't require the signature DnaJ domain. Furthermore, the His-Pro-Asp tripeptide that is essential for stimulating HSP70 adenosine triphosphatase diverges in candidate orthologues, including human DnaJB13. Video microscopy reveals hesitance in bend initiation and propagation as well as irregular stalling and stroke switching despite fairly normal waveform. The in vivo evidence suggests that the evolutionarily conserved HSP40 specifically transforms multiple spoke proteins into stable conformation capable of mechanically coupling the CP with dynein motors. This enables 9 + 2 cilia and flagella to bend and switch to generate alternate power strokes and recovery strokes.
Introduction
The axonemes in cilia and flagella are microtubule-based super complexes constructed from hundreds of different polypeptides. In general, proteins belonging to each molecular complex are synthesized and assembled into precursors in the cell body before they are delivered into flagella by intraflagellar transport toward the tip of flagella (Fowkes and Mitchell, 1998; Qin et al., 2004). One central question that remains to be answered is how these precursors are assembled into the macromolecular frameworks, which not only support these organelles but, in the case of 9 + 2 cilia, act as eukaryotic nanomachines that generate powerful propulsive force with alternate power strokes and recovery strokes in the viscous aqueous environment.
Molecular chaperones responsible for various protein-folding events are among the top contenders for the assembly of axonemes. Chaperones, including heat shock protein (HSP) 60, 70, and 90, are present in cilia and flagella (Bloch and Johnson, 1995; Stephens and Lemieux, 1999; Seixas et al., 2003). Also present are J proteins (Ostrowski et al., 2002; Pazour et al., 2005; Satouh et al., 2005; Yang et al., 2005), the obligatory cochaperones that assist HSP70 ATPases in recruiting protein substrates and stimulating ATP hydrolysis with the signature DnaJ domain (for review see Craig et al., 2006).
The disparate numbers and locations of HSP70s and J proteins, however, suggest that the dynamic interplay and the functional mechanism of the chaperone machinery in these organelles may differ from the norm. For example, proteomic studies of Chlamydomonas reinhardtii flagella revealed multiple peptide hits for at least three HSP70s but only a single J protein (Pazour et al., 2005). The most well characterized of the HSP70s, HSP70A may be enriched at the tip of flagella (Bloch and Johnson, 1995), where assembly of axonemes primarily occurs (Rosenbaum and Child, 1967; Dentler and Rosenbaum, 1977; Johnson and Rosenbaum, 1992), implicating HSP70A in ciliogenesis. In addition, a fraction of HSP70A is constitutively associated with the C1b central pair (CP) projection that anchors enzymes for ATP synthesis (Mitchell et al., 2005). Ironically though, the sole J protein is located in a different complex, the T-shaped radial spokes (RS) that were known for motility control rather than protein folding (Yang et al., 2005).
The spoke J protein is RS protein (RSP) 16, one of the spoke-specific components. It is also present in the RS of sperm flagella in Ciona intestinalis, a deuterostome (Satouh et al., 2005). Notably, both contain all of the molecular modules characteristic of type II J proteins (HSP40s), including the signature DnaJ domain with the His-Pro-Asp (HPD) tripeptide essential for stimulating HSP70 ATPase activities, the G/F region, and the DnaJ C domain for peptide binding and dimerization, but both lack the cysteine-rich domain in type I HSP40 (Fig. 1 A). In addition, RSP16 undergoes homodimerization (Yang et al., 2005) as expected of HSP40s, which assume a U-shape–like dimeric conformation critical for protein binding (Sha et al., 2000; Borges et al., 2005). Consistent with the transient interaction between J proteins and client substrates, RSP16 homodimer is transported into flagella separately from the 12S spoke precursors, which are first preassembled in the cell body and contain the other identified RSPs (Qin et al., 2004; Yang et al., 2005). The major distinction is that J proteins usually dissociate from client polypeptides after folding, whereas RSP16 becomes integrated into the mature 20S spoke complex in the axonemes.
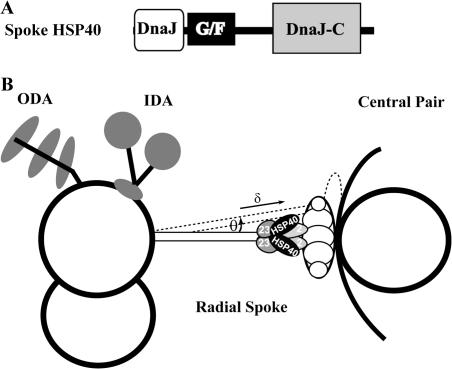
Figure 1.
Schematic picture depicting spoke HSP40 and the radial spoke in the 9 + 2 axoneme. (A) Spoke HSP40 contains the signature domains of the type II HSP40. (B) A model highlights the predicted position of HSP40 relative to RS, CP, and the dynein motors. Outer and inner dynein arms (ODA and IDA) power the interdoublet sliding that is subsequently converted to bend and oscillatory beating. It is postulated that the conversion is locally controlled by transient interactions of RS with CP resulting in tilt (θ), lengthening (δ), and strain of the engaged RS (Warner and Satir, 1974). The shape and size of the spoke HSP40 homodimer are largely based on the U-shaped dimeric type II HSP40 SIS1 (3 × 7 × 9 nm; Sha et al., 2000) relative to the ∼40-nm-long RS (Witman et al., 1978). Dimeric HSP40 possibly interacts with RSP2 and/or RSP23 (2 and 23, gray ovals) underneath the bulbous spokehead. The latter two are depicted as homodimers as well, based on the predicted resemblance of their DPY-30 domain to the homodimerized RIIa domain of cAMP-dependent protein kinase. The unfilled circles in the spokehead represent the five head proteins.
Comparison of protein defects in spoke mutants suggests that spoke HSP40 likely interacts with RSP2, a DPY-30 domain protein, and RSP23, a nucleoside diphosphate kinase (Fig. 1 B). The levels of these three proteins are diminished in the C. reinhardtii RSP2 mutant pf24, in which the spokehead and head end of the stalk appear defective, whereas they are normal in mutants lacking only the spokehead (Huang et al., 1981; Patel-King et al., 2004; Yang et al., 2004, 2005). These findings, along with immunogold electron microscopy (Satouh et al., 2005), suggest that the trios are stalk proteins, possibly forming a subcomplex located at the head–stalk juncture of the spoke.
The addition of HSP40 underneath the spokehead suggests that HSP40 is involved in either the assembly or function of the head domain. The importance of spokeheads and RS is verified by the C. reinhardtii mutants. Flagella defective in spokeheads only or in entire spokes are paralyzed similarly (Huang et al., 1981), suggesting that spokeheads mediate a central function of the entire complex. Notably, RS tilt and lengthen (Fig. 1 B, θ and δ) slightly only at the bend of cilia, suggesting that RS engage CP transiently and strain occurs during the engagement (Warner and Satir, 1974; Goodenough and Heuser, 1985). It is postulated that the transient engagement is a part of the mechanical feedback converting dynein-driven interdoublet sliding into local bend formation and propagation (Warner and Satir, 1974) or switching opposing active outer doublets to generate oscillatory beating with planar waveform (Satir and Matsuoka, 1989; Yagi et al., 1994; Sakakibara et al., 2004; Yokoyama et al., 2004; Lechtreck and Witman, 2007; Lindemann, 2007; Lindemann and Mitchell, 2007). Possibly, the engagement enables the distribution of the signal between the asymmetrical CP through RS and specific subsets of outer doublets (Mitchell, 2003; Wargo and Smith, 2003). Genetic and biochemical evidence suggests that the CP and RS constitute a control system governing dynein motors through dynein regulatory complex on the outer doublets as well (Huang et al., 1982; Piperno et al., 1994). The prediction was further supported by the structural contiguity among these molecular complexes by electron microscopy and tomography (Gardner et al., 1994; Nicastro et al., 2006). In addition, the second messengers that change flagellar beating may partly act through the control system (for review see Porter and Sale, 2000).
Despite the predicted key role and mechanism, motile 9 + 0 cilia and reactivated paralyzed flagella indicate that CP–RS is not required for the oscillatory beating or asymmetrical waveform (Omoto et al., 1996; Wakabayashi et al., 1997; Yagi and Kamiya, 2000). The question is what the proposed control system of CP–RS actually contributes to a basic nine outer doublets that could already beat. Studies of suppressors and in vitro reactivation primarily revealed their role in generating asymmetrical waveform for powerful propulsive force (Brokaw 1982; Hosokawa and Miki-Noumura, 1987; White et al., 2005), although mutant axonemes with impaired CP or RS could be reactivated to beat with asymmetrical waveform under altered conditions (Wakabayashi et al., 1997). Conversely, phenotypes of various CP and RS mutants, ranging from paralysis, jiggling, partial swimming, or reduced beat frequency (Dymek et al., 2004; Yokoyama et al., 2004; Mitchell et al., 2005; Yang and Yang, 2006; Lechtreck and Witman, 2007; for review see Smith and Yang, 2004), argue for a broader role but have not yet shed light on the proposed mechanism. One major challenge for interpreting the motility mutants is that the molecules involved in the CP–RS interactions have not been identified. Furthermore, most of the existing mutants of single-gene mutations are defective in multiple, possibly coassembled, proteins, resulting in loss of an entire complex, a part of the complexes, or absence of engagement. The ideal mutants are defective in a single protein and retain partial CP–RS interactions.
