Abstract
The peripheral nuclear lamina, which is largely but not entirely associated with inactive chromatin, is considered to be an important determinant of nuclear structure and gene expression. We present here an inducible system to target a genetic locus to the nuclear lamina in living mammalian cells. Using three-dimensional time-lapse microscopy, we determined that targeting of the locus requires passage through mitosis. Once targeted, the locus remains anchored to the nuclear periphery in interphase as well as in daughter cells after passage through a subsequent mitosis. Upon transcriptional induction, components of the gene expression machinery are recruited to the targeted locus, and we visualized nascent transcripts at the nuclear periphery. The kinetics of transcriptional induction at the nuclear lamina is similar to that observed at an internal nuclear region. This new cell system provides a powerful approach to study the dynamics of gene function at the nuclear periphery in living cells.
Introduction
The paradoxical nature of the mammalian nucleus as a compartmentalized, yet dynamic structure has been well established based on studies of nuclear domains (for review see Spector, 1993, 2006) and nuclear protein dynamics (for review see Misteli, 2001). The nuclear envelope marks the nuclear periphery in mammalian cells and is composed of an outer and inner nuclear membrane interrupted in places by nuclear pore complexes (for reviews see Goldman et al., 2002; Hetzer et al., 2005). A nuclear lamina consisting of type V intermediate filament proteins called lamins and other lamin-associated proteins underlies the inner nuclear membrane (for review see Goldman et al., 2002). The lamin proteins form a meshwork between the inner nuclear membrane and chromatin. Lamins play an important role in nuclear envelope assembly/disassembly during mitosis and are considered to be an important determinant of nuclear architecture and gene expression (for review see Goldman et al., 2002). Microscopic analyses of fixed cells (Belmont et al., 1993) and in vitro binding assays with lamins (Taniura et al., 1995) have suggested that lamins interact with chromatin either directly or indirectly via lamina-associated polypeptide (LAP) 2 (for reviews see Goldman et al., 2002; Schirmer and Foisner, 2007). In Drosophila melanogaster, distinct regions of chromatin have been shown to directly interact with the nuclear lamina (Marshall et al., 1996; Zhao et al., 1996; Pickersgill et al., 2006). Most interestingly, cells expressing a mutant form of lamin A (LADelta50) associated with Hutchinson-Gilford progeria syndrome exhibit a loss of perinuclear heterochromatin, further emphasizing the role of the nuclear lamina in chromatin organization and function (Shumaker et al., 2006).
The chromosomes in an interphase nucleus occupy a discrete spatially defined subvolume of the nucleus called the chromosome territories (for review see Cremer and Cremer, 2001). In mammalian cells, chromosomes show probabilistic (Parada et al., 2003), yet nonrandom radial positions with gene-rich chromosomes at the nuclear interior and gene-poor chromosomes at the periphery (Croft et al., 1999; Cremer et al., 2001). However, the broad morphological classification of chromatin into euchromatin (less-condensed or open chromatin) and heterochromatin (more-condensed or closed chromatin) does not always correlate with gene activity (Gilbert et al., 2004).
The view that individual genes show preferred nuclear positions (i.e., interior vs. peripheral) based on their transcriptional status has been supported by a variety of evidence that the nuclear envelope is primarily a silent compartment. However, recent findings in Saccharomyces cerevisiae and D. melanogaster indicate that the nuclear periphery can be associated with both transcriptionally active and inactive genes (Andrulis et al., 1998; Gerasimova et al., 2000; Ishii et al., 2002; Brickner and Walter, 2004; Casolari et al., 2004; Gartenberg et al., 2004; Mendjan et al., 2006; Pickersgill et al., 2006; Taddei et al., 2006). Studies of fixed mammalian cells have indicated the positioning of several genes to more internal nuclear regions upon activation (for review see Lanctot et al., 2007). However, evidence for gene activation at the nuclear periphery has been observed during differentiation of specialized cell types such as T or B lymphocytes and erythroid cells (for reviews see Brown and Silver, 2007; Lanctot et al., 2007). Because differentiation is associated with rather global changes in chromatin condensation, it is difficult to distinguish the direct cause/effect relationship between gene activity and nuclear position. Hence, it has been difficult to directly test the effects of nuclear position on transcriptional inducibility of a specific locus in differentiated cells.
Although lamins are known to play a role in establishing nuclear architecture, the spatial and temporal targeting of chromatin to the nuclear periphery has not been observed in living mammalian cells. Initially, lac operator–repressor interactions were used to tag DNA and visualize chromatin organization in vivo (for review see Belmont, 2001). Subsequently, this approach has been used extensively to study large-scale unfolding of chromatin structure (Tumbar et al., 1999) and long-range directional movement of a specific chromatin site (Chuang et al., 2006) and to visualize gene expression in living cells (Tsukamoto et al., 2000; Janicki et al., 2004).
