Abstract
Endosomal sorting complex required for transport III (ESCRT-III) proteins function in multivesicular body biogenesis and viral budding. They are recruited from the cytoplasm to the membrane, where they assemble into large complexes. We used “deep-etch” electron microscopy to examine polymers formed by the ESCRT-III proteins hSnf7-1 (CHMP4A) and hSnf7-2 (CHMP4B). When overexpressed, these proteins target to endosomes and the plasma membrane. Both hSnf7 proteins assemble into regular approximately 5-nm filaments that curve and self-associate to create circular arrays. Binding to a coexpressed adenosine triphosphate hydrolysis–deficient mutant of VPS4B draws these filaments together into tight circular scaffolds that bend the membrane away from the cytoplasm to form buds and tubules protruding from the cell surface. Similar buds develop in the absence of mutant VPS4B when hSnf7-1 is expressed without its regulatory C-terminal domain. We demonstrate that hSnf7 proteins form novel membrane-attached filaments that can promote or stabilize negative curvature and outward budding. We suggest that ESCRT-III polymers delineate and help generate the luminal vesicles of multivesicular bodies.
Introduction
Multivesicular bodies (MVBs) are mid-stage endosomes that contain intraluminal vesicles (ILVs). ILVs are generated by invagination and scission from the limiting membrane of the endosome. Ultimately, most ILVs are delivered to lysosomes, enabling degradation of transmembrane proteins and lipids (Katzmann et al., 2002; Gruenberg and Stenmark, 2004; for reviews see Babst, 2005; Hurley and Emr, 2006). However, in certain specialized cells, MVBs also fuse with the plasma membrane to secrete their ILVs as entities called exosomes, which subserve a variety of important functions in the immune system and elsewhere (Stoorvogel et al., 2002; Thery et al., 2002; Fevrier and Raposo, 2004). A great deal of attention has recently focused on understanding how proteins are sorted into MVBs and how ILVs actually form.
Among the proteins involved in creating ILVs are at least 18 that were identified via genetic studies of vacuolar protein sorting in the yeast Saccharomyces cerevisiae. Interfering with the function of any these proteins leads to missorting of cargo normally destined for the ILV, causing it to accumulate on the limiting membranes of abnormal compartments that form adjacent to the yeast vacuole (its lysosome equivalent), the so-called class E compartments (Raymond et al., 1992). Homologues of these class E proteins are present in mammalian cells and their role in the formation of MVBs appears to be well conserved (for reviews see Hurley and Emr, 2006; Williams and Urbe, 2007). Most of the class E proteins have also been implicated in the topologically similar process of viral budding (Demirov and Freed, 2004; Morita and Sundquist, 2004).
All but one of the class E proteins are intrinsically soluble, cycling on and off the membrane as peripheral membrane proteins. Most are components of three large complexes, termed endosomal sorting complexes required for transport (ESCRTs), including ESCRT-I, -II, and -III (for reviews see Babst, 2005; Hurley and Emr, 2006; Slagsvold et al., 2006; Williams and Urbe, 2007). These complexes are thought to cooperate with each other (and with additional factors) to promote cargo selection and ILV formation. Signals that bring ESCRT complexes to the membrane include ubiquitin and phosphatidylinositol-3-phosphate, with current models suggesting that ESCRT complexes are recruited sequentially to ubiquitinated cargo on the endosomal membrane (for review see Hurley and Emr, 2006). It is not yet clear how ESCRT complexes orchestrate ILV formation but it is thought that they mostly dissociate from the invaginating membrane before the ILV is released because none end up at a high concentration within the lumen of the vesicle or viral particle (for review see Olver and Vidal, 2007). The release of ESCRT complexes from the membrane is probably driven by the AAA+ (ATPases associated with a variety of cellular activities) protein VPS4, of which there are two isoforms in mammalian cells, VPS4A and VPS4B/SKD1 (Babst et al., 1998).
ESCRT-I and -II are stable heterooligomeric complexes (Katzmann et al., 2001; Babst et al., 2002b). Each has recently been crystallized both in pieces and as a complex (Hierro et al., 2004; Teo et al., 2004, 2006; Kostelansky et al., 2006, 2007) and a reasonable understanding of how these complexes interface with ubiquitin, the membrane, and other components of the MVB sorting machinery is emerging (for reviews see Hurley and Emr, 2006; Williams and Urbe, 2007).
Less is known about the organization and interactions of proteins within the ESCRT-III complex. This complex consists of several related proteins that are recruited from the cytosol to the endosomal membrane, where they assemble into large detergent insoluble polymers (Babst et al., 2002a). Yeast has six ESCRT-III proteins, each a 200–250-residue protein with basic N-terminal and acidic C-terminal halves. Four of these proteins are thought to form the core ESCRT-III complex (Snf7p, Vps20p, Vps2p, and Vps24p), whereas two others (Did2p and Vps60p) associate with the core complex and may play regulatory roles (Babst et al., 2002a; Nickerson et al., 2006). The ESCRT-III family is expanded to 11 proteins in humans, which are referred to either as human orthologues of their yeast counterparts or as charged MVB proteins (CHMPs). Although each of the six ESCRT-III proteins in yeast is needed for normal MVB biogenesis, there may be some functional redundancy among the 11 human proteins. Understanding how ESCRT-III components work together and why so many related proteins are needed for MVB biogenesis clearly requires additional insight into the organization and function of the complex or complexes that they form.
A recent crystal structure of part of the ESCRT-III protein hVps24 (CHMP3) showed that it consists of an ∼7-nm-long α-helical hairpin buttressed by three shorter helices (Muziol et al., 2006). Based on homology, it seems likely that other ESCRT-III proteins share a similar structural core. Clues about how ESCRT-III proteins assemble into large complexes come from the several types of contacts between hVps24 protomers in the crystal, any or all of which could be involved in ESCRT-III polymerization.
Recent studies support the idea that individual ESCRT-III proteins have an intrinsic ability to bind to membranes that is regulated by autoinhibitory sequences located near their C termini (Muziol et al., 2006; Zamborlini et al., 2006; Shim et al., 2007). In one case (the crystallized fragment of hVps24), basic residues spread along a gently curved surface have been shown to be necessary for membrane binding (Muziol et al., 2006) and similar charge-based interactions may be at least partially responsible for association of the other ESCRT-III proteins with membranes. Binding to the membrane is thought to coincide with assembly of the ESCRT-III complex, leading to a model in which “closed” ESCRT-III proteins are soluble, whereas “open” ESCRT-III proteins on the membrane are available for interaction with other ESCRT components. Experimentally, membrane binding and complex assembly can be elicited by deleting C-terminal autoinhibitory sequences (Shim et al., 2007). Such deletions are thought to mimic the action of cellular ESCRT-III binding partners. One example of a binding partner that recruits and activates an ESCRT-III protein is the ESCRT-II subunit Vps25 (EAP25 in mammalian cells), which binds to the N-terminal half of Vps20 (Teo et al., 2004; Yorikawa et al., 2005; Langelier et al., 2006). Other examples of interacting proteins opening ESCRT-III proteins seem likely to emerge.