We reason that reverse genetics of spoke HSP40 could address this problem because the molecule is not coassembled with the other RSPs in the cell body and the spoke complex is quite stable (Yang et al., 2005). An RNAi strategy is taken to determine the roles of spoke HSP40 in the assembly and function of RS complex, in ciliogenesis, and in flagellar HSP70s. The phenotypes from the seemingly single-protein defect provide the in vivo evidence supporting the central role of the control system and shed light on molecular chaperones in cilia and flagella.
Results
Generation of RSP16 RNAi strains
Vector-based RNAi was performed to knock down RSP16. To create a hairpin construct, RSP16 genomic DNA from exon 6 to 7 and the corresponding cDNA were ligated in opposite orientation flanking a 400-bp Arabidopsis thaliana EARLI1 gene (Bubier and Schläppi, 2004). The hairpin fragment was inserted into the pSI103 plasmid upstream of the paromomycin (PMM)-resistant aphVIII cassette (Fig. 2 A). The rationale of this design was to use the promoter activity (Fig. 2 A, double-headed arrows) of the Rubisco 3′ untranslated region in the cassette (Sizova et al., 2001) to drive the transcription of the hairpin-containing 400-bp double-stranded RNA and two loops from the sixth intron and antisense strand of EARLI1 fragment. The hairpin construct was confirmed by restriction digest and sequencing.
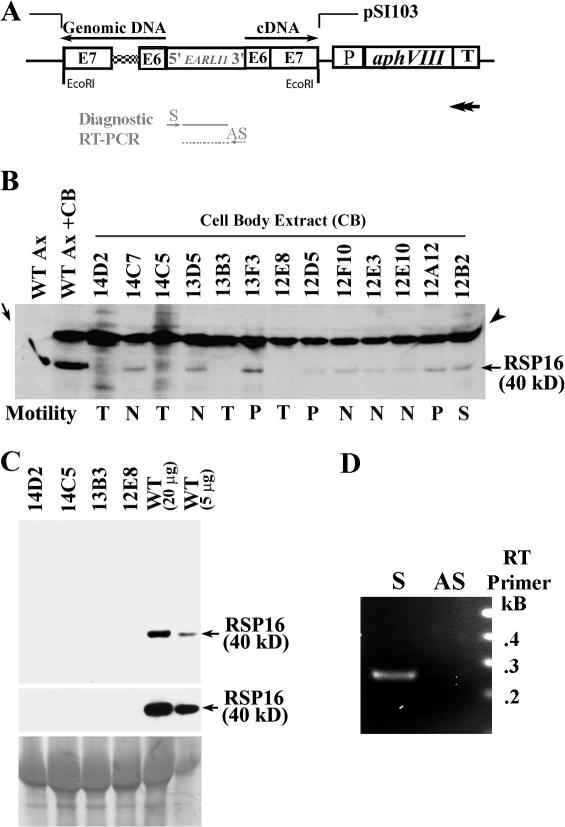
Figure 2.
Vector-based RNAi of RSP16. (A) The design of the RNAi construct. The RSP16 genomic DNA and cDNA fragments spanning exons 6 and 7 were ligated in an opposite orientation (black arrows) at each end of a spacer DNA that was PCR amplified from the A. thaliana EARLI1 gene. The hairpin fragment was cloned into the EcoRI site of the pSI103 vector upstream of the AphVIII expression cassette. The promotor activity in the opposite strand of the Rubisco 3′ untranslated region (T) should drive the expression (double-headed arrow) of the inverted sequence. Gray arrows and lines are reverse transcription primers and RT-PCR products for diagnosis of the transcript by RT-PCR. Dashed lines indicate the control, which detects amplification of genomic DNA. (B) Western blot screening RSP16 in the cell body extract from transformants displaying flagellar phenotypes. N, no flagella; P, paralyzed flagella; T, twitching flagella; and S, swimmers (control). The 50-kD cell body protein (arrowhead), which is also recognized by anti-RSP16 serum, served as a loading indicator and RSP16 in axonemes (arrow) was the positive control. Note that the 40-kD RSP16 was not discernable in the four twitching clones. (C) Western blots showed that the RSP16 band easily detected in the 20- and 5-μg wild-type axonemes was not detectable in ∼20-μg axonemes from the twitching clones. (bottom) Ponceau stain. (D) Transcription of hairpin construct was detected by RT-PCR of the exogenous loop using total RNA prepared from12E8 and the sense reverse transcription primer that could anneal to the predicted transcript. Control was using antisense primer (AS) that only annealed to genomic DNA.
The construct was transformed alone or cotransformed with pSI103 into wild-type cc124 cells and the transformants selected by PMM resistance. The resistant clones were first screened for flagellar phenotypes because presence of RNAi transgenes in C. reinhardtii frequently failed to cause sufficient protein reduction (Rohr et al., 2004). All of the colonies from the single transformation swam like the parental strain, whereas in the cotransformation group, four flagellar phenotypes could be distinguished among 400 resistant strains collected from two independent experiments, including three clones that had entirely paralyzed flagella (Fig. 2 B, P), four that had flagella that twitched actively (Fig. 2 B, T), and at least five that didn't have flagella (Fig. 2 B, N). The cell body extract from these strains and a swimmer (Fig. 2 B, S) was assessed by Western blots. The RSP16-enriched wild-type axoneme was the positive control (Fig. 2 B, arrow). Despite the pronounced 50-kD protein (Fig. 2 B, arrowhead) and others that were also recognized by anti-RSP16 serum, the 40-kD RSP16 was not discernable in the four clones with twitching flagella but was detectable in the other transformants. Importantly, RSP16 was not detectable in ∼20-μg axonemes from all four twitching clones in contrast to the obvious bands in 20- or 5-μg wild-type axonemes (Fig. 2 C, compare top and middle blots). Quantification of RSP16 levels in the cell body was not suitable because RSP16 could not be detected reliably because of a weak RSP16 signal and the strong 50-kD bands. Furthermore, some cells, like 12D5, may have less RSP16 polypeptide (Fig. 2 B), but the reduction was not enough to affect the amount in axonemes. To test if the hairpin construct was transcribed in the RSP16− clones as designed, RT-PCR was performed to amplify the exogenous antisense EARLI1 loop rather than the double-stranded region that was the target of Dicer (Meister and Tuschl, 2004) and was derived from the endogenous gene. Total RNA used as a template was prepared from 12E8 and treated with DNase to degrade residual genomic DNA. The product was present but faint if reverse transcription was primed by the sense primer (Fig. 2 A, S) that annealed to the antisense EARLI1 sequence in the transcript (Fig. 2 D). Absence of the band in the control using antisense primer (Fig. 2 A, AS) indicated that the product was amplified from a transcript, not DNA from the hairpin construct.
To test that the hairpin transcript is correlated with cleavage and decay of endogenous mRNA (Orban and Izaurralde, 2005), RT-PCR of 12E8 and wild type using the reverse transcription primer annealing specifically near polyA of RSP16 messages was performed (Fig. 3 A). The 35-cycle amplification was chosen to detect RSP16 messages. The 3′ UTR fragment was amplified from both the wild-type and 12E8 reverse transcription sample but not from both controls lacking reverse transcription (Fig. 3 B), which indicated that genomic DNA was degraded. Exon 7, which was included in the double-strand construct, was amplified as well but less efficiently from 12E8 than wild type (Fig. 3 C, top). Full-length message was only amplified (Fig. 3 C, bottom) from the wild-type reverse transcription sample, as previously reported (Yang et al., 2005), and was not amplified from 12E8. Collectively, the RT-PCR results indicated that the hairpin construct was transcribed, RSP16 messages were cleaved at the region where small RNA complemented, and decay of cleaved mRNA fragments was incomplete.
Figure 3.
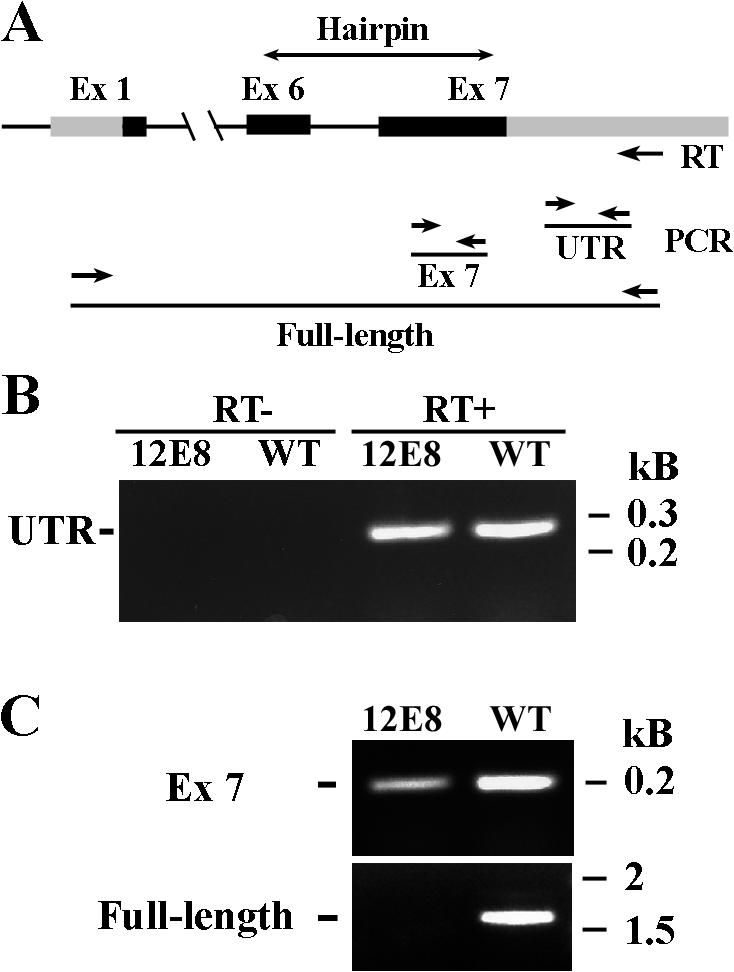
RT-PCR revealed that the endogenous RSP16 messages were transcribed and cleaved. (A) The schematic depicts the location of the primers (arrows) and products from the RT-PCR. (B) 3′ UTR was amplified from wild type and 12E8. No products were present in the control lacking reverse transcription. (C) RT-PCR of the seventh exon from 12E8 was much less robust than the wild type. Full-length cDNA was amplified from wild-type reverse transcription but not from 12E8.