To target a genetic locus to the nuclear lamina, and to visualize the spatial and temporal window of targeting, we developed a cumate-inducible nuclear lamina–targeting system. This system is based on a previously characterized U2OS-2-6-3 stable cell line developed in our laboratory to visualize gene expression in living cells (Janicki et al., 2004). Although this cell line contains a 200-copy tandem array of a reporter transgene, extensive analysis has indicated that it faithfully reports real-time readout of transcription (Janicki et al., 2004) and mRNP trafficking (Shav-Tal et al., 2004) after the expected trends of these processes based on molecular analyses. Using this newly developed system, we directly visualized that targeting of a genetic locus to the nuclear lamina required passage through mitosis. Upon transcriptional induction, components of the gene expression machinery were recruited to the targeted locus and nascent transcripts were visualized at the transcription site. This new cell system is a powerful tool to study the dynamics of gene function at the nuclear periphery during normal physiology and in disease states, such as envelopathies, in living cells.
Results
Development of an inducible nuclear lamina–targeting gene expression system
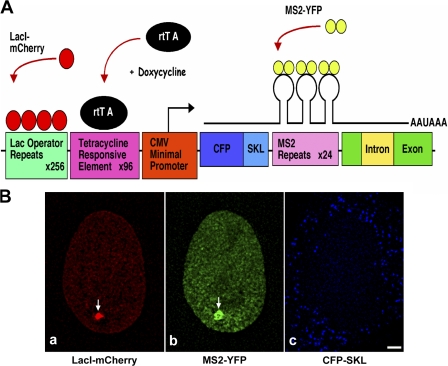
To address the influence of the nuclear lamina on gene expression in living human cells, we developed an inducible nuclear lamina–targeting gene expression system. For this, we took advantage of the existing U2OS-2-6-3 stable cell line developed in our laboratory to visualize gene expression in living cells at the level of DNA, RNA, and protein (Janicki et al., 2004) and modified it to suit our targeting strategy. The 20-kb plasmid used for visualization of gene expression consists of 256 copies of lac operator repeats, 96 copies of tetracycline response elements (TREs), a minimal cytomegalovirus promoter (pCMV), CFP fused to the peroxisomal-targeting signal SKL (CFP-SKL), 24 MS2 translational operators (MS2 repeats), a rabbit β-globin intron/exon module, and a cleavage/polyadenylation signal (Fig. 1 A, modified from Janicki et al. [2004]). 200 copies of this gene expression plasmid were stably integrated as a 4-Mb transgene array in the 1p36 region of human U2OS-2-6-3 cells. The genetic locus can be visualized by a lac repressor–mCherry fusion protein (Fig. 1 B, a), RNA transcripts by the MS2 coat–YFP fusion protein (Fig. 1 B, b), and the protein product as a CFP-SKL protein (Fig. 1 B, c) that is targeted to cytoplasmic peroxisomes.
Figure 1.
Visualization of gene expression. (A) Schematic representation of the gene expression plasmid (modified from Janicki et al. [2004]). The cassette consists of 256 copies of lac operator repeats, 96 copies of TREs, a minimal CMV promoter, CFP fused to a peroxisomal targeting signal SKL (CFP-SKL), 24 MS2 translational operators (MS2 repeats), a rabbit β-globin intron/exon module, and a cleavage/polyadenylation signal. LacI-mCherry binds to the lac operator repeats resulting in visualization of the DNA. Tet-On (rtTA) expression in the presence of Dox induces gene expression from the minimal CMV promoter. MS2-YFP (YFP fused to the MS2 coat protein) binds to the MS2 repeats, allowing visualization of the transcribed mRNA. (B) 200 copies of the gene expression cassette in A are stably integrated as a transgene array at the 1p36 region in human U2OS cells, referred to as U2OS-2-6-3 cells (Janicki et al., 2004). Transient expression of LacI-mCherry, Tet-On in the presence of Dox, and MS2-YFP in U2OS-2-6-3 cells allowed visualization of DNA (a, arrow), RNA (b, arrow), and CFP-SKL protein (c, blue). Bar, 5 μm.