Once assembled into complexes on the membrane, it is not yet clear how ESCRT-III proteins contribute to luminal vesicle formation or viral budding. Genetic studies in the yeast S. cerevisiae suggest that they act late in the process, after ESCRT-I and ESCRT-II. Missing from current models is an understanding of which factors drive the required membrane deformation and eventual separation of ILVs from the limiting membrane of the endosome (for reviews see Hurley and Emr, 2006; Williams and Urbe, 2007). In the present study, we find that the ESCRT-III proteins hSnf7-1/CHMP4A and hSnf7-2/CHMP4B assemble into circular membrane-associated polymers that can be engaged to deform the membrane to which they are attached. We propose that membrane-associated ESCRT-III polymers similar to these may contribute to delineating and generating vesicles within the lumen of the MVB.
Results
hSnf7 assembles into homopolymeric filaments on the membrane
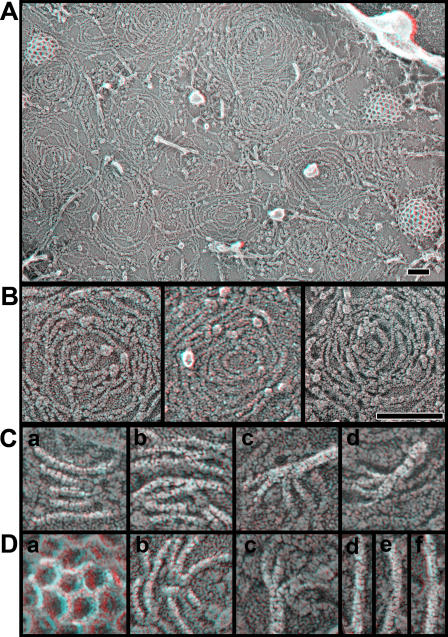
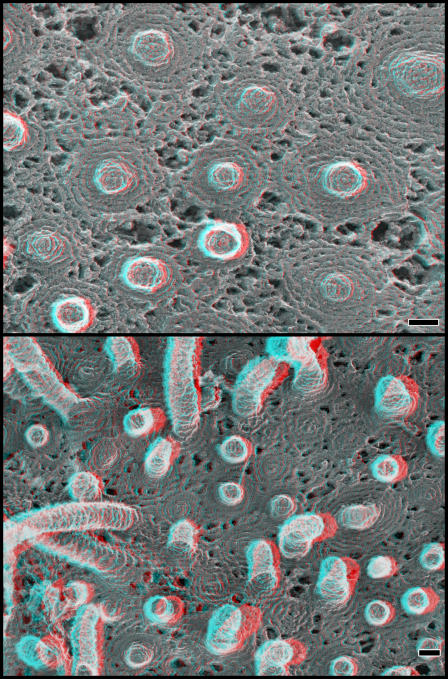
To study the organization of ESCRT-III–containing polymers by quick-freeze deep-etch EM (DEEM), we took advantage of our previous observation that overexpressed hSnf7 (CHMP4) proteins accumulate in patches on or adjacent to the plasma membrane as well as on internal, mostly endosomal, compartments (Lin et al., 2005; Shim et al., 2007). The plasma membrane is more accessible to DEEM than internal organelles because cells need only be “unroofed” to obtain expansive images of it and structures attached to it (Heuser, 2000a,b). The anaglyph in the top of Fig. 1 shows a typical image of the inner surface of the plasma membrane of a COS-7 cell, which in this case is transiently transfected with a plasmid encoding full-length FLAG-tagged hSnf7-1 (CHMP4A). Visible on the membrane are the usual cortical cytoskeletal components, including actin filaments and polygonal clathrin lattices, but, in addition, there are abundant filaments that are curved and interconnected to form a variety of circular arrays. In some areas, these new filaments intermingle with actin and clathrin, whereas in other areas they are so abundant that they displace these normal residents of the plasma membrane. It is important to note that transiently transfected COS-7 cells produce ESCRT-III proteins at levels that greatly exceed the concentration of their endogenous counterparts (unpublished data), creating a situation in which we can study the behavior of transfected proteins without significant contributions from endogenous proteins or binding partners.
Figure 1.
hSnf7 proteins form curved filaments on the plasma membrane. Shown in 3D are anaglyphs of the inside of the plasma membrane of COS-7 cells expressing the constructs indicated. Use view glasses for the 3D structure (left = red). (A) Plasma membrane of cell expressing FLAG hSnf7-1. (B) Higher magnification views of membrane coated with filaments of FLAG hSnf7-1 (left), FLAG hSnf7-2 (center), and FLAG hSnf7-1 (right). Bars, 100 nm. (C) High magnification views of FLAG hSnf7-1 filaments. (D) Clathrin lattice (a), two additional panels of FLAG hSnf7-1 filaments (b and c), and three views of actin filaments (d, e, and f). Filaments in C and D have been highlighted for clarity. Panels in C and D are each 100 nm across.
Higher magnification DEEM views of cells expressing hSnf7-1 or the related hSnf7-2 (CHMP4B) show that the novel filaments have a unit diameter of ∼5-6 nm (including the ∼2-nm thickness of the platinum replica that coats them) and appear to be tightly associated with the plasma membrane (Fig. 1 B). Transverse striations can be seen faintly in many regions along the filaments. When most distinct, these striations repeat every ∼4 nm (Fig. 1 C, a). The filaments frequently associate with each other laterally, either forming focal contact points between single filaments (Fig. 1 C, b; and Fig. 1 D, b) or generating wider strands that contain two or more intertwined filaments. Larger bundles containing four or more strands tend to be less tightly associated with the membrane but are split into single-diameter filaments at the membrane (Fig. 1 C, c and d; and Fig. 1 D, c). Sometimes filaments branch without apparently changing their diameter, which suggests that there are numerous ways in which hSnf7 protomers interact with each other. The length of the hSnf7 filaments offers a straightforward explanation for why overexpressed hSnf7-1 remains insoluble after treatment with nondenaturing detergents (Lin et al., 2005; Shim et al., 2007). The connections between adjacent filaments may contribute to stabilizing the circular patterns formed by the filaments. Particles corresponding at least in part to transmembrane proteins appear to be confined both at the center of the hSnf7 arrays and between the filaments, which suggests that one function of hSnf7 polymers may be to control the distribution of proteins in the membrane in a manner similar to that previously proposed for actin filaments close to the plasma membrane (Morone et al., 2006).
Several observations indicate that these novel filaments are predominantly homopolymers of hSnf7. Most striking is their altered appearance when GFP is fused to the C terminus of hSnf7-1 (Fig. 2). Filaments built from hSnf7-1–GFP are wider and bumpier than those formed by hSnf7-1 alone, as would be expected from doubling the mass of each subunit (both hSnf7-1 and GFP are ∼25 kD in mass). Additional support for the idea that the filaments are primarily polymers of hSnf7 includes the fact that the filaments are present only in a subset of cells after transient transfection with hSnf7 and the fact that the filaments can be decorated specifically with gold-labeled antibody recognizing an epitope tag on hSnf7 (see Fig. 4, top; and not depicted).
Figure 2.
Filaments containing hSnf7-1–GFP show their GFP. Adding GFP to the C terminus of hSnf7-1 (hSnf7-1–GFP) creates bumpy, tightly wound filaments on the inner surface of the plasma membrane. Bar, 100 nm.