The twitching phenotype was distinct. Despite the different degrees of defects, the spoke mutants pf14, 1, 17, and 24 had flaccid flagella that bent slowly and occasionally (Huang et al., 1981; for review see Kamiya, 2002), whereas the RSP16− twitching flagella appeared to be much more active but still insufficient to support cell procession. Interestingly, we found that the mutants that resembled RSP16− flagella best were two pf6 alleles that lacked the C1a CP projection. Notably though, a portion of cells swam regardless of a point mutation or deletion in the PF6 gene (Dutcher et al., 1984; Rupp et al., 2001).
No additional protein defects detected in the RSP16− axonemes
To assess the assembly of RS and the other major complexes, Western analyses of axonemes purified from two of the twitching strains (12E8 and 13B3) and the two paralyzed strains (12D5 and 13F3) were performed (Fig. 4). IC140 of inner dynein arm I1 served as a loading control. Spokeless pf14 axoneme was the negative control. RSP16 was present in the paralyzed flagella of 12D5 and 13F3. Importantly, although RSP16 was absent in the twitching flagella, the other RSPs in the head and stalk were normal, which was revealed by monospecific (Fig. 4 A, left and right) or antispoke complex antibodies (Fig. 4 A, middle). The faster migration of RSP11 and 16 (Fig. 4 A, left) in 12D5 strain was caused by slight irregular electrophoresis as revealed by protein staining. In particular, RSP2 and 23, which were diminished along with RSP16 in RSP2 mutant pf24, were normal in 12E8 (Fig. 4 A, right). To test whether RSP16 was involved in the conversion of 12S cytoplasmic precursor complex into mature 20S spoke complex, RS were extracted with 0.5 M KI from 12E8 and sedimented through 5–20% sucrose gradient. Western analyses showed that 12E8 RS lacking RSP16 still sediment at 20S (Fig. 4 B), similar to wild-type RS (Yang et al., 2001). Western analyses probing proteins from outer dynein arm (IC69), inner dynein arm (IC140), and CP (cpc1, HSP70, and pf20) indicated that these major axonemal complexes were normal (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200705069/DC1).
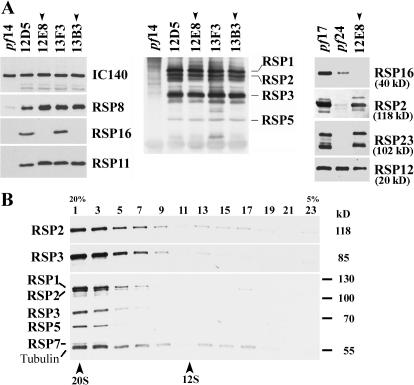
Figure 4.
RSP16 depletion did not result in gross assembly defect of the RS complex. (A) Western analyses of axonemes using specific RSP antibodies (left and right) and an anti-spoke complex antibody (middle) revealed that in 12E8 and 13B3 (arrowheads), the RSPs in the spokehead (RSP1) and spokestalk were normal, whereas RSP16 was not detectable. These proteins were normal in entirely paralyzed flagella (12D5 and 13F3). The spokeless mutant pf14 was a negative control. RSP2 and 23, which were diminished as RSP16 in pf24 axonemes, were also normal in 12E8 (right). The blot was probed sequentially for RSP23 and RSP2. RSP12, positive control. (B) Western blots probed with anti-RSP2, anti-RSP3, and anti-spoke complex showed that RS in 12E8 KI extract sediment in 20S fractions in 5–20% sucrose gradient as wild-type spokes but distinct from the 12S spoke precursors.
Collectively, these data showed that the RSP16− twitching flagella had no obvious defects in any major axonemal complexes, including RS.
Twitching flagella are caused by depletion of RSP16
To test whether the twitching flagella were caused by the lack of RSP16 or by accidental insertional mutagenesis, two independent approaches were taken.
First, 12E8 was backcrossed with wild-type strain cc620. 10 progenies were randomly selected for analyses of motility and flagellar RSP16 (Fig. 5 A). Six had twitching flagella and lacked RSP16, whereas the rest swam like wild-type cells and contained the same amount of RSP16 as wild type. Among the twitching group, only three were resistant to PMM. Thus, the phenotype of twitching flagella cosegregated with RSP16 depletion and was not correlated with the cotransformed pSI103. Consistently, transformation with the hairpin construct alone did not result in any RSP16− strains.
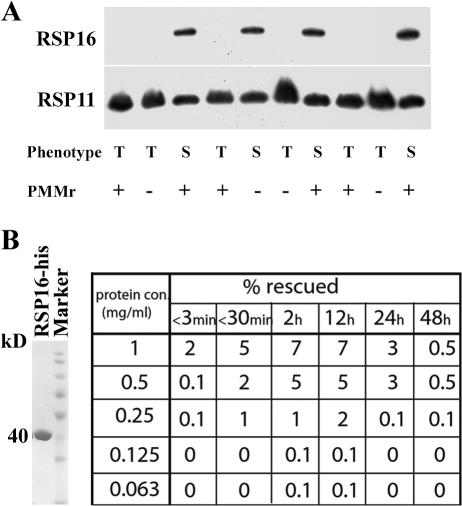
Figure 5.
Backcross and rescue with recombinant proteins indicated that RSP16 depletion resulted in twitching flagella. (A) RSP16− 12E8 was backcrossed with the wild-type strain cc620. 10 randomly picked progenies were scored for motility, antibiotic resistance, and the presence of RSP16 in flagella. Swimming cells (S) contained RSP16, whereas those with twitching flagella (T) did not. Absence of RSP16 was not correlated with PMM resistance. RSP11, loading control. (B) Coomassie protein gel showed Ni-NTA–purified recombinant RSP16-6his used for rescue of 12E8 (left). Approximately 2% of cells that were electroporated with 1 mg/ml of recombinant RSP16 became swimmers upon electroporation. The percentage of swimmers gradually increased within the first 12 h and decreased in 24 h or later. The efficiency of rescue was concentration dependent.
Independently, we adopted protein electroporation that was successfully used to rescue dynein mutants with recombinant dynein light chains (Hayashi et al., 2002). If RSP16 depletion caused twitching flagella, the RSP16− 12E8 cells would start swimming as RSP16 was restored. To test this, His-tagged RSP16 was purified from bacteria with Ni-NTA affinity chromatography (Fig. 5 B, left; Yang et al., 2005) and electroporated into 12E8 cells. Immediately after electroporation, swimmers were visible among jiggling cells (Fig. 5 B, right). The rescue efficiency (Fig. 5 B, % rescue) improved with increased protein concentration and time for up to 12 h after electroporation. Fewer swimmers were observed the next day, likely because of electroporation-related cell death. Swimming cells were never observed in controls electroporated with buffer only or with recombinant RSP11.
DnaJ domain in RSP16 was not required for motility rescue
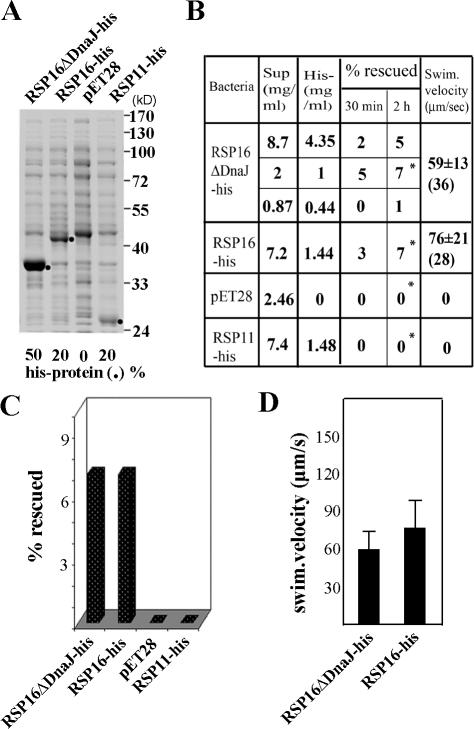
To test whether the signature DnaJ domain was required, we engineered a construct expressing RSP16ΔDnaJ-his lacking the corresponding N-terminal 70-aa residues. The soluble truncated protein in the bacterial extract (Fig. 6 A, left) precipitated upon Ni-NTA purification (not depicted). Thus, electroporation was performed using soluble fractions of bacterial extract. Interestingly, the supernatant containing recombinant RSP16-his or RSP16ΔDnaJ-his rescued the twitching phenotype but supernatant alone or supernatant with RSP11-his did not (Fig. 6, B and C, compare data with asterisks for similar amount of molecules). The velocity of swimmers rescued by ΔDnaJ was slightly slower (Fig. 6 D; P < 0.001). However, both groups were comparable to the electroporated wild-type cells (mean, 74.0 ± 27.5 μm/s from 100 cells) but all were significantly slower than the nonelectroporated wild-type control (111.2 ± 21.4 μm/s from 34 cells; P = 0.000). The lower velocity may be related to cell death from electroporation. See Videos 1 and 2 (available at http://www.jcb.org/cgi/content/full/jcb.200705069/DC1) for rescue with RSP16ΔDnaJ-his and control RSP11-his.