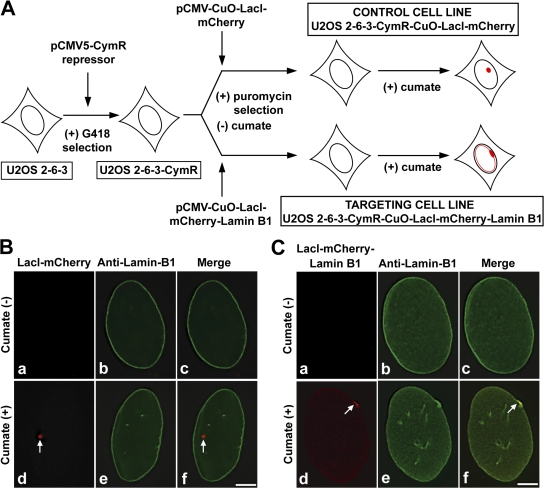
We used the U2OS-2-6-3 cells to develop cumate-inducible cell lines stably expressing lacI-mCherry or lacI-mCherry–lamin B1. The targeting fusion protein (lacI-mCherry–lamin B1) associates with the stably integrated locus in U2OS-2-6-3 cells via a lac operator–repressor interaction and with the nuclear lamina via a lamin B1 protein. The cumate expression system has two components: a repressor plasmid (pCMV5-CymR repressor) and a cumate-inducible response plasmid (pCMV–cumate operator [CuO]). The response plasmid has a CMV promoter and a CymR repressor binding site (CuO). Development of stable cumate-inducible clones required two rounds of stable selection (Fig. 2 A). U2OS-2-6-3 cells were transfected with pCMV5-CymR repressor plasmid (Fig. 2 A) and stable G418-resistant clones were screened by transient transfection with pCMV-CuO–LacI-mCherry in the presence or absence of cumate to obtain a tightly regulated cell line. This first stable cell line was referred to as U2OS-2-6-3–CymR (Fig. 2 A). Subsequently, U2OS-2-6-3–CymR was transfected with either pCMV-CuO–LacI-mCherry (control) or pCMV-CuO–LacI-mCherry–Lamin B1 (targeting) plasmids and stable puromycin resistance lines were screened in the absence or presence of cumate to obtain tightly regulated cell lines. These stable cumate-inducible cell lines were referred to as U2OS-2-6-3–CymR–CuO–LacI-mCherry (Fig. 2 A, control cell line) or U2OS-2-6-3–CymR–CuO–LacI-mCherry–lamin B1 (Fig. 2 A, targeting cell line).
Figure 2.
Generation and characterization of control and targeting cell lines. (A) Parental U2OS-2-6-3 cells were stably transfected with pCMV5-CymR repressor plasmid and selected in the presence of G418 to obtain the first stable line U2OS-2-6-3–CymR constitutively expressing the CymR repressor. U2OS-2-6-3–CymR cells were then transfected in parallel with one of two cumate-inducible plasmids, pCMV-CuO–LacI-mCherry (control) or pCMV-CuO–LacI-mCherry–Lamin B1 (targeting), and selected in the presence of puromycin and absence of cumate to obtain two cumate-inducible stable lines, namely U2OS-2-6-3–CymR–CuO–LacI-mCherry (control cell line) and U2OS-2-6-3–CymR–CuO–LacI-mCherry–lamin B1 (targeting cell line). Both puromycin-resistant lines were screened to obtain cumate-inducible control cell line (red, nontargeted locus) and targeting cell line (red, lamina-targeted locus). (B and C) Immunofluorescence analysis of control and targeting cell lines in the absence (a–c) or presence (d–f) of cumate. Anti–lamin B1 immunolabeling marks the nuclear periphery (B [b, c, e, and f]) and C [b, c, e, and f] green). (B) In the control cell line, when lacI-mCherry was not expressed the locus was not visible (a and c). In the presence of cumate and lacI-mCherry expression, the locus was visualized (d and f, red, arrow) internal to the nuclear periphery (f). (C) In the targeting cell line, in the absence of lacI-mCherry–lamin B1 (targeting fusion) expression there was no signal for the locus (a and c). Cumate-induced targeting fusion expression resulted in targeting the locus to the lamina (d, red, arrow) and the targeted locus colocalized with endogenous lamin B1 (f, yellow, arrow). Bars, 5 μm.
To further evaluate how tightly the stable clones were regulated by cumate and to investigate whether the locus is targeted to the nuclear periphery upon cumate-induced expression of the targeting protein, both the control and targeting cells were treated with (+) or without (−) cumate for 12 h. The cells were then fixed in formaldehyde and immunostained with anti–lamin B1 antibody to mark the nuclear periphery. In the absence of cumate, CymR repressor binds to CuO located between the promoter and the fusion cassette and sterically represses transcription of the fusion cassette. Absence of the locus-mCherry (red) signal in both the control (Fig. 2 B, a and c) and targeting cells (Fig. 2 C, a and c) when cumate was not added to the medium indicated that the cell lines were tightly regulated. However, upon addition of cumate, the cumate-bound repressor leaves CuO and repression is relieved, initiating expression from the CMV promoter in both control and targeting cells. 12 h after cumate induction, in the control cells expressing lacI-mCherry (control fusion) the locus was visualized as a spherical dot (Fig. 2 B,d and f, red, arrows) internal to the nuclear periphery (Fig. 2 B, f, green). In contrast, in the targeting cells expressing lacI-mCherry–lamin B1 (targeting fusion) the locus was targeted to the nuclear lamina, appearing as a flattened signal plastered against the nuclear periphery (Fig. 2 C, d, red, arrow), and the targeted locus colocalized with endogenous lamin B1 (Fig. 2 C, f, yellow, arrow). Importantly, the targeted locus was easily visible even in the background of peripheral endogenous lamin B1 (Fig. 2 C, e). To further establish that the targeted locus at the nuclear periphery was indeed the targeting fusion bound to the lac operator DNA repeats (genetic locus), we transiently expressed a YFP–Tet-on fusion (YFP-rtTA) in targeting cells. The cells were induced with cumate for targeting fusion expression and then with doxycycline (Dox) to induce binding of the YFP-rtTA protein to the TREs within the genetic locus. The YFP-rtTA protein was enriched at the site of the lacI-mCherry signal, confirming that this was the genetic locus (Fig. S1, arrow, available at http://www.jcb.org/cgi/content/full/jcb.200706060/DC1).