A difference between the filaments formed by hSnf7-1 with no tag or a small epitope tag and those formed by hSnf7-1–GFP is that the latter are frequently tightly associated with each other, often to the point of creating compact circular arrays that appear as confluent domains of protein. Such tight structures are not seen in cells overexpressing hSnf7-1 without GFP (compare Figs. 1 and Figs.2). Their formation is not caused by the dimerization of GFP because mutating GFP to reduce its affinity for itself (Snapp et al., 2003) does not change their appearance (not depicted). Fusing GFP to the C terminus of hSnf7-1 must therefore expose something within hSnf7-1 that enhances lateral interactions between filaments.
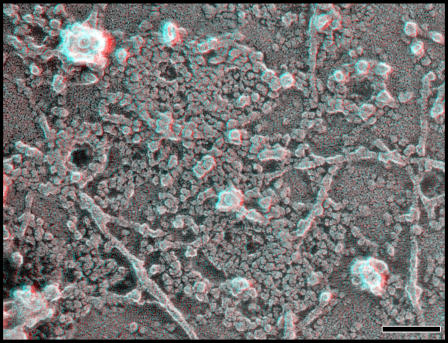
Two limitations to imaging unroofed cells are, first, that we can only examine the bottom or ventral surface of the cell where changes in the shape of the membrane are constrained and, second, that we have to lyse the cells during sample preparation. To look instead at the dorsal or top surfaces of nondisrupted cells, we switched to freeze-drying and platinum-replicating whole cells. As expected, cells overexpressing hSnf7-1 display areas with subtle circular patterns on their top surfaces that are comparable in size and organization to the curved filament arrays seen on the plasma membrane of unroofed cells (Fig. 3, top).
Figure 3.
hSnf7 filaments on the top surface of the cell. (top) Patterns created by hSnf7-1 filaments on the outer surface of whole cells. Shown is the top surface of a COS-7 cell transfected with FLAG hSnf7-1, fixed, and replicated without disruption. Note the subtle circular patterning of particles within the membrane. (middle) Views of hSnf7-1 filaments in the subplasmalemmal “membrane skeleton” revealed by extracting fixed whole cells with detergent. The cell on the left expresses FLAG hSnf7-1, whereas the one on the right does not. (bottom left) Fixed and extracted cell expressing higher levels of FLAG hSnf7-1. (bottom right) Fixed and extracted cell expressing hSnf7-1–mGFP. Bars, 100 nm.
To study the top surface of the cell in more detail, we applied a method for imaging the cell cortex in which cells are chemically fixed and then treated with a mixture of Triton X-100 and saponin to extract plasmalemmal lipids (Heuser and Kirschner, 1980). In nontransfected cells, this procedure provides views of a subplasmalemmal cytoskeleton that has been converted by chemical fixation into a microtrabecular meshwork (Fig. 3, middle, see cell on the right; Heuser, 2002). Cells expressing hSnf7-1 additionally contain characteristic domains of interconnected, roughly circular arrays of filaments (Fig. 3, middle and bottom). These filaments can be immunodecorated with a gold-conjugated antibody against a FLAG epitope tag on hSnf7-1, confirming that they contain hSnf7-1 (not depicted).
Both on unroofed plasma membranes and along the top surface of hSnf7-expressing cells, there is considerable variation in the curvature of individual filaments and the spacing between adjacent filaments that create a range of hSnf7-based arrays. A consequence of this variation that is most apparent when viewing the tops of cells is that in the closely spaced arrays, central rings tend to rise above those at the periphery (Fig. 3, bottom left). This phenomenon is particularly striking with polymers formed from GFP-tagged hSnf7-1 (Fig. 3, bottom right).
An important functional difference between hSnf7-1 with and without GFP fused to its C terminus is that overexpressed hSnf7-1–GFP strongly inhibits MVB maturation and viral budding, whereas hSnf7-1 lacking GFP does not (von Schwedler et al., 2003). This could be caused by the attached GFP perturbing the protein's normal closed conformation, effectively locking hSnf7 into an open state. Alternatively, the GFP might interfere with recruitment of specific C-terminal binding partners. Either way, we wondered whether there might be a correlation between the arrangement of hSnf7 filaments, membrane eversion, and the normal functioning of ESCRT-III proteins in the MVB pathway. We therefore decided to study the effects of manipulating hSnf7's C terminus by either adding binding partners or deleting domains.
A hydrolysis-defective mutant of VPS4B binds to hSnf7 polymers and promotes membrane eversion
Among proteins that interact with ESCRT-III family members, the most general appears to be the AAA+ ATPase VPS4 (of which there are two isoforms in mammalian cells, VSP4A and VPS4B/SKD1). This ATPase binds to most if not all of the individual ESCRT-III proteins, albeit with varying affinity (von Schwedler et al., 2003; Scott et al., 2005b; Lottridge et al., 2006; Tsang et al., 2006; Zamborlini et al., 2006; Shim et al., 2007). VPS4 is thought to hydrolyze ATP and disassemble ESCRT-III complexes, most likely by unfolding and removing individual protein subunits from the polymeric ESCRT-III complex. Recent studies reveal that VPS4 binds directly to a microtubule interacting and trafficking–interacting motif present near the C termini of CHMP1, CHMP2, and CHMP3 (Obita et al., 2007; Stuchell-Brereton et al., 2007). Other sequences are likely to be responsible for VPS4 binding to hSnf7 (CHMP4) proteins.
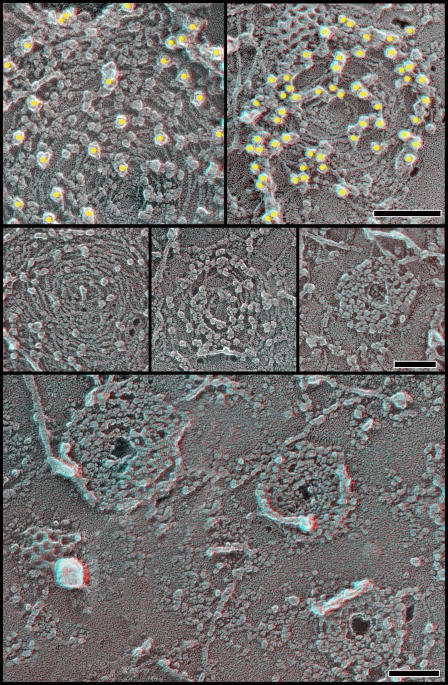
To gain insight into how VPS4 might affect the hSnf7 polymers examined here, we coexpressed a “substrate trap” mutant of VPS4B, VPS4B(E235Q), with hSnf7-1 in COS-7 cells. This mutant form of VPS4B is unable to hydrolyze ATP because of a change in its Walker B motif and therefore binds tightly to its protein substrates (Hanson and Whiteheart, 2005). We found that VPS4B(E235Q) with or without a GFP tag accumulated on hSnf7 filaments, where it appeared as a large particle along the cytoplasmic surface of the filaments (Fig. 4). These particles were recognized by antibodies against epitope tags or GFP attached to VPS4, which confirms that, as expected, they are VPS4B(E235Q) (Fig. 4, top). Their sizes range from ∼10 to ∼16 nm, which is significantly larger than expected for a VPS4B-GFP monomer and could instead be consistent with models of VPS4B operating as an oligomer (Scott et al., 2005a). Although the density of VPS4B particles varied widely, there were many arrays on which the particles were essentially confluent. These VPS4B oligomers not only obscured the underlying hSnf7-1 filaments but also tightened the arrays to create compact circular structures on the membrane.