Figure 6.
The DnaJ domain in recombinant RSP16 was dispensable for rescue of RSP16− cells. (A) Coomassie protein gel showed the bacterial extract used for electroporation. Purified RSP16ΔDnaJ-his precipitated and, thus, was not suitable for the experiment. The extract was prepared from IPTG-induced bacteria expressing truncated RSP16 (RSP16ΔDnaJ-his), RSP16-his, and the control RSP11-his (dots) or pET28(a) vector only. The percentage of recombinant protein was based on digital quantification of the protein gel. (B) The table lists the protein concentration and the results obtained at 30 min and 2 h after electroporation. Extract containing RSP16, both with or without the DnaJ domain, restored swimming ability to some 12E8 cells. No swimmers were found in the negative controls. Cells were digitally recorded 2 h after electroporation. Randomly selected swimmers (numbers indicated in parentheses) were analyzed for velocity. (C and D) For clarity, the percentage of swimmers (asterisks in B) was plotted as a histogram (C), as was velocity of swimmers rescued by similar concentration of recombinant proteins (D). The RSP16ΔDnaJ-his group was slightly slower than the intact RSP16 group (error bars represent SD; P < 0.001).
Diverged HPD tripeptide among candidate spoke HSP40 orthologues
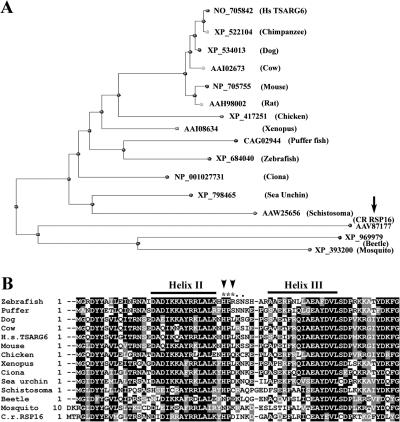
The dispensable DnaJ domain suggested that the conserved features for stimulating ATPase may diverge among the spoke HSP40 orthologues. Previous BlastP with C. reinhardtii or C. intestinalis spoke HSP40 showed that DnaJB13 (TSARG6) enriched in mammalian testis may be the orthologues (Satouh et al., 2005; Yang et al., 2005). Reverse Blastp using human DnaJB13 revealed a phylogenic tree of ∼288 HSP40s. Notably, grouped into a branch were C. intestinalis HSP40, RSP16, and one TSARG6-like HSP40 per species ranging from single cell organisms to vertebrates (Fig. 7 A). Interestingly, the sequence at the exposed loop between helix II and III in the DnaJ domain (Szyperski et al., 1994), including HPD tripeptide and the subsequent R and N (Fig. 7 B, dots) residues that were crucial for stimulating HSP70s (Genevaux et al., 2002), diverged significantly (Fig. 7 B). Actually, HPD in this region was only present in C. reinhardtii, C. intestinalis, and sea urchin. The divergence was significant because among 19 human type I and II J proteins (HSP40s), DnaJB13 was the only one lacking HPD (Qiu et al., 2006). Collectively, these results suggested that the members in the HSP40 subfamily lacking the HPD domain are RSP16 orthologues crucial for flagellar motility and that the feature for stimulating HSP70s is not absolutely required for spokes.
Figure 7.
HPD residues crucial for stimulation of HSP70 ATPase activity diverged in candidate orthologues of spoke HSP40. (A) Blastp in NCBI with human TSAR6 revealed a branch of an HSP40 phylogenic tree. The branch contained C. reinhardtii RSP16 (arrow), C. intestinalis spoke HSP40, and one TSAR6-like molecule from evolutionarily diverse organisms. Partial or redundant sequences were removed for clarity. (B) Multiple sequence alignment of DnaJ domains in TSAR6-like molecules from A demonstrated extensive sequence homology. However, the residues in the loop between the sequence corresponding to helixes II and III diverged significantly. Notably, the HPD tripeptides (asterisks) were only retained in the spoke HSP40 from C. reinhardtii, C. intestinalis, and sea urchin. H, D (arrowheads), and the two subsequent residues (dots) for stimulating HSP70 enzymatic activities diverged. The identical and similar residues are shaded in black and gray, respectively.
Morphology of HSP40− RS
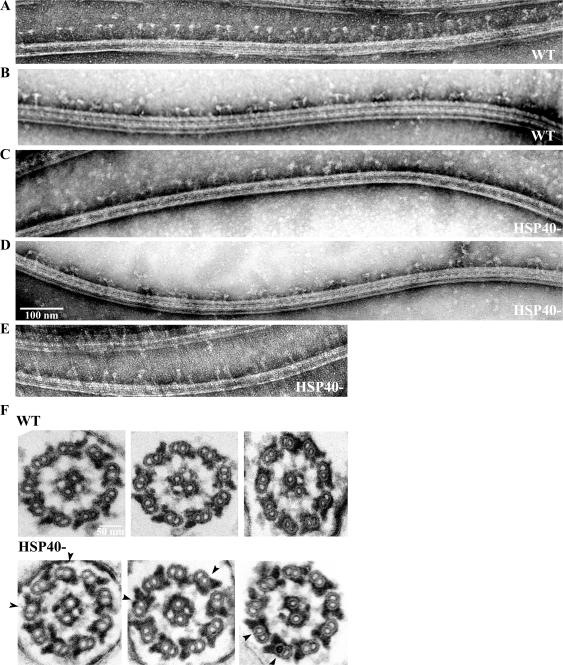
To assess the morphology, transmission electron microscopy was performed. Negative-stained RS from splayed wild-type flagella were easily recognized based on doublets of the T-shaped structure with a condensed electron-translucent spokehead (Fig. 8 A). Sometimes spokeheads appeared less typical (Fig. 8 B). Nonetheless the spokeheads remained distinctive. RSP16− axonemes did splay well; however, we had not encountered the typical T-shaped spokes. The 3 best images among 11 revealed that spoke-like doublets tended to flop or appear stretched, whereas the spokehead region seemed less distinctive (Fig. 8, C–E). However, the variation may be caused by the force that ripped the associated outer doublets, the orientation as splayed doublets settled on the grids, or how the metal stain was deposited.
Figure 8.
Electron microscopy of negative-stained splayed or sectioned flagella. Doublets of the typical well-defined T-shaped RS with a compact spokehead and a thin stalk vertical to outer doublets (A) or less typical spokes (B) identified in wild-type samples. In RSP16− flagella, the doublets of spoke-like images were present but the head region appeared less defined and the stalks tilted more frequently (C–E). (F) Cross sections showed wild-type RS with a bulbous spokehead abutted against CP and a long stalk associated to outer doublets (top). The RSP16− RS appeared less homogenous. A few spokes in each section often appeared with a Y, J, or stunted T shape with a shorter stalk and a deformed head farther away from the CP apparatus (arrowheads).
To evaluate spoke morphology in original context, sectioned flagella were observed. In wild-type flagella, RS anchored at the outer doublets projecting inward with the bulbous spokeheads abutted against CP (Fig. 8 F, top). HSP40− sections could be differentiated because of less homogeneous RS. Some spokes were indistinguishable from wild-type ones, whereas one to three spokes in most sections had a wider or bent spokehead and a shorter spokestalk, rendering Y-, J-, or stunted T-shaped spokes that were located a greater distance from the CP than those in wild type (Fig. 8 F, bottom, arrowheads). The preferential orientation of the asymmetrical CP toward a specific outer doublet in disintegrating axonemes and bending flagella (Wargo and Smith, 2003; Mitchell and Nakatsugawa, 2004) was not so obvious in isolated flagella (unpublished data). Overall, there were no obvious defects in composition and morphology.