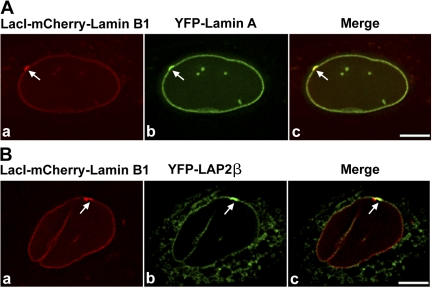
To ascertain whether the targeting fusion exhibited properties similar to endogenous lamin B1, we examined its localization in regard to the localization of lamin B–interacting proteins, such as lamin A and LAP2β. We transiently expressed YFP–lamin A or YFP–LAP2β (Fig. 3, A or B) in targeting cells and induced targeting fusion expression for 12 h with cumate. The targeted locus was enriched for YFP–lamin A (Fig. 3 A, b and c, arrows) and YFP–LAP2β (Fig. 3 B, b and c, arrows), indicating that the targeting fusion behaved analogous to endogenous lamin B1 protein.
Figure 3.
Lamin B–interacting proteins are enriched at the targeted locus at the nuclear periphery. (A and B) Targeting cells were transiently transfected with YFP-LA or YFP–LAP2β and induced for 12 h with cumate to express the targeting fusion protein (red). YFP-LA and YFP–LAP2β were enriched at the targeted loci (A [b and c] and B [b and c], arrows) at the nuclear periphery. Panel a shows the targeted locus. Bars, 5 μm.
Targeting the locus to the nuclear lamina requires passage through mitosis
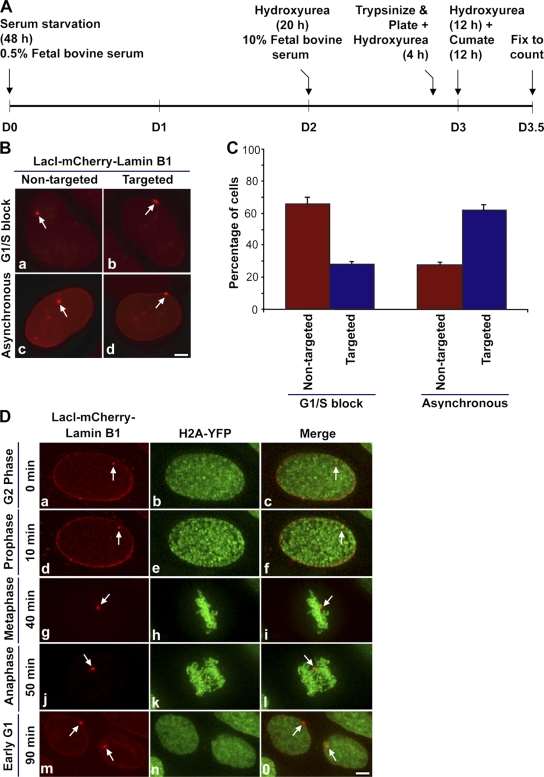
Having obtained cumate-inducible stable cell lines, we were interested in determining the temporal window during which the locus is targeted to the nuclear periphery. A cumate-induction time course study showed that the percentage of targeted cells increased with the passage of time, indicating that targeting could be a postmitotic event (unpublished data). To test this possibility, we performed a G1/S-phase block by treating the targeting cells with hydroxyurea (Fig. 4 A). If cells required passage through mitosis for the locus to be targeted to the nuclear lamina, cells blocked at G1/S and induced with cumate would have a high percentage of nontargeted cells (locus internal to the nuclear lamina) compared with targeted cells (locus targeted to the nuclear lamina). However, in the absence of a G1/S block, the cumate-induced targeting cells would divide asynchronously and, therefore, more targeted loci should be observed.
Figure 4.