Figure 4.
hSnf7/CHMP4 filaments bind VPS4B(E235Q). Anaglyphs of plasma membranes from COS cells expressing FLAG hSnf7-1 and VPS4B(E235Q)-GFP. (top) Immunodecoration with antibodies against FLAG tag on FLAG hSnf7-1 (left) and GFP in VPS4B(E235Q)-GFP (right). Yellow circles have been superimposed on the 18-nm gold particles for clarity. Note that gold particles obscure individual VPS4B particles only in the right panel. (middle) Circles of hSnf7-1 filaments with an increasing number of VPS4B(E235Q)-GFP particles bound. (bottom) Low magnification survey view of COS-7 cell plasma membrane showing hSnf7-1 arrays heavily decorated with VPS4B(E235Q)-GFP. Note that all three have small central holes. Bars, 100 nm.
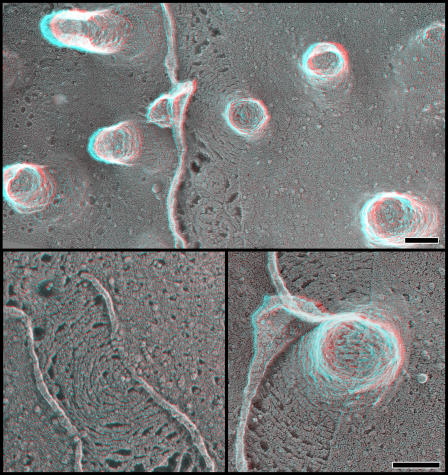
Importantly, the change in the hSnf7 filament arrays caused by binding to VPS4(E235Q) was accompanied by the appearance of distortions in the membrane at the center of the arrays. This could be glimpsed in views of unroofed plasma membranes, where dense hSnf7 arrays had small central gaps or holes that appeared slightly everted (presumably limited by the fact that the ventral surface of the cell cannot extend far before reaching the glass on which the cells are growing; Fig. 4). Much more obvious changes could be seen on the tops of freeze-dried whole cells. There, it was apparent that coexpressing hSnf7 and VPS4B(E235Q) led to the formation of buds and occasionally tubules that extended out from the cell (Fig. 5). These measured ∼100–120 nm in diameter and extended to varying heights. They could easily be distinguished from microvilli by their comparatively large size because microvilli are only ∼50 nm in diameter. Similar tubules developed on cells coexpressing hSnf7-2 and VPS4B(E235Q)-GFP as well as hSnf7-1 and VPS4B(E235Q)-myc (unpublished data).
Figure 5.
Buds and tubules protrude from the top surface of cells coexpressing hSnf7-1 and VPS4B(E235Q)-GFP. (top) Overview of fixed whole cell. (bottom) Higher magnification views of selected buds and tubules showing the range of observed structures. Bars, 100 nm.
We examined the protein scaffold lining these tubules by again extracting fixed cells with detergents, which revealed closely spaced and highly regular subplasmalemmal filaments with a unit diameter similar to the filaments seen on top of cells expressing only hSnf7 (Fig. 6). These filaments could be immunodecorated with antibodies against epitope tags on either hSnf7 or VPS4B (unpublished data). Note that there are no visible VPS4B(E235Q) particles on the detergent-extracted scaffolds because these are external views of the filaments and would not be expected to show proteins attached to the opposite cytoplasmic surface.
Figure 6.
Protein scaffolds line buds and tubules in cells coexpressing hSnf7-1 and VPS4B(E235Q)-GFP. Fixed whole cells extracted with detergents after fixation show submembranous skeleton. (top) Uniform budlike structures on region of a cell. (bottom) Buds and tubules of varying lengths along the surface of another cell. Bars, 100 nm.
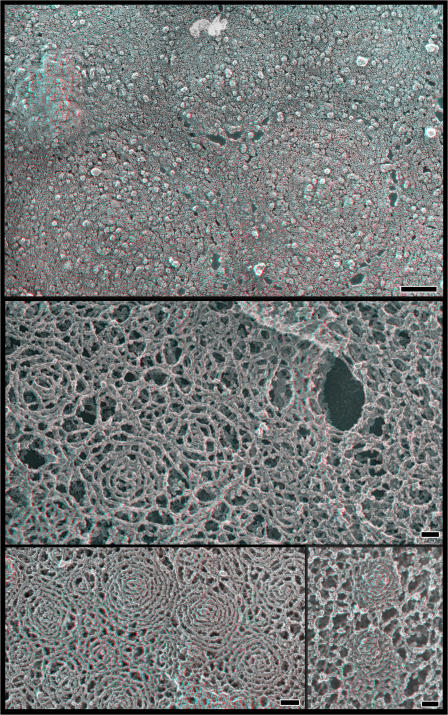
The subplasmalemmal scaffolds could also be seen to good advantage in replicas of unextracted cells when and where the membrane happened to peel back during the freeze-drying process (Fig. 7). This is a fortuitous occurrence that provides clean and undistorted views of submembranous protein lattices (unpublished data). In the case of cells expressing hSnf7 and VPS4B(E235Q), these views confirm that hSnf7 filaments extend beyond the perimeter of the buds and tubules to create a circular base. Examination of the spacing between adjacent filaments shows that the filaments are tightly and regularly packed within and directly around the everting tubules and may become less interconnected as the lattice extends further away from the tubule. Not all of the lattices have central eversions, perhaps because those that recruit less VPS4B do not undergo the required structural change.
Figure 7.
Spontaneous tears along the top surface of fixed whole cells reveal the fine structure of the underlying membrane skeleton (with no detergent treatment). (top) Survey view. (bottom) Higher magnification views. Bars, 100 nm.
Polymers formed by an hSnf7-1 fragment suggest a role for the N-terminal region in bud formation
To further explore the relationship between hSnf7 and membrane eversion, we studied the effects of removing the C-terminal half of the hSnf7-1 protein. We used an N-terminal hSnf7-1 fragment (residues 1–116) that we previously showed associated well with the plasma membrane (Lin et al., 2005). This fragment retains the core α-helical hairpin predicted by the CHMP3/hVps24 crystal structure (Muziol et al., 2006) but is constitutively open and does not interact with VPS4B (Lin et al., 2005). Strikingly, whole cells expressing hSnf7(1–116) have regions on their dorsal surfaces that are abundantly studded with distinctive buds (Fig. 8). These look similar to the buds seen on cells coexpressing full-length hSnf7 and VPS4B(E235Q), although they are narrower in diameter (∼80 vs. ∼100–120 nm) and only rarely elongate into tubules. Again, concentrated at the centers or apices of these buds are particles that probably correspond to trapped transmembrane proteins. Not surprisingly, plasma membrane buds were easier to find in cells that expressed a mutant form of hSnf7(1–116) in which a cysteine replaces serine at residue two because this mutant undergoes palmitoylation and is more efficiently targeted to the plasma membrane than normal (Lin et al., 2005).
Figure 8.
hSnf7-1 N-terminal fragment (1–116) drives formation of everting buds. (left) Top surface of a fixed whole cell expressing hSnf7-1(1–116). (top right) Filaments and large particles on the inner surface of the plasma membrane of cells expressing hSnf7-1(1–116). (right, middle) Freeze fracture image of membrane from cells expressing hSnf7-1(1–116) grown on sapphire. (bottom right) Selected views from fixed whole cells extracted with detergents after fixation. Bars, 100 nm.