Distinct motility phenotypes of RSP16− flagella
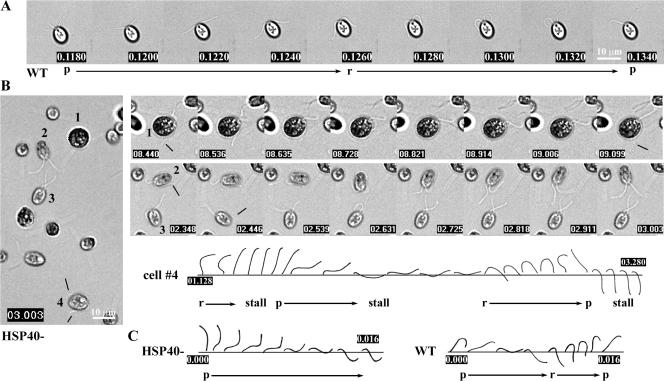
To characterize the defective motility, high-speed video microscopy of HSP40− and wild-type cells were compared (Fig. 9). Images of wild-type flagella beating at ∼60 Hz were captured at a frame rate of 500 Hz to show breast stroke–like asymmetrical waveform in 9–10 frames. As shown previously (Ringo, 1967; Brokaw and Luck, 1983; Brokaw and Kamiya, 1987), for power stroke (Fig. 9 A, p), flagella bent near flagellar base but the rest remained fairly straight to generate maximal forward propulsive force (Fig. 9 A and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200705069/DC1). For recovery stroke (Fig. 9 A, r) that restored flagella to the original position, bend propagated from base to tip, bringing flagella near the cell body to reduce backward movement. Importantly, recording of biflagellate cells clearly showed that power stroke started before recovery stroke completed, rendering a curved flagellar tip (Fig. 9 A, first and last panels).
Figure 9.
Video microscopy demonstrated the uncoupled power and recovery strokes of HSP40− twitching flagella. (A) Images recorded at 500 Hz revealed the asymmetrical waveforms of power (p) and recovery strokes (r) in each beat cycle of a forward-swimming wild-type cell. Note that the power stroke bends near the flagellar base before the recovery stroke is completed, rendering a curved flagellar tip (first and last panels). (B) HSP40− jiggling cells were taken at a frame rate of ∼12 Hz. The four cells that were not stuck to the slide (left) were further analyzed. (1) Two flagella crossing each other at the anterior end of the cell body after sequentially undergoing a sluggish full recovery stroke without the subsequent power stroke. The next powerful stroke without subsequent recovery stroke sent one flagellum underneath the cell body (lines point to the flagellar tips). (2) One flagellum stalled at the beginning of an active stroke. Upon completion, both flagella recovered to the normal position. (3) Both flagella beat alternately first but then stalled at the end of recovery stroke, rendering crossed flagella as well. (4) A traced diagram showed that one flagellum, after stalling, bent near the base but stalled again in the middle of the power stroke. After wavering, the flagellum underwent recovery stroke followed by a swift power stroke and stalled. (C) Recording at 500 Hz showed that swift power strokes also bend near the base but ∼50% slower than wild type. The recording was stamped in seconds.
The HSP40− cells were first filmed at 10–12 Hz for an ∼2-Hz irregular twitching rate. In general, flagella were found in various positions (Fig. 9 B; and compare Videos 4 and 5, available at http://www.jcb.org/cgi/content/full/jcb.200705069/DC1) in contrast to the hands-up position of most paralyzed CP and RS mutants (Lechtreck and Witman, 2007; Video 4). Notably, two flagella often crossed each other in front of the cell body (Fig. 9 B, cell 3), a pattern not seen in wild-type cells but observed in CP kinesin-knockdown cells (Yokoyama et al., 2004). Analyses of the time-lapse images of three cells and flagella traced from the fourth cell as examples demonstrated the actual anomaly. First, the two flagella were not coordinated (Fig. 9 B, cells 1–3). Second, bend initiation between strokes was often delayed. Consequently recovery strokes were fully completed so that flagella passed midline and often times crossed each other (Fig. 9 B, cells 1 and 3). Normally, power strokes occurring before the completion of recovery strokes prevented such incidence in wild-type cells (Fig. 9 A). Delayed recovery strokes made flagella appear obstructed by the cell body (Fig. 9 B, cells 1, 2, and 4, lines point to the flagellar tip). Third, stalling occurred randomly, between or during strokes, and could last for seconds. The stalled flagella appeared to be struggling. Most interestingly, those stalled in the middle of the power stroke often aborted the stroke and switched to the recovery stroke (Fig. 9 B, cell 2, second and third panels; and cell 4). Fourth, bend propagation rate varied. The sluggish ones, often of recovery strokes, captured at 12 Hz (Fig. 9 B, cells 1 and 4), showed similar bend propagation to the wild type, albeit slower. For analyzing the occasional fast strokes (Fig. 9 B, cell 2, frames 3 and 4; cell 3, frames 1 and 2; and cell 4), HSP40− cells were recorded at a 500-Hz frame rate. A power stroke was completed in 0.016 s, about half as slow as the wild type, and occurred primarily near the base (Fig. 9 C). The crossed flagella, sometimes with a seemingly large reverse bend (Fig. 9 C, first two frames), were first interpreted as a symmetric waveform but were actually crossed because of overextended recovery strokes of asymmetrical waveform based on frame-by-frame analyses. Collectively, the video microscopy showed uncoordinated and uncoupled power strokes and recovery strokes, and the waveform was predominantly asymmetrical.
Discussion
It has been assumed that RS and CP confer the motile capacity of 9 + 2 axonemes that 9 + 0 axonemes lack and are preferred wherever powerful propulsion is desired. By taking reverse genetics, removing spoke HSP40 without grossly affecting the composition of RS or axonemes, this study suggests that one HSP40 is positioned to fine tune the RS, enabling the structural complex to coordinate dynein-driven interdoublet sliding to produce alternate strokes of 9 + 2 cilia and flagella.
RNAi and rescue with recombinant proteins
Three independent lines of evidence indicate that absence of RSP16 (the spoke HSP40) results in the twitching flagella. First, two HSP40− clones were recovered from >200 transformants in each of two independent experiments. Second, the twitching phenotype from transformants and backcross progenies are strictly linked to the absence of RSP16 but irrelevant to the cotransformed antibiotic-resistant plasmid (Figs. 2 B and 5 A). Most importantly, recombinant RSP16, but not control protein or buffer alone, restores the motility with little delay (Figs. 5 B and Figs.6).
Two properties in RSP16− cells are unusual for RNAi in C. reinhardtii (Rohr et al., 2004; Schroda, 2006). First, the phenotypes of the four strains have remained stable without selection pressure for 2 yr. Second, RSP16 is undetectable in axonemes. Great care was taken to demonstrate the transcription of hairpin construct and cleavage of endogenous RSP16 mRNA in RSP16− cells by using DNase-treated RNA template and proper controls. The cleavage is likely mediated by the complex RNAi mechanism recently recognized in C. reinhardtii (Molnar et al., 2007). Comprehensive characterization of the RNAi mechanism in C. reinhardtii may shed light on effective knockdown in this study.
Rescue with recombinant proteins is a compromise for the dominant-negative nature of RNAi. As in transformation with DNA, only a small fraction of electroporated cells can take in a sufficient amount of proteins and survive. The rescue is transient as well. Nonetheless, the few rescued swimming cells are easily noted amid paralyzed cells. This method provides an alternative for confirming motility defects from RNAi.
Unique roles and mechanisms of HSP40/HSP70 in cilia and flagella
RSP16 is certainly an unusual HSP40. In addition to being a constitutive component of a structural complex unrelated to protein-folding machinery, its DnaJ domain is not essential and the HPD tripeptide diverges in candidate orthologues in higher deuterostome. Consistently, recombinant RSP16 fails to help HSP70 refold denatured luciferase (unpublished data). The normal length of RSP16− twitching flagella and immediate rescue of motility by electroporated protein also indicates that spoke HSP40 is not involved in ciliogenesis or elaborate assembly process but specifically serves the purpose of RS. These results indicate that the role of spoke HSP40s is not to help HSP70s to fold a broad spectrum of polypeptides with the signature domain at the N terminus. Rather, it is to bind and buttress the structural complex through the dimeric C terminus. Nonetheless, it is premature to conclude that J domain is useless or to exclude HSP70. The sequences of DnaJ domain are highly homologous among candidate orthologues and the truncated spoke HSP40 is less soluble. We speculate that J domain, although not essential, may recruit additional molecules, such as HSP70, to help the binding of electroporated or endogenous RSP16 with RS.
This conclusion raises questions related to flagellar chaperones, especially HSP70 ATPase, which can't fold protein efficiently by itself (Laufen et al., 1999). Are there other J proteins in these organelles? Are flagellar HSP70s dependent on J proteins after all and what could HSP70s do without an HSP40? Some flagellar HSP70s, such as HSP70A in CP (Mitchell et al., 2005), may not necessarily mediate constant protein folding as RSP16 does and do not rely on J proteins. It is likely that the properties of some chaperone molecules are used differently in cilia and flagella.
Final touch of RS by HSP40
Assembly of 12S precursors, 12S to 20S conversion, and incorporation of 20S spokes into axonemes are independent of HSP40. Binding of HSP40 occurs near the end of the assembly process. The predicted location of RSP16 in RS is in contrast to broad client polypeptides of HSP40s. Notably, the theoretical isoelectric point of proteins near the spokehead are around five or fewer, in contrast to seven for HSP40, the most basic RSP (Yang et al., 2006). C. intestinalis spoke HSP40 was particularly basic as well (Satouh et al., 2005). Thus, unlike the hydrophobic interaction of HSP40s and client polypeptides (for review see Craig et al., 2006), ionic interaction may also be involved in the specific binding of spoke HSP40.
The binding does not obviously change conformation or morphology of RS. The result is reasonable considering the dimeric HSP40 is only 1/15 of the ∼1,200-kD spoke particles (Padma et al., 2003). In addition, HSP40s are known to bind and release client peptides rather than to alter protein conformations. Perhaps spoke HSP40 may simply secure molecular interaction of the spokehead region.