Targeting the locus to the nuclear lamina requires passage through mitosis. (A) G1/S-phase block using hydroxyurea in targeting cell line from day 0 (D0) to day 3.5 (D3.5). (B) Representative images of LacI-mCherry–Lamin B1–expressing cells used for counting to quantify G1/S-blocked nontargeted (a, red, arrow) and targeted (b, red, arrow) cells and asynchronous nontargeted (c, red, arrow) and targeted (d, red, arrow) cells. Bar, 5 μm. (C) Quantification of the percentage of nontargeted and targeted cells in G1/S-blocked and asynchronous cells. The graphical data represents the mean and standard deviation of three experiment (n = 100 nuclei per experiment). Approximately 10% of the cells did not exhibit a nuclear mCherry signal. (D) Still images (a–o) from time series (0–90 min, G2 to early G1 phase) of targeting clone H2A-YFP going through mitosis. Targeting cells stably expressing histone H2A-YFP (green) were induced to express LacI-mCherry–Lamin B1 (red, arrow). Images are projections of deconvolved z stacks taken after every 10 min. Bar, 5 μm. See video 1 (available at http://www.jcb.org/cgi/content/full/jcb.200706060/DC1) for the entire time series.
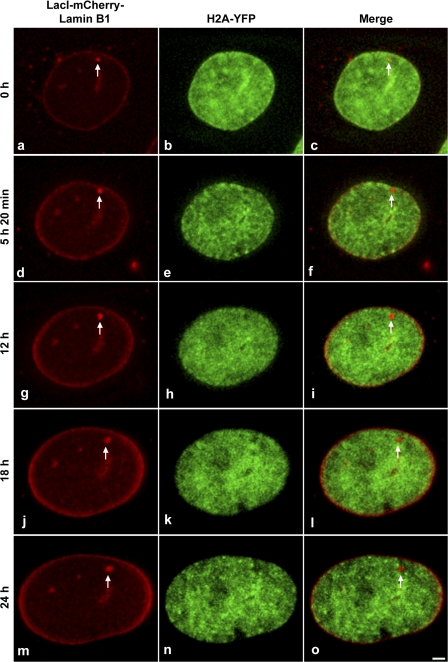
Fig. 4 B shows representative images of cells used for counting targeted loci in G1/S-blocked or asynchronous cells. When cells were blocked in G1/S phase, there was an approximately twofold increase in the number of nontargeted cells as compared with targeted cells (Fig. 4 C). However, when cells were allowed to divide asynchronously there was a twofold increase in the number of targeted cells compared with nontargeted cells. These results indicated that targeting likely required passage through cell division. Furthermore, and consistent with this possibility, analysis of the dynamics of loci associated with the targeting fusion protein for up to 24 h, during interphase and in the absence of hydroxyurea, did not result in the observation of targeted loci that were stably associated with the nuclear periphery (Fig. 5). Interestingly, at the 5-h 20-min time point the locus shown in Fig. 5 transiently contacts the peripheral lamina, creating tension at the point of contact (Fig. 5, d, arrow); however, it subsequently moves away from the periphery. These results indicate that binding of the lacI-mCherry–lamin B1 targeting fusion to the locus is not sufficient for its stable targeting to the peripheral nuclear lamina during interphase.
Figure 5.
Targeting of lamin B1–associated loci does not take place during interphase. Targeting cells stably expressing histone H2A-YFP (green) were induced to express LacI-mCherry–Lamin B1 (red, arrow) and cells with a nontargeted lamin B1–bound locus were selected for imaging. Still images (a–o) are shown from an interphase time series (0–24 h, early G1 to late G2 phase). Images are projections of deconvolved z stacks taken every 20 min. Bar, 5 μm.