Further examination of cells expressing hSnf7(1–116) (with either cysteine or the wild-type serine at residue two) revealed that fine filaments are visible on the plasma membrane. These can be seen on the inner surface of the membrane in unroofed cells (Fig. 8, top right) as well as in freeze-fracture images of cells grown on sapphire (Fig. 8, middle right) and on top of fixed whole cells that have been extracted with detergent (Fig. 8, bottom right). In many cases, the filaments associate laterally with each other and curve into circular arrays. The fact that the pattern of these filament arrays is particularly evident in freeze-fracture images demonstrates that the protein directly or indirectly affects the structure of the membrane bilayer. A peculiarity of the hSnf7(1–116) filaments visible in unroofed cells is that they are often decorated on their cytoplasmic surfaces with many copies of an unusually large particle (>20 nm in diameter). Whether these particles are normal binding partners for the ESCRT-III machinery (i.e., ESCRT-I and ESCRT-II complexes) or are something unrelated has yet to be determined.
VPS4B(E235Q) mutant accumulates in rings on membranes of cells even with no exogenous ESCRT-III expression
To determine whether filaments or scaffolds anything like those shown in the previous section form in cells expressing only endogenous levels of ESCRT-III components, we finally examined stable tetracycline-inducible HEK293 cell lines in which the ESCRT pathway can be inhibited by regulated expression of VPS4B(E235Q)-GFP (Lin et al., 2005). A few hours after adding tetracycline (when endosome biogenesis and viral budding are already impaired and endogenous ESCRT proteins are associated with VPS4B(E235Q); Lin et al., 2005), we prepared unroofed plasma membranes for DEEM, taking advantage of the fact that VPS4B(E235Q) can also be found on both endosomes and the plasmalemma (Lin et al., 2005; Booth et al., 2006). On these membranes, immunoreactive VPS4B(E235Q)-GFP again appears as particles 10–16 nm in diameter (Fig. 9). These particles typically appear in chains, as if they are decorating an underlying threadlike polymer on the membrane (likely one composed of endogenous ESCRTs, although we cannot rule out other possibilities). The chains of VPS4B(E235Q) particles often curve to form circles ∼100 nm or more in diameter, reminiscent of individual rings created by overexpressed full-length hSnf7. Sometimes, the circles of mutant VPS4B particles surround circular or helical arrays of filaments that distort the plasma membrane, pushing it outwards. One possibility is that these represent endogenous ESCRT-III filaments “corralled” by mutant VPS4. Alternatively, these could be coat components of an unidentified virus that is intrinsic to our cultured cells and whose budding has been blocked by expression of the mutant VPS4.
Figure 9.
VPS4B(E235Q)-GFP forms large corral-like rings on the plasma membrane of cells expressing only endogenous ESCRT-III proteins. (top) Overview of plasma membrane from a cell expressing only VPS4B(E235Q)-GFP. A GFP tag on VPS4 was used to selectively immunodecorate these particles with 18 nm gold. (bottom) Selected higher magnification views of rings formed by VPS4B(E235Q)-GFP. Bars: (top) 500 nm; (bottom) 100 nm.
Discussion
MVB biogenesis is governed at least in part by transient recruitment of ESCRT complexes and associated proteins to the limiting membrane of the endosome. Among the proteins involved in formation of ILVs must be factors that select membrane and cargo for incorporation into the ILV and factors that promote the membrane deformation needed to drive ILV formation. Here, we found that the ESCRT-III proteins hSnf7-1 (CHMP4A) and hSnf7-2 (CHMP4B) assemble into filamentous polymers on membranes. The circles formed by these filaments may contribute both to defining the contents of a nascent ILV and deforming the membrane to create it. Insight into the function of ESCRT-III proteins has been slow in coming (Russell et al., 2006) and the novel properties of the subset of ESCRT-III proteins described here should form the basis for further exploration of their role in ILV formation.
Structure of hSnf7 (CHMP4) polymers, novel filaments on the membrane
The most significant observation in this study is that hSnf7 proteins assemble into novel filaments that attach to the membrane and are capable of distorting it. Although we cannot define the arrangement of protein subunits within these filaments, some possibilities can be gleaned from the recent crystal structure of a fragment of another ESCRT-III protein, hVps24 (CHMP3) (Muziol et al., 2006). The core of this ESCRT-III protein is an ∼7-nm-long α-helical hairpin, which in the crystal binds to a partner subunit to form an antiparallel dimer. To fit such dimers into the smallest filaments seen here, the long axes of the helical hairpins would have to run roughly parallel to the filament. This could be accomplished by a simple tip-to-tip interaction between dimers as occurs in the crystal or by opening up the hairpins to allow other types of intersubunit interactions not seen in the crystal. The variety of interconnections that we see between adjacent hSnf7 filaments indicates that there is more than one way for these subunits to interact with each other, and coassembly of hSnf7 with other ESCRT-III proteins is likely to yet further expand the ways in which the subunits associate with each other. Our finding that filaments also form when the N-terminal half of hSnf7-1 (which contains little besides the α-helical hairpin) is expressed (Fig. 9) suggests that the N-terminal helical hairpin domain by itself is sufficient to allow polymer assembly.
Once assembled and attached to a membrane, hSnf7 filaments have an intrinsic tendency to curve (Figs. 1–3). This appears to derive from the shape of the subunits and/or the way in which they interact with each other. The degree of curvature is not fixed because the path taken by individual filaments varies widely (e.g., compare the curvature of a filament near the center of an array with that of one at its periphery). Interfilament contacts may contribute to stabilizing or even creating the circular arrays. When the filaments are tightly associated with each other and the membrane, their shape could play a role in deforming the membrane, as will be further discussed in the final section of the Discussion.
Relevance of hSnf7/CHMP4 polymers to a native ESCRT-III complex
It is important to emphasize that the polymers studied in this paper arise from overexpressed proteins and do not necessarily correspond to normal intermediates in the ESCRT pathway. Both up- and downstream components of the ESCRT pathway have been bypassed or overwhelmed by hSnf7 overexpression, allowing unopposed growth of hSnf7 polymers that may exaggerate or even change their normal effects on membranes. There are, however, several reasons to believe that these overgrown polymers reveal important characteristics of ESCRT-III proteins that are relevant to understanding how these proteins contribute to MVB biogenesis. These include the facts that the filaments (a) are highly organized, (b) bind specifically to relevant proteins such as VPS4B (Figs. 4–7), (c) bind tightly to both the plasmalemma and endosomes (this paper; Lin et al., 2005; Shim et al., 2007), (d) do not form nonspecific aggregates with other cellular proteins, (e) share properties with complexes of endogenous ESCRT proteins trapped by inhibiting the pathway in yeast, including being detergent insoluble and extremely large, and (f) resemble structures trapped by expressing mutant VPS4B(E235Q) on its own (Fig. 9). The fact that individual hSnf7 proteins homooligomerize into regular structures leads us to suggest that ESCRT-III complex could be either a defined heteropolymer, as is currently assumed, or a series of interconnected homopolymers. Our preliminary studies of other ESCRT-III proteins support the idea that each can form homopolymers that when opened or activated recruit other ESCRT-III components (Shim et al., 2007). Further exploration of the structure of complexes containing multiple ESCRT-III proteins and their effects on membranes will clearly be an important next step.