Roles of RS
Despite inconsistent predictions regarding the roles of CP and RS, the simplest, yet inclusive, model is that the CP–RS system governs the rudimentary motile machinery of nine outer doublets (for review see Kamiya, 2002) to produce regulated powerful beating by acting as a mechanochemical transducer (for review see Smith and Yang, 2004). Faulty mechanical transduction could adequately explain the HSP40− flagellar phenotype.
In contrast to the paralyzed pf17 flagella in which spokeheads are absent, the active twitching flagella suggest that HSP40− 20S RS actually could engage CP but the engagement is insufficient or inappropriate. Regardless of the abnormal crossing of two flagella, frame-by-frame image analysis revealed no major changes in the asymmetrical waveform. Instead, the anomaly includes delay in bend initiation and propagation as well as sporadic stalling. These observations indicate that the primary defect is the timing rather than the order of sequential dynein activation around the circumstance and along the length of axonemes. The shaking of stalled flagella shows that forces are evident but cannot be coordinated, supporting the predicted antagonistic dynein-driven forces that are supposed to be coordinated by CP–RS but become apparent when CP–RS is defective (Yagi et al., 1994; Sakakibara et al., 2004; Lindemann, 2007). The untimely stroke switching, perhaps caused by the unregulated competing force, is consistent with the demonstration of interdoublet sliding triggered by forced bend (Morita and Shingyoji, 2004). The waveform, which sometimes deviated from the typical ones, (Fig. 9 C) is likely also because of the occasional uncoordinated force.
The mechanistic flaw of HSP40− spokes is unclear. It is possibly at the cyclic engagement and disengagement of CP–RS (Warner and Satir, 1974) or at the subsequent transduction that possibly redistributes t-force or maneuvers outer doublets leading to oscillation of flagellar diameter (Sakakibara et al., 2004;Lindemann, 2007). HSP40− RS that deform more readily may not execute at either level consistently and precisely. Perhaps the peptide binding regions of dimeric HSP40 bundle the multiple spoke proteins into a stable distal end (Fig. 1) that exposes the proper interface, provides the appropriate elasticity and rigidity for distortion or force distribution, or prevents the spokehead from falling apart upon encountering strain.
The prediction may be partly applicable to the twitching flagella of pf6, which lacks C1a projection and the corresponding five polypeptides (Dutcher et al., 1984; Rupp et al., 2001; Wargo et al., 2005). Importantly, some pf6 cells swim but HSP40− cells can't, suggesting that C1a projection and additional CP components, such as kinesin- or hydin-associated complexes (Yokoyama et al., 2004; Lechtreck and Witman, 2007), are involved in the same regulatory pathway as spoke HSP40. Perhaps any defect interfering with the transduction between CP and dynein motors will be manifested as twitching flagella of various severities. Collectively, the evidence suggests that the coordination of dyneins by CP–RS is so fundamental that it not only dictates bend formation and propagation (Warner and Satir, 1974) but also the switching of active dyneins to generate coupled power strokes and recovery strokes of planar waveform characteristics of 9 + 2 axonemes. Although other elements of the pathway may play a decisive role in stroke switching (Yokoyama et al., 2004; Lechtreck and Witman, 2007; Lindemann, 2007) and waveform determination, HSP40 at the head–stalk juncture is central to the fundamental mechanical feedback mediated by intermittently interacting CP–RS for timely coordinated activation of dynein motors. The timely coordination is likely crucial for effective beating that requires consistency in repeated bend formation and smooth propagation of bend at high frequency. The HSP40 specifically committed to RS at the end of spoke assembly is rightfully retained for the 9 + 2 nanomachine throughout evolution.
Materials and methods
Strains and culture conditions
C. reinhardtii strains, including wild-type strains cc124 (−) and cc620 (+) and the defined RS mutants pf14, pf17, and pf24 and the CP mutant pf18 cc1036(+), were acquired from the Chlamydomonas Genetics Center. The pf28pf30 strain lacks both the 20S outer arm dynein and inner arm dynein I1 as described previously (Piperno, et al., 1990). All cells were grown in liquid Tris-acetate-phosphate (TAP) medium under aerated photoheterotrophic growth condition in 14/10 light/dark cycle (Witman, 1986).
Molecular biology
Construction of plasmids with an inverted repeat.
A 400-bp PCR product, serving as the spacer of the hairpin, was amplified from the A. thaliana EARLI1 gene (Bubier and Schläppi, 2004) using the following primer pair with built-in double restriction sites (in italics): 5′-tCCATGGgaagatctaccttaaccgctgcaacag-3′ (NcoI and BglII) and 5′-tCATATGgggtaccgaagaacactgagagatatc-3′ (NdeI and KpnI). The spacer was cloned into pGEM-Teasy vector (Promega) and subsequently released by EcoRI digest. The band-purified fragment was ligated into the same site in the Maa7/× IR vector (obtained from J. Rohr and H. Cerruti, University of Nebraska, Lincoln, NE; Rohr et al., 2004), resulting in the following multiple cloning sites flanking the spacer: 5′-EcoRI, SpeI, NcoI, and BglII; and 3′-KpnI, NdeI, and EcoRI.
To construct inverted sequences, cDNA and genomic DNA fragment of RSP16 were ligated sequentially into the cloning sites flanking the EARLI1 spacer in an opposite orientation. A cDNA fragment of ∼500 bp was amplified from the construct pET28-RSP16-his (Yang et al., 2005) by using the following primer pairs with built-in restriction sites: sense 16s5, aggtacccttctgttcagcggcgagtacgtggag (KpnI); and antisense 16s3, acatatgggcggaaggaagaacgcgctgcgtag (NdeI). The cDNA fragment was ligated into the same restriction sites downstream of the spacer. Genomic DNA of ∼750 bp was amplified from wild-type genomic DNA by using the following primer pairs: sense 16gs5, aagatctaccatggcttctgttcagcggcg (BglII); and antisense 16s3. The PCR product was ligated into pGEM-Teasy vector first and subsequently released by BglII and SpeI digest. The genomic fragment was cloned into the same sites upstream of the spacer fragment. The hairpin construct was transformed into GC10 competent cell (GeneChoice, Inc.) and cultured for 24 h at 30°C. The hairpin fragment containing cDNA, spacer, and genomic DNA was released by EcoRI digest and ligated into pSI103 vector at the same site (Sizova et al., 2001) for transformation rescue.
Construction of expression vectors.
The coding sequence for RSP16ΔDnaJ-his construct was amplified using the pET28-RSP16-6his expression vector (Yang et al. 2005) as a template and the following primer pairs: sense T16S-NcoI, cccatggcgaaggggttttatgacctatacggag; and antisense 16fix3′EcoRI, ggggaattcctataccgcttgaacagcc. The 860-bp PCR product was directionally cloned into the same sites of the expression vector pET28(a). The RSP11− 6his expression vector was described previously (Yang and Yang, 2006).
Expression vectors were transformed into BL21(DE3) cells (EMD) and protein expression was induced with 1 mM IPTG at room temperature for 3–5 h. Soluble proteins were purified by Ni-NTA agarose (QIAGEN) in the native condition based on the manufacturer's instructions.
RT-PCR.
Reverse transcription with gene-specific reverse transcription primers was catalyzed by SuperScript III reverse transcription (Invitrogen) using total RNA extracted from 5–10 × 106 liquid-cultured cells with TRIZOL LS reagent (Invitrogen) and DNase treated with TURBO DNA-free kit (Ambion). The procedures followed the instruction of manufacturers. For amplifying the RSP16 transcript, the 3′ end reverse transcription primer was agctcaggctgacggcatagaag. PCR primer pairs for 3′ UTR fragment were the following: sense, aggcgctggataaggacatgtgtac; and antisense, agaagcagggtgctgaagattactcc; for exon 7: sense, catcggcaccacgcttgatatcc; and antisense, gtagcagcatcttttgcgtctcc; and for complete cDNA: sense, cggcgagctttgggcgattggta; and antisense agctcaggctgacggcatagaag.
For amplifying the spacer sequence in the hairpin transcript, reverse transcription primers were the following: sense, gttgcaacccaagtcctaagcacaag; and control antisense, ccaaaccttggatgagcgagcaaca. PCR primer pairs were the following: sense, aacccaagtcctaagcacaagcct; and antisense, agcaacatggttgagctgatggctg.
Biochemistry
Axonemal extraction and purification of 20S RS were performed as described previously (Yang et al., 2001).
Cell body extract.
Cells cultured on TAP plates were harvested and washed once with TAP medium. The cell pellets were suspended at a 1:1 ratio with 10 mM Hepes, pH 7.4, containing the following protease inhibitors (buffer A): one tablet of Complete protease inhibitor cocktail (Roche), 50 μg/ml aprotinin (Sigma-Aldrich), and 40 μl of saturated PMSF (Sigma-Aldrich) in isopropanol per 7 ml of buffer. The cell suspension was briefly sonicated until 95% of the cells were lysed (Sonicator W-225R; Ultrasonics Inc.). After centrifugation at 15,000 g for 15 min, the supernatant was diluted with 4 vol of buffer A and fixed by the addition of 5× sample buffer for SDS-PAGE.
Antibodies.