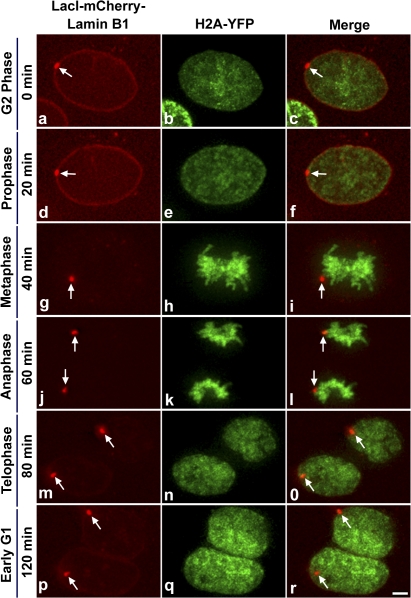
Prior studies have shown that lamin B1 is targeted to the nuclear envelope in late telophase and is polymerized as part of the nuclear lamina during early G1 (Moir et al., 2000). Furthermore, chromatin is most dynamic during early G1 phase of the cell cycle before the establishment of chromosome territories (Chubb et al., 2002; Walter et al., 2003). Based on these observations, we hypothesized that the best opportunity for the locus to be targeted could be during mitosis, at the transition from telophase to early G1 phase. To test this hypothesis we followed the dynamics of the locus during mitosis in the targeting cell line stably expressing histone H2A-YFP in the absence of hydroxyurea. To minimize phototoxicity associated with cell imaging, we reduced the time window of imaging by roughly preselecting cells based on cell size (large nucleus with a spherical nontargeted locus indicative of a cell in G2 phase) before imaging, we used short exposure times and rapid shutters, and we placed a UV-blocking filter in the light path of our imaging system. Fig. 4 D shows still images (a–o) from a time series (0–90 min, G2 to early G1 phase) of the targeting line H2A-YFP progressing through mitosis (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200706060/DC1). At 0 min, the locus was spherical (Fig. 4 D, a and c, red, arrows) and localized away from the nuclear lamina. Chromatin marked by H2A-YFP (Fig. 4 D, b and c, green) was homogenously distributed within the large nucleus, which is indicative of a G2 cell. At the 10-min time point, chromatin began to condense, which is indicative of cells in early prophase (Fig. 4 D, e and f, green). The locus was still spherical and was not targeted to the intact lamina (Fig. 4 D, d and f, red, arrows). By 20 min, the locus was still visible as a round spot, whereas the lamina started to dissociate and the chromosomes exhibited a prometaphase configuration (Video 1). By 30 min, the locus continued to appear as a round spot, whereas the lamina was completely dissociated and the chromosomes began to congress toward the metaphase plate (Video 1). At 40 min, the locus (Fig. 4 D, g and i, red, arrows) was observed aligned at the metaphase plate (Fig. 4 D, h and i, green). At the 50-min time point, chromatids (Fig. 4 D, k and l, green) started moving toward the poles and loci appeared as coaligned doublets (Fig. 4 D, j and l, red, arrows) that were slightly stretched along their long axis, possibly because of torsional force generated during chromosome segregation at anaphase. By 70 min, which marks late telophase, both loci were targeted to the nuclear lamina (Video 1). At, the 80- and 90-min time points, which mark early G1 phase, chromatin was more diffusely distributed throughout the daughter nuclei (Fig. 4 D, n and o, green; and Video 1) and the targeted loci were anchored to the nuclear lamina (Fig. 4 D, m and o, red, arrows; and Video 1). Although in the 2D projection it appears that only one locus is at the edge of the nucleus, the targeting of both loci is obvious when examined in individual Z sections (Fig. S2 and Video 2). Thus, our live cell data showed that the temporal window for targeting the locus to the nuclear lamina was during the transition from late telophase to early G1 phase of the cell cycle.
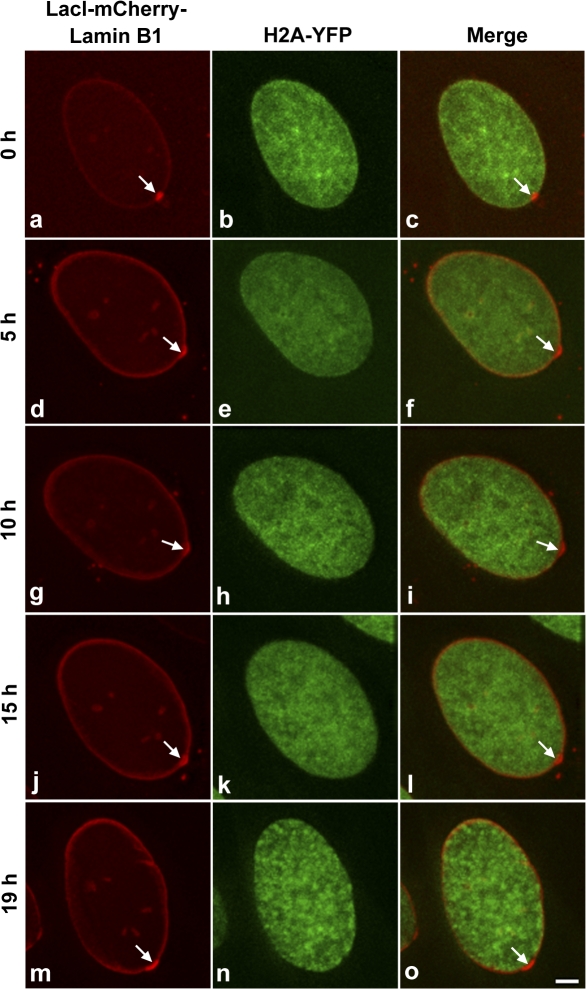
Next, we were interested in examining the stability of the targeted locus with the nuclear periphery during interphase. As shown in Fig. 6, the targeted locus was stably anchored to the peripheral lamina throughout the 19-h period of interphase observation. Subsequently, we examined cells containing a targeted locus as the cells progressed through mitosis, and we found that targeted loci remained targeted in daughter cells (Fig. 7 and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200706060/DC1).