Although we focused our study on plasma membrane polymers for technical reasons (e.g., ease of visibility), there is good evidence that ESCRT proteins can function on the plasma membrane as well as on the endosome. Most proteins involved in MVB biogenesis (including hSnf7 and VPS4) have been implicated in the budding of several enveloped viruses, which at least in some cases happens on the plasma membrane (von Schwedler et al., 2003; Fisher et al., 2007; Fraile-Ramos et al., 2007). Moreover, several recent studies have shown that there are endosome-like domains in the plasma membrane, which in turn has led to the proposal that exosome-like vesicles might bud directly from the plasmalemma (Booth et al., 2006; Nydegger et al., 2006). Finally, the ESCRT machinery has recently been shown to be necessary for completing cytokinesis, again requiring function at the plasma membrane (Carlton and Martin-Serrano, 2007; Morita et al., 2007). Thus, at least in certain situations, the ESCRT machinery, probably including the ESCRT-III complex, is recruited to and operational at the plasma membrane.
ESCRT-III scaffolds and membrane deformation
Current thinking is that one way in which proteins deform membranes is by attaching tightly to them and imposing their intrinsic geometry upon them. Well-studied examples of this include the curved polymers formed by coat proteins (i.e., the binding of clathrin lattices to the plasmalemma during coated pit formation; Heuser, 1989a; Hinrichsen et al., 2006) and viral structural proteins (i.e., the binding of viral matrix proteins to the plasmalemma to form viral particles; Karacostas et al., 1989; Morita and Sundquist, 2004) as well as the more localized deformations associated with curved helical proteins containing Bin/amphiphysin/Rvs (BAR) domains and related structural motifs (Peter et al., 2004; McMahon and Gallop, 2005; Itoh and De Camilli, 2006; Shimada et al., 2007).
In the case of the ESCRT pathway and ILV formation, there has been to date little structural evidence for involvement of complexes with the ability to distort the membrane. Our images suggest that ESCRT-III subunits assembled into filaments and circular lattices may be able to drive and/or stabilize negative membrane curvature such as is needed to generate vesicles that bud away from the cytoplasm. The mechanism for these effects could be similar to that used by BAR domain–containing proteins (Peter et al., 2004; McMahon and Gallop, 2005), with the shape of the ESCRT-III polymers promoting negative rather than positive curvature. The idea that proteins with an “inverse” BAR domain shape might promote negative curvature (and membrane evolution) has been proposed (McMahon and Gallop, 2005) and, very recently, demonstrated in a study of two actin binding proteins involved in the formation of filopodia, missing in metastasis, and IRSp53 (Mattila et al., 2007). These proteins contain a gently curved helical domain and create everting tubules lined on their interior with the proteins. It is not difficult to imagine that dimers of the α-helical hairpin at the core of ESCRT-III subunits would use a positively charged, gently convex surface (Muziol et al., 2006) to create the kind of membrane distortions described in the present paper.
There are several questions that need to be answered before we can propose a specific model for the role of ESCRT-III polymers in MVB biogenesis. The most important is when and where the polymers assemble and are subsequently removed. Defining when ESCRT components dissociate from each other and the membrane is particularly important because their continued presence on the membrane of a vesicle after it is released from the limiting membrane would trap them inside the lumen of the vesicle. This is inconsistent with current data showing that ESCRT proteins are not present at a high concentration in either viral particles or ILVs and exosomes (for review see Olver and Vidal, 2007). One way out of this dilemma would be for ESCRT-III polymers to initiate membrane deformation but remain only at the perimeter or evolving neck of the vesicle, perhaps as a result of ongoing and closely coupled VPS4-driven disassembly of the ESCRT complexes. Changes in the lipid composition of the evolving vesicle could play a role in vesicle evolution, as has been previously suggested for lysobisphosphatidic acid, a cone-shaped lipid known to be present at high concentrations in ILVs (Matsuo et al., 2004).
The Snf7 subfamily of ESCRT-III proteins may have a unique and particularly important role in the steps leading to creation of ILVs, particularly if they recruit other ESCRT-III proteins and their specific binding partners to join the polymers they form. Indeed, evidence for a special role for hSnf7 comes from studies of viral budding, where recent data shows that the late domain-interacting protein Alix only functions in viral release when its ability to recruit hSnf7 is intact (Fisher et al., 2007).
Current understanding of how membrane budding into the MVB is driven is clearly limited. Although ESCRT-III proteins have been implicated in this process, evidence for their involvement has been indirect and primarily based on the development of a class E compartment in yeast when ESCRT-III function is impaired. The images presented here of hSnf7 show that even without upstream regulatory factors, this protein is capable of forming uniform circular structures that can be induced to drive formation of everting buds and tubules. These images are the first to show that an ESCRT-III polymer is associated with changes in the curvature of a membrane. Important future steps will be to compare the structure of other ESCRT-III homo- and heteropolymers with those of hSnf7 to determine how these polymers interface with other components of the ESCRT machinery and finally to visualize the actual events that occur on isolated, functional MVBs as they actively form and involute their ILVs.
Materials and methods
Plasmids
pcDNA3.1-FLAG-hSnf7-1 full length (residues 1–222), pcDNA3.1-FLAG-hSnf7-1 (residues 1–116) with residue two either Ser or Cys, pEGFP–hSnf7-1–GFP, pEGFP-VPS4B(E235Q)-GFP, pEGFP-VPS4B-GFP, and pcDNA4-VPS4B(E235Q)-His6myc were used as described previously (Lin et al., 2005). Note that VPS4B is synonymous with SKD1. pcDNA3.1-FLAG-hSnf7-2 full length (residues 1–222) was used as described previously (Shiels et al., 2007). Finally, a QuikChange site-directed mutagenesis kit (Stratagene) was used to mutate leucine 221 of GFP to lysine in hSnf7-1–GFP to create hSnf7-1–mGFP (Snapp et al., 2003).
Cell growth
COS-7 cells were grown in DME (Invitrogen) containing 5% fetal bovine serum (Invitrogen), 5% supplemented calf serum (Thermo Fisher Scientific), and 2 mM l-glutamine. TREx-HEK293 cells expressing VPS4B(E235Q)-GFP were grown in DME containing 10% tetracycline-free fetal bovine serum (Atlanta Biologics), 2 mM l-glutamine, 5 μg/ml blasticidin, and 100 μg/ml zeocin as described previously (Lin et al., 2005). Expression of VPS4B(E235Q) in these cells was induced by adding 0.5 μg/ml tetracycline for 4–6 h.
Transfections and sample preparation for DEEM
For DEEM, cells were grown on 3 × 3-mm glass (or where indicated, sapphire) coverslips. COS-7 cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and were used 18–24 h after transfection. To prepare unroofed cells, coverslips were briefly rinsed in serum-free, Hepes-buffered Ringer solution (30 mM Hepes, pH 7.4, 100 mM NaCl, and 2 mM CaCl2) and then unroofed using a brief pulse of ultrasound as described previously (Heuser, 2000a) in an intracellular buffer (30 mM Hepes, pH 7.2, 70 mM KCl, 5 mM MgCl2, and 3 mM EGTA). Samples were immediately fixed in the same buffer containing 2% glutaraldehyde or 4% paraformaldehyde. To prepare whole cells, cells on coverslips were simply fixed in Ringer solution containing 2% glutaraldehyde or 4% paraformaldehyde. To extract fixed whole cells with detergent, fixed coverslips were incubated for at least 2 h in buffer containing 1% Triton X-100 (Sigma-Aldrich) and 0.1% saponin.