Anti-HSP70A monoclonal antibody was purchased from Affinity BioReagents (MA3-006). Polyclonal rabbit antibodies for RSP1, 2, 3, and 6 (obtained from D. Diener and J.L. Rosenbaum, Yale University, New Haven, CT); for RSP23 (obtained from S.M. King, University of Connecticut, Storrs, CT); for RSP8, 11, 12, and 16 (Patel-King et al. 2004; Qin et al., 2004; Yang et al., 2005, 2006); for CP protein CPC1 (obtained from D.R. Mitchell, State University of New York Upstate Medical University, Syracuse, NY; Zhang and Mitchell, 2004) and PF20 (obtained from E.F. Smith, Dartmouth College, Hanover, NH; Smith and Lefebvre 1997); and for inner dynein subunits p28 (Sigma-Aldrich) and IC140 (Yang and Sale, 1998) were described previously. Anti–outer dynein IC69 was provided by D.R. Mitchell. Chicken yolk antibody was raised against the 20S RS complex (Yang et al., 2005).
Genetics
Transformation.
The RSP16 hairpin construct was transformed or cotransformed with the pSI103 plasmid that confers PMM resistance (Zorin et al., 2005) into C. reinhardtii wild-type strain cc124(−) using the glass beads method (Kindle, 1990). Cells were plated on TAP plates with 10 μg/ml PMM after recovery under light overnight. Single colonies that formed after 4–5 d were saved on TAP plates and suspended later in TAP medium in 96-well plates for motility analysis using stereo microscopy.
Backcross.
The RNAi strain was crossed with wild-type strain cc620(+) according to the standard procedure (Yang and Yang, 2006), except that 2.5% agar (A-7921; Sigma-Aldrich) plates were used for zygote maturation and tetrad dissection.
Electron microscopy
Negative staining of splayed axoneme was performed as previously described (Yang et al., 2001). For sectioning, flagella were prepared for transmission electron microscopy as previously described (Mitchell and Sale, 1999). Silver sections were cut with a diamond knife using an ultramicrotome (Ultracut E; Reichert) and collected on 200 mesh copper grids. Silver sections were stained with 1% uranyl acetate and Reynold's lead citrate. Images were taken at 25–100 K using a transmission electron microscope (H-600; Hitachi) operating at 75 kV.
Protein electroporation
Electroporation was performed as previously described (Hayashi et al., 2002) with minor modifications. In brief, autolysin-treated cells were washed with solution E (0.2 mM ATP, 0.8 mM imidazole, 0.1 mM CaCl2, 0.5 mM 2-mercaptoethanol, and 60 mM sucrose, pH 7.5) and suspended with the same buffer into 1–2 × 108 cells/ml. Aliquots of suspensions were mixed with purified proteins in solution E or equal volumes of bacterial lysate in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0). A 125-μl aliquot was placed in a 2-mm electroporation cuvette (model 620; BTX Technologies, Inc.) and subjected to an electric pulse (ECM 600 electroporation apparatus; BTX Technologies, Inc.) of 500 V/capacitance and resistance, 1,200 μF capacitance, 24 Ohms, and 240 V. The cells were then allowed to recover in a water bath at room temperature for 30 min. The mixture was further diluted with 300 μl TAP medium and then transferred into a 1.5-ml tube for continued recovery under light. After 1 h, the cells were gently spun down by centrifugation and washed three times with TAP buffer. Cell suspension was observed immediately or at indicated time points after electroporation by compound light microscopy at 100×. The percentage of rescued cells was calculated as the ratio of swimming cells out of a total of ∼1,000 cells observed from randomly selected fields.
Motility assessment
For initial motility analysis, the mutant cells were digitally recorded under 400× bright field light microscopy using a charge-coupled device camera (CoolSNAP ES; Photometrics) at a maximum rate of ∼12 frames per second and a Plan Apo 40× objective with a 0.95 NA (Nikon). RNAi cells were observed after electroporation using 100× bright field light microscopy and were recorded similarly. Mean velocity of swimming cells was determined by tracing swimming paths of 20–30 swimming cells using MetaMorph 1.02 software (MDS Analytical Technologies) as previously described (Yang and Yang, 2006). The statistical analysis was performed using SPSS 10.0 (SPSS, Inc.) for Windows at one-way analysis of variance. For capturing fast movement, a high speed video camera (MotionPro HP-3; Redlake) was used at a rate of 500 frames per second.
Online supplemental material
Fig. S1 shows Western analyses of axonemes from various strains, which showed that RSP16 depletion did not affect the assembly of dyneins or the CP apparatus. Video 1 Shows that some HSP40− cells became swimming after electroporation with bacterial extract containing truncated recombinant RSP16 lacking the DnaJ domain. Video 2 shows that HSP40− cells remained jiggling after electroporation with control bacterial extract containing recombinant RSP11. Video 3 shows high-speed video microscopy of wild-type C. reinhardtii cells, which revealed the waveform of power strokes and recovery strokes in each beat cycle at the rate of ∼60 Hz. Video 4 shows that the flagella of spoke HSP40− cells twitched irregularly at the rate of ∼2 Hz. Video 5 shows that the mutant pf17 that lack radial spokeheads had paralyzed flagella that waved occasionally. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200705069/DC1.
Supplemental Material
Acknowledgments
We wish to thank J. Rohr and Dr. H. Cerutti (University of Nebraska) for providing plasmids used in hairpin construction; Drs. N. Wilson (University of Okalahoma) and J. Pan (Chinghwa University) for advice on RNAi; Drs. E.F. Smith (Dartmouth College), D.R. Mitchell (State University of New York Upstate Medical University), S.M. King (University of Connecticut Health Center), D. Diener and J.L. Rosenbaum (Yale University) for kindly providing antibodies; Drs. G.B. Witman and K. Lechtreck for comments on motility analyses and RT-PCR; and Drs. W.S. Sale and M. Wirschell (Emory University) for suggestions on the manuscript.
This work is supported by National Institutes of Health grant GM-068101 (to P. Yang).
Abbreviations used in this paper: CP, central pair; HPD, His-Pro-Asp; HSP, heat shock protein; PMM, paromomycin; RS, radial spokes; RSP, RS protein.
References
- Bloch, M.A., and K.A. Johnson. 1995. Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J. Cell Sci. 108:3541–3545. [DOI] [PubMed] [Google Scholar]
- Borges, J.C., H. Fischer, A.F. Craievich, and C.H. Ramos. 2005. Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J. Biol. Chem. 280:13671–13681. [DOI] [PubMed] [Google Scholar]
- Brokaw, C.J. 1982. Introduction: generation of the bending cycle in cilia and flagella. Prog. Clin. Biol. Res. 80:137–141. [DOI] [PubMed] [Google Scholar]
- Brokaw, C.J., and D.J. Luck. 1983. Bending patterns of Chlamydomonas flagella I. Wild-type bending patterns. Cell Motil. 3:131–150. [DOI] [PubMed] [Google Scholar]
- Brokaw, C.J., and R. Kamiya. 1987. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil. Cytoskeleton. 8:68–75. [DOI] [PubMed] [Google Scholar]
- Bubier, J., and M. Schläppi. 2004. Cold induction of EARLI1, a putative Arabidopsis lipid transfer protein, is light and calcium dependent. Plant Cell Environ. 27:929–936. [Google Scholar]
- Craig, E.A., P. Huang, R. Aron, and A. Andrew. 2006. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156:1–21. [DOI] [PubMed] [Google Scholar]
- Dentler, W.L., and J.L. Rosenbaum. 1977. Flagellar elongation and shortening in Chlamydomonas. III. Structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J. Cell Biol. 74:747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher, S.K., B. Huang, and D.J. Luck. 1984. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 98:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek, E.E., P.A. Lefebvre, and E.F. Smith. 2004. PF15p is the Chlamydomonas homologue of the Katanin p80 subunit and is required for assembly of flagellar central microtubules. Eukaryot. Cell. 3:870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowkes, M.E., and D.R. Mitchell. 1998. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell. 9:2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, L.C., E. O'Toole, C.A. Perrone, T. Giddings, and M.E. Porter. 1994. Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J. Cell Biol. 127:1311–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux, P., F. Schwager, C. Georgopoulos, and W.L. Kelley. 2002. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics. 162:1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough, U.W., and J.E. Heuser. 1985. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100:2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M., H.A. Yanagisawa, M. Hirono, and R. Kamiya. 2002. Rescue of a Chlamydomonas inner-arm-dynein-deficient mutant by electroporation-mediated delivery of recombinant p28 light chain. Cell Motil. Cytoskeleton. 53:273–280. [DOI] [PubMed] [Google Scholar]
- Hosokawa, Y., and T. Miki-Noumura. 1987. Bending motion of Chlamydomonas axonemes after extrusion of central-pair microtubules. J. Cell Biol. 105:1297–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., G. Piperno, Z. Ramanis, and D.J. Luck. 1981. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Z. Ramanis, and D.J. Luck. 1982. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 28:115–124. [DOI] [PubMed] [Google Scholar]
- Johnson, K.A., and J.L. Rosenbaum. 1992. Polarity of flagellar assembly in Chlamydomonas. J. Cell Biol. 119:1605–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, R. 2002. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 219:115–155. [DOI] [PubMed] [Google Scholar]
- Kindle, K.L. 1990. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 87:1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufen, T., M.P. Mayer, C. Beisel, D. Klostermeier, A. Mogk, J. Reinstein, and B. Bukau. 1999. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. USA. 96:5452–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.F., and G.B. Witman. 2007. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell Biol. 176:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann, C.B. 2007. The geometric clutch as a working hypothesis for future research on cilia and flagella. Ann. NY Acad. Sci. 1101:477–493. [DOI] [PubMed] [Google Scholar]