Figure 6.
The targeted locus is stably anchored to the nuclear lamina during interphase. Targeting cells stably expressing histone H2A-YFP (green) were induced to express LacI-mCherry–Lamin B1 (red, arrow) and cells with a targeted locus were selected for imaging. Still images (a–o) are shown from an interphase time series (0–19 h, G1 to G2/M phase). Images are projections of deconvolved z stacks taken every 20 min. Bar, 5 μm.
Figure 7.
Upon passage through mitosis, a targeted locus remains targeted in daughter cells. Targeting cells stably expressing histone H2A-YFP (green) were induced to express LacI-mCherry–Lamin B1 (red, arrow) and cells with a targeted locus were selected for imaging. Still images (a–r) are shown from time series (0–120 min, G2 to early G1 phase) of targeting cells stably expressing H2A-YFP progressing through mitosis. Images are projections of deconvolved z stacks taken every 20 min. Bar, 5 μm.
Transcriptional induction at the nuclear periphery
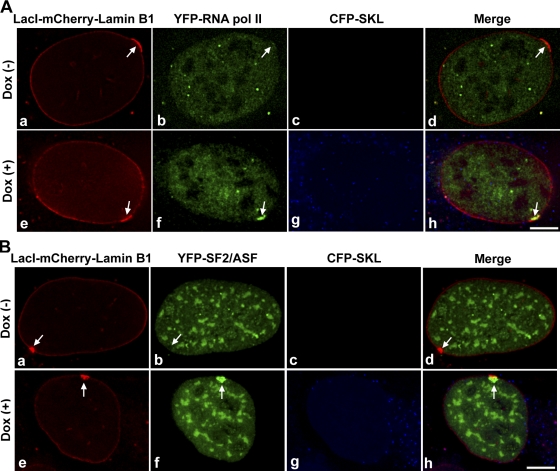
Next, we addressed whether components of the gene expression machinery, such as RNA polymerase II (RNA pol II) and the pre-mRNA splicing factor SF2/ASF, were recruited to the targeted locus at the nuclear lamina. To address this possibility, we took advantage of the transcriptional inducibility of the locus by Dox. We transiently coexpressed Tet-On with either YFP–RNA pol II or YFP-SF2/ASF (Fig. 8, A or B) in targeting cells and induced targeting-fusion expression for 12 h with cumate (Fig. 8, A or B). In the absence of Dox (Fig. 8, A [a–d] or B [a–d]), YFP–RNA pol II and YFP-SF2/ASF were not recruited to the targeted loci (Fig. 8, A [b and d] or B [b and d], no green enrichment, arrows) and there was no CFP-SKL protein at the cytoplasmic peroxisomes (Fig. 8, A [c and d] or B [c and d]). However, in the presence of Dox (Fig. 8, A [e–h] or B [e–h]) for 5 h, YFP–RNA pol II and YFP-SF2/ASF were recruited to the targeted loci (Fig. 8, A [f] and B [f], arrows) at the nuclear lamina, and CFP-SKL protein was observed at the cytoplasmic peroxisomes (Fig. 8, A [g and h] or B [g and h]). Quantitation showed that ∼76% (n = 50) of the control loci, 68% (n = 50) of lamin B1–bound internal loci in targeting cells, and 64% (n = 50) of targeted loci in targeting cells showed recruitment of YFP–RNA pol II in the presence of Dox. Also, ∼80% (n = 50) of the control cells, 76% (n = 50) of lamin B1–bound internal loci in targeting cells, and 74% (n = 50) of targeted loci in targeting cells showed recruitment of YFP-SF2/ASF in the presence of Dox. In the absence of Dox, ∼96% (n = 50) of the control or targeting loci did not show recruitment of YFP–RNA pol II or YFP-SF2/ASF. Because control and targeting cells showed a relatively similar percentage of recruitment of the gene expression machinery, association with the nuclear lamina does not appear to preclude accessibility to the gene expression machinery.
Figure 8.
Components of the gene expression machinery are recruited to the lamina-targeted locus upon gene activation. Recruitment of YFP–RNA pol II (A) or YFP-SF2/ASF (B) to targeted and activated locus at the lamina. Targeting cells were transiently transfected with pTet-On and YFP–RNA pol II or YFP-SF2/ASF (green), induced for 12 h with cumate to express the targeting fusion protein (red, arrows), and then incubated in the absence of Dox (A [a–d] or B [a–d]) or presence of Dox (A [e–h] or B [e–h]) for 5 h. In Dox (−) condition, YFP–RNA pol II or YFP-SF2/ASF was not recruited (A [b and d] or B [b and d], arrows) and CFP-SKL protein was not observed (A [c and d] or B [c and d]). In Dox (+) condition, YFP–RNA pol II and YFP-SF2/ASF were recruited (A [f] or B [f], arrows) and localized with the locus (A [h] or B [h], arrows) and CFP-SKL protein was observed at cytoplasmic peroxisomes (A [g and h] or B [g and h]). Bars, 5 μm.