Immunogold antibody decoration
For antibody decoration, samples fixed in formaldehyde were quenched in 50 mM NH4Cl, 50 mM lysine, and 50 mM glycine. Samples were then blocked in 30 mM Hepes, pH 7.4, 100 mM NaCl, and 2 mM CaCl2 containing 1% bovine serum albumin, incubated in primary antibody for 1 h, washed, incubated with 18-nm gold-conjugated goat anti–rabbit or anti–mouse, washed again, and postfixed in buffer containing 2% glutaraldehyde. Primary antibodies used were rabbit anti-GFP and rabbit and mouse (M2) anti-FLAG (Sigma-Aldrich).
Freezing, replicating, and imaging samples
Coverslips with samples to be frozen were washed in water and then quick-frozen by abrupt application (slamming) of the coverslip onto a liquid helium–cooled copper block using a cryopress (Heuser et al., 1979, 1989b). Coverslips were stored in liquid nitrogen until mounting in a freeze-etch device (Oerlikon Balzers), where they were immediately warmed to −80°C, freeze-dried for 15 min in vacuo, and replicated with 2 nm of platinum, which was vacuum evaporated onto them from 24° above horizontal while they rotated at 20 rpm. For freeze-fracture of cells grown on sapphire, the coverslip was frozen upside down, stored in liquid nitrogen, mounted in the freeze-etch device, warmed to −105°C, fractured by popping the sapphire off with the knife, etched by sitting for 2 min in vacuo, and then replicated as above.
In all cases, replicas were separated from the coverslips by flotation on concentrated hydrofluoric acid, and washed briefly with 4% sodium hypochlorite (bleach) and several rinses of water before being retrieved on 400-mesh Formvar-coated grids (Electron Microscopy Sciences). These were then viewed with a standard transmission EM (JEOL) operating at 100 kV and imaged at two different degrees of tilt (±10°) with standard EM film. Thereafter, the stereo pairs of film were aligned by superimposition on a copy stand and rerecorded as 4,492 × 3,328-pixel (17-Mp) digital images with a digital single-lens reflex camera (EOS-1Ds Mark II; Canon). Finally, the digital image pairs were converted (one to red and the other to green), layered on top of each other with the screen blending mode in Photoshop (Adobe), aligned to each other, and, where necessary, adjusted to equalize their brightness and contrast to create the final 3D anaglyphs shown here (Heuser, 2000b).
Acknowledgments
We thank Lisa Kimpler for help in preparing transfected cells; Soomin Shim, Lisa Kimpler, and members of the Hanson and Heuser laboratories for helpful discussions; Lucy Loultcheva, Jennifer Scott, and Tanya Tenkova for help in preparing anaglyphs; and our many colleagues at Washington University and elsewhere who have generously shared their insight as this work has evolved.
This work was supported by grants from the National Institutes of Health to J.E. Heuser (R01GM029647), the American Heart Association to P.I. Hanson (07501782Z), the W.M. Keck Foundation to P.I. Hanson, and an Andrew and Virginia Craig Faculty Fellowship to P.I. Hanson.
Abbreviations used in this paper: BAR, Bin/amphiphysin/Rvs; CHMP, charged MVB; DEEM, deep-etch EM; ESCRT, endosomal sorting complex required for transport; ILV, intraluminal vesicle; MVB, multivesicular body.
References
- Babst, M. 2005. A protein's final ESCRT. Traffic. 6:2–9. [DOI] [PubMed] [Google Scholar]
- Babst, M., B. Wendland, E.J. Estepa, and S.D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., D.J. Katzmann, E.J. Estepa-Sabal, T. Meerloo, and S.D. Emr. 2002. a. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 3:271–282. [DOI] [PubMed] [Google Scholar]
- Babst, M., D.J. Katzmann, W.B. Snyder, B. Wendland, and S.D. Emr. 2002. b. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289. [DOI] [PubMed] [Google Scholar]
- Booth, A.M., Y. Fang, J.K. Fallon, J.M. Yang, J.E. Hildreth, and S.J. Gould. 2006. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 172:923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton, J.G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 316:1908–1912. [DOI] [PubMed] [Google Scholar]
- Demirov, D.G., and E.O. Freed. 2004. Retrovirus budding. Virus Res. 106:87–102. [DOI] [PubMed] [Google Scholar]
- Fevrier, B., and G. Raposo. 2004. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 16:415–421. [DOI] [PubMed] [Google Scholar]
- Fisher, R.D., H.Y. Chung, Q. Zhai, H. Robinson, W.I. Sundquist, and C.P. Hill. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 128:841–852. [DOI] [PubMed] [Google Scholar]
- Fraile-Ramos, A., A. Pelchen-Matthews, C. Risco, M.T. Rejas, V.C. Emery, A.F. Hassan-Walker, M. Esteban, and M. Marsh. 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell. Microbiol. 9:2955–2967. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J., and H. Stenmark. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5:317–323. [DOI] [PubMed] [Google Scholar]
- Hanson, P.I., and S.W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529. [DOI] [PubMed] [Google Scholar]
- Heuser, J. 1989. a. Effects of cytoplasmic acidification on clathrin lattice morphology. J. Cell Biol. 108:401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser, J.E. 1989. b. Development of the quick-freeze, deep-etch, rotary-replication technique of sample preparation for 3-D electron microscopy. Prog. Clin. Biol. Res. 295:71–83. [PubMed] [Google Scholar]
- Heuser, J. 2000. a. The production of ‘cell cortices’ for light and electron microscopy. Traffic. 1:545–552. [DOI] [PubMed] [Google Scholar]
- Heuser, J.E. 2000. b. Membrane traffic in anaglyph stereo. Traffic. 1:35–37. [DOI] [PubMed] [Google Scholar]
- Heuser, J. 2002. Whatever happened to the ‘microtrabecular concept’? Biol. Cell. 94:561–596. [DOI] [PubMed] [Google Scholar]
- Heuser, J.E., and M.W. Kirschner. 1980. Filament organization revealed in platinum replicas of freeze-dried cytoskeletons. J. Cell Biol. 86:212–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser, J.E., T.S. Reese, M.J. Dennis, Y. Jan, L. Jan, and L. Evans. 1979. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J. Cell Biol. 81:275–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro, A., J. Sun, A.S. Rusnak, J. Kim, G. Prag, S.D. Emr, and J.H. Hurley. 2004. Structure of the ESCRT-II endosomal trafficking complex. Nature. 431:221–225. [DOI] [PubMed] [Google Scholar]
- Hinrichsen, L., A. Meyerholz, S. Groos, and E.J. Ungewickell. 2006. Bending a membrane: how clathrin affects budding. Proc. Natl. Acad. Sci. USA. 103:8715–8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, J.H., and S.D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, T., and P. De Camilli. 2006. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta. 1761:897–912. [DOI] [PubMed] [Google Scholar]
- Karacostas, V., K. Nagashima, M.A. Gonda, and B. Moss. 1989. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc. Natl. Acad. Sci. USA. 86:8964–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., M. Babst, and S.D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., G. Odorizzi, and S.D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893–905. [DOI] [PubMed] [Google Scholar]