- Lindemann, C.B., and D.R. Mitchell. 2007. Evidence for axonemal distortion during the flagellar beat of Chlamydomonas. Cell Motil. Cytoskeleton. 64:580–589. [DOI] [PubMed] [Google Scholar]
- Meister, G., and T. Tuschl. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature. 431:343–349. [DOI] [PubMed] [Google Scholar]
- Mitchell, B.F., L.B. Pedersen, M. Feely, J.L. Rosenbaum, and D.R. Mitchell. 2005. ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol. Biol. Cell. 16:4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D.R. 2003. Orientation of the central pair complex during flagellar bend formation in Chlamydomonas. Cell Motil. Cytoskeleton. 56:120–129. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.R., and W.S. Sale. 1999. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 144:293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D.R., and M. Nakatsugawa. 2004. Bend propagation drives central pair rotation in Chlamydomonas reinhardtii flagella. J. Cell Biol. 166:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, A., F. Schwach, D.J. Studholme, E.C. Thuenemann, and D.C. Baulcombe. 2007. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 447:1126–1129. [DOI] [PubMed] [Google Scholar]
- Morita, Y., and C. Shingyoji. 2004. Effects of imposed bending on microtubule sliding in sperm flagella. Curr. Biol. 14:2113–2118. [DOI] [PubMed] [Google Scholar]
- Nicastro, D., C. Schwartz, J. Pierson, R. Gaudette, M.E. Porter, and J.R. McIntosh. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 313:944–948. [DOI] [PubMed] [Google Scholar]
- Omoto, C.K., T. Yagi, E. Kurimoto, and R. Kamiya. 1996. Ability of paralyzed flagella mutants of Chlamydomonas to move. Cell Motil. Cytoskeleton. 33:88–94. [DOI] [PubMed] [Google Scholar]
- Orban, T.I., and E. Izaurralde. 2005. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 11:459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski, L.E., K. Blackburn, K.M. Radde, M.B. Moyer, D.M. Schlatzer, A. Moseley, and R.C. Boucher. 2002. A proteomic analysis of human cilia: identification of novel components. Mol. Cell. Proteomics. 1:451–465. [DOI] [PubMed] [Google Scholar]
- Padma, P., Y. Satouh, K. Wakabayashi, A. Hozumi, Y. Ushimaru, R. Kamiya, and K. Inaba. 2003. Identification of a novel leucine-rich repeat protein as a component of flagellar radial spoke in the Ascidian Ciona intestinalis. Mol. Biol. Cell. 14:774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King, R.S., O. Gorbatyuk, S. Takebe, and S.M. King. 2004. Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol. Biol. Cell. 15:3891–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., N. Agrin, J. Leszyk, and G.B. Witman. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Z. Ramanis, E.F. Smith, and W.S. Sale. 1990. Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J. Cell Biol. 110:379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., K. Mead, M. LeDizet, and A. Moscatelli. 1994. Mutations in the “dynein regulatory complex” alter the ATP-insensitive binding sites for inner arm dyneins in Chlamydomonas axonemes. J. Cell Biol. 125:1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M.E., and W.S. Sale. 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151:F37–F42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., D.R. Diener, S. Geimer, D.G. Cole, and J.L. Rosenbaum. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X.B., Y.M. Shao, S. Miao, and L. Wang. 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63:2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringo, D.L. 1967. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J. Cell Biol. 33:543–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr, J., N. Sarkar, S. Balenger, B.R. Jeong, and H. Cerutti. 2004. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 40:611–621. [DOI] [PubMed] [Google Scholar]
- Rosenbaum, J.L., and F.M. Child. 1967. Flagellar regeneration in protozoan flagellates. J. Cell Biol. 34:345–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp, G., E. O'Toole, and M.E. Porter. 2001. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell. 12:739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara, H.M., Y. Kunioka, T. Yamada, and S. Kamimura. 2004. Diameter oscillation of axonemes in sea-urchin sperm flagella. Biophys. J. 86:346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir, P., and T. Matsuoka. 1989. Splitting the ciliary axoneme: implications for a “switch-point” model of dynein arm activity in ciliary motion. Cell Motil. Cytoskeleton. 14:345–358. [DOI] [PubMed] [Google Scholar]
- Satouh, Y., P. Padma, T. Toda, N. Satoh, H. Ide, and K. Inaba. 2005. Molecular characterization of radial spoke subcomplex containing radial spoke protein 3 and heat shock protein 40 in sperm flagella of the ascidian Ciona intestinalis. Mol. Biol. Cell. 16:626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda, M. 2006. RNA silencing in Chlamydomonas: mechanisms and tools. Curr. Genet. 49:69–84. [DOI] [PubMed] [Google Scholar]
- Seixas, C., C. Casalou, L.V. Melo, S. Nolasco, P. Brogueira, and H. Soares. 2003. Subunits of the chaperonin CCT are associated with Tetrahymena microtubule structures and are involved in cilia biogenesis. Exp. Cell Res. 290:303–321. [DOI] [PubMed] [Google Scholar]
- Sha, B., S. Lee, and D.M. Cyr. 2000. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure. 8:799–807. [DOI] [PubMed] [Google Scholar]
- Sizova, I., M. Fuhrmann, and P. Hegemann. 2001. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 277:221–229. [DOI] [PubMed] [Google Scholar]
- Smith, E.F., and P.A. Lefebvre. 1997. PF20 gene product contains WD repeats and localizes to the intermicrotubule bridges in Chlamydomonas flagella. Mol. Biol. Cell. 8:455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E.F., and P. Yang. 2004. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton. 57:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, R.E., and N.A. Lemieux. 1999. Molecular chaperones in cilia and flagella: implications for protein turnover. Cell Motil. Cytoskeleton. 44:274–283. [DOI] [PubMed] [Google Scholar]
- Szyperski, T., M. Pellecchia, D. Wall, C. Georgopoulos, and K. Wuthrich. 1994. NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2-108) containing the highly conserved J domain. Proc. Natl. Acad. Sci. USA. 91:11343–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi, K., T. Yagi, and R. Kamiya. 1997. Ca2+-dependent waveform conversion in the flagellar axoneme of Chlamydomonas mutants lacking the central-pair/radial spoke system. Cell Motil. Cytoskeleton. 38:22–28. [DOI] [PubMed] [Google Scholar]
- Wargo, M.J., and E.F. Smith. 2003. Asymmetry of the central apparatus defines the location of active microtubule sliding in Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA. 100:137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo, M.J., E.E. Dymek, and E.F. Smith. 2005. Calmodulin and PF6 are components of a complex that localizes to the C1 microtubule of the flagellar central apparatus. J. Cell Sci. 118:4655–4665. [DOI] [PubMed] [Google Scholar]
- White, D., S. Aghigh, I. Magder, J. Cosson, P. Huitorel, and C. Gagnon. 2005. Two anti-radial spoke monoclonal antibodies inhibit Chlamydomonas axonemal motility by different mechanisms. J. Biol. Chem. 280:14803–14810. [DOI] [PubMed] [Google Scholar]
- Warner, F.D., and P. Satir. 1974. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J. Cell Biol. 63:35–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B. 1986. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134:280–290. [DOI] [PubMed] [Google Scholar]
- Witman, G.B., J. Plummer, and G. Sander. 1978. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76:729–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi, T., and R. Kamiya. 2000. Vigorous beating of Chlamydomonas axonemes lacking central pair/radial spoke structures in the presence of salts and organic compounds. Cell Motil. Cytoskeleton. 46:190–199. [DOI] [PubMed] [Google Scholar]
- Yagi, T., S. Kamimura, and R. Kamiya. 1994. Nanometer scale vibration in mutant axonemes of Chlamydomonas.Cell Motil. Cytoskeleton. 29:177–185. [DOI] [PubMed] [Google Scholar]
- Yang, C., and P. Yang. 2006. The flagellar motility of Chlamydomonas pf25 mutant lacking an AKAP-binding protein is overtly sensitive to medium conditions. Mol. Biol. Cell. 17:227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C., M.M. Compton, and P. Yang. 2005. Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Mol. Biol. Cell. 16:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., and W.S. Sale. 1998. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol. Biol. Cell. 9:3335–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., D.R. Diener, J.L. Rosenbaum, and W.S. Sale. 2001. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 153:1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., C. Yang, and W.S. Sale. 2004. Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot. Cell. 3:72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., D.R. Diener, C. Yang, T. Kohno, G.J. Pazour, J.M. Dienes, N.S. Agrin, S.M. King, W.S. Sale, R. Kamiya, et al. 2006. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119:1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, R., E. O'Toole, S. Ghosh, and D.R. Mitchell. 2004. Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. USA. 101:17398–17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., and D.R. Mitchell. 2004. Cpc1, a Chlamydomonas central pair protein with an adenylate kinase domain. J. Cell Sci. 117:4179–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorin, B., P. Hegemann, and I. Sizova. 2005. Nuclear-gene targeting by using single-stranded DNA avoids illegitimate DNA integration in Chlamydomonas reinhardtii. Eukaryot. Cell. 4:1264–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.