- Kostelansky, M.S., J. Sun, S. Lee, J. Kim, R. Ghirlando, A. Hierro, S.D. Emr, and J.H. Hurley. 2006. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 125:113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky, M.S., C. Schluter, Y.Y. Tam, S. Lee, R. Ghirlando, B. Beach, E. Conibear, and J.H. Hurley. 2007. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetrAm. Cell. 129:485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier, C., U.K. von Schwedler, R.D. Fisher, I. De Domenico, P.L. White, C.P. Hill, J. Kaplan, D. Ward, and W.I. Sundquist. 2006. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J. Virol. 80:9465–9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., L.A. Kimpler, T.V. Naismith, J.M. Lauer, and P.I. Hanson. 2005. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 280:12799–12809. [DOI] [PubMed] [Google Scholar]
- Lottridge, J.M., A.R. Flannery, J.L. Vincelli, and T.H. Stevens. 2006. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. USA. 103:6202–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, H., J. Chevallier, N. Mayran, I. Le Blanc, C. Ferguson, J. Faure, N.S. Blanc, S. Matile, J. Dubochet, R. Sadoul, et al. 2004. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 303:531–534. [DOI] [PubMed] [Google Scholar]
- Mattila, P.K., A. Pykalainen, J. Saarikangas, V.O. Paavilainen, H. Vihinen, E. Jokitalo, and P. Lappalainen. 2007. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain–like mechanism. J. Cell Biol. 176:953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, H.T., and J.L. Gallop. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438:590–596. [DOI] [PubMed] [Google Scholar]
- Morita, E., and W.I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425. [DOI] [PubMed] [Google Scholar]
- Morita, E., V. Sandrin, H.Y. Chung, S.G. Morham, S.P. Gygi, C.K. Rodesch, and W.I. Sundquist. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morone, N., T. Fujiwara, K. Murase, R.S. Kasai, H. Ike, S. Yuasa, J. Usukura, and A. Kusumi. 2006. Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J. Cell Biol. 174:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol, T., E. Pineda-Molina, R.B. Ravelli, A. Zamborlini, Y. Usami, H. Gottlinger, and W. Weissenhorn. 2006. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 10:821–830. [DOI] [PubMed] [Google Scholar]
- Nickerson, D.P., M. West, and G. Odorizzi. 2006. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 175:715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydegger, S., S. Khurana, D.N. Krementsov, M. Foti, and M. Thali. 2006. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita, T., S. Saksena, S. Ghazi-Tabatabai, D.J. Gill, O. Perisic, S.D. Emr, and R.L. Williams. 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 449:735–739. [DOI] [PubMed] [Google Scholar]
- Olver, C., and M. Vidal. 2007. Proteomic analysis of secreted exosomes. Subcell. Biochem. 43:99–131. [DOI] [PubMed] [Google Scholar]
- Peter, B.J., H.M. Kent, I.G. Mills, Y. Vallis, P.J. Butler, P.R. Evans, and H.T. McMahon. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 303:495–499. [DOI] [PubMed] [Google Scholar]
- Raymond, C.K., I. Howald-Stevenson, C.A. Vater, and T.H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 3:1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, M.R., D.P. Nickerson, and G. Odorizzi. 2006. Molecular mechanisms of late endosome morphology, identity and sorting. Curr. Opin. Cell Biol. 18:422–428. [DOI] [PubMed] [Google Scholar]
- Scott, A., H.Y. Chung, M. Gonciarz-Swiatek, G.C. Hill, F.G. Whitby, J. Gaspar, J.M. Holton, R. Viswanathan, S. Ghaffarian, C.P. Hill, and W.I. Sundquist. 2005. a. Structural and mechanistic studies of VPS4 proteins. EMBO J. 24:3658–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A., J. Gaspar, M.D. Stuchell-Brereton, S.L. Alam, J.J. Skalicky, and W.I. Sundquist. 2005. b. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. USA. 102:13813–13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels, A., T.M. Bennett, H.L. Knopf, K. Yamada, K. Yoshiura, N. Niikawa, S. Shim, and P.I. Hanson. 2007. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am. J. Hum. Genet. 81:596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, S., L.A. Kimpler, and P.I. Hanson. 2007. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 8:1068–1079. [DOI] [PubMed] [Google Scholar]
- Shimada, A., H. Niwa, K. Tsujita, S. Suetsugu, K. Nitta, K. Hanawa-Suetsugu, R. Akasaka, Y. Nishino, M. Toyama, L. Chen, et al. 2007. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 129:761–772. [DOI] [PubMed] [Google Scholar]
- Slagsvold, T., K. Pattni, L. Malerod, and H. Stenmark. 2006. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 16:317–326. [DOI] [PubMed] [Google Scholar]
- Snapp, E.L., R.S. Hegde, M. Francolini, F. Lombardo, S. Colombo, E. Pedrazzini, N. Borgese, and J. Lippincott-Schwartz. 2003. Formation of stacked ER cisternae by low-affinity protein interactions. J. Cell Biol. 163:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoorvogel, W., M.J. Kleijmeer, H.J. Geuze, and G. Raposo. 2002. The biogenesis and functions of exosomes. Traffic. 3:321–330. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton, M.D., J.J. Skalicky, C. Kieffer, M.A. Karren, S. Ghaffarian, and W.I. Sundquist. 2007. ESCRT-III recognition by VPS4 ATPases. Nature. 449:740–744. [DOI] [PubMed] [Google Scholar]
- Teo, H., O. Perisic, B. Gonzalez, and R.L. Williams. 2004. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell. 7:559–569. [DOI] [PubMed] [Google Scholar]
- Teo, H., D.J. Gill, J. Sun, O. Perisic, D.B. Veprintsev, Y. Vallis, S.D. Emr, and R.L. Williams. 2006. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 125:99–111. [DOI] [PubMed] [Google Scholar]
- Thery, C., L. Zitvogel, and S. Amigorena. 2002. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2:569–579. [DOI] [PubMed] [Google Scholar]
- Tsang, H.T., J.W. Connell, S.E. Brown, A. Thompson, E. Reid, and C.M. Sanderson. 2006. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 88:333–346. [DOI] [PubMed] [Google Scholar]
- von Schwedler, U.K., M. Stuchell, B. Muller, D.M. Ward, H.Y. Chung, E. Morita, H.E. Wang, T. Davis, G.P. He, D.M. Cimbora, et al. 2003. The protein network of HIV budding. Cell. 114:701–713. [DOI] [PubMed] [Google Scholar]
- Williams, R.L., and S. Urbe. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8:355–368. [DOI] [PubMed] [Google Scholar]
- Yorikawa, C., H. Shibata, S. Waguri, K. Hatta, M. Horii, K. Katoh, T. Kobayashi, Y. Uchiyama, and M. Maki. 2005. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem. J. 387:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini, A., Y. Usami, S.R. Radoshitzky, E. Popova, G. Palu, and H. Gottlinger. 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. USA. 103:19140–19145. [DOI] [PMC free article] [PubMed] [Google Scholar]