Because we observed that the gene expression machinery was recruited to the targeted locus, we were next interested in directly visualizing transcription at the targeted locus. To do so, we took advantage of the fact that the mRNA that is transcribed from the genetic locus contains 24 tandem MS2 translational operators (MS2 repeats; Janicki et al., 2004). As the MS2 coat protein directly binds to these repeats, we were able to use an MS2-YFP fusion protein to localize RNA transcripts at the locus in living cells. To directly visualize transcription, the control or targeting cells were transiently transfected with a dual promoter vector expressing both Tet-On and MS2-YFP simultaneously (Fig. 9, A and B, pVitro2–Tet-On + MS2-YFP, green), as shown in representative images. The cells were first induced for 12 h with cumate for control and targeting fusion expression (Fig. 9, A and B, red, arrows), and then incubated in the absence of Dox (Fig. 9, A [a–c] and B [a–c]) or presence of Dox (Fig. 9, A [d–f] and B [d–f]) for 5 h. In the absence of Dox, transcription was not induced and MS2-YFP did not accumulate at the site of nontargeted (Fig. 9, A, b and c, arrows) or targeted loci (Fig. 9 B, b and c, arrows). Upon induction of transcriptional activity with Dox (Fig. 9, A or B), MS2-YFP accumulated at both the nontargeted and targeted active loci (Fig. 9, A [e] and B [e], arrows). The partial colocalization of the active loci with MS2-YFP (Fig. 9, A [f] and B [f], arrows) was caused by the highly decondensed nature of the loci. Quantification showed that ∼90% (n = 100) of the control or targeting loci were transcriptionally inactive in the absence of Dox. However, ∼70% (n = 100) of the targeted loci in targeting cells, 76% (n = 100) of lamin B1–bound internal loci in targeting cells, and ∼90% (n = 100) of the control loci were transcriptionally active based on MS2-YFP accumulation in the presence of Dox. These results indicated that although association of lamin B with a locus, internal or targeted, had some effect on the percentage of loci that were able to be transcriptionally induced, a significant percentage of gene loci anchored to the nuclear lamina could be induced. However, because the transcriptional analysis was performed 12 h after cumate-induced expression of lacI-mCherry–lamin B1, it is not possible, based on fixed-cell analysis, to distinguish whether Tet-On–induced transcription initiated before or after the locus reached the nuclear lamina.
Figure 9.
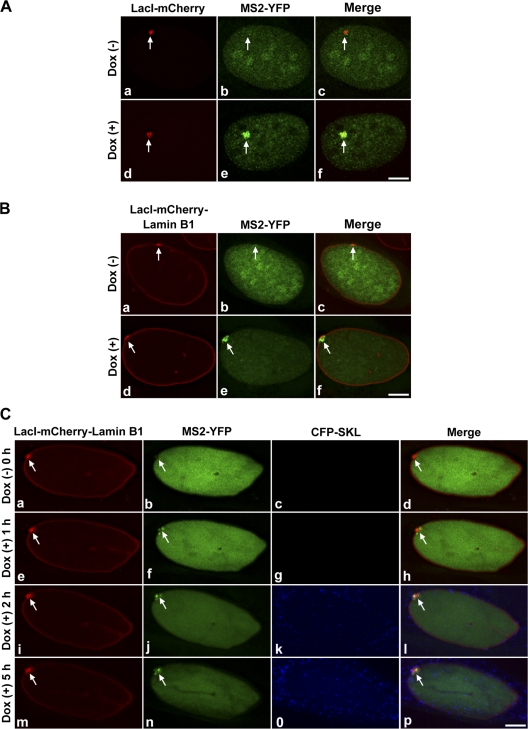
Transcription of the targeted locus at the nuclear lamina. (A and B) Representative images used for counting cells to quantify the percentage of cells showing transcription in control or targeting conditions. Control cells or targeting cells were transiently transfected with pVitro2–Tet-on + MS2-YFP (green), induced for 12 h with cumate for targeting fusion expression (red, arrows), and then incubated in the absence (a–c) or presence (d–f) of Dox for 5 h. In the Dox (+) condition, MS2-YFP accumulated at the site of the targeted locus (B, e, green enrichment, arrow) and MS2-YFP localized with the locus (B, f, arrow). In the presence of Dox, ∼90% of control cells and ∼70% of targeting cells were transcriptionally active. Arrows in A (b) and B (b) represent absence of accumulation of MS2-YFP. n = 100 nuclei per experiment for each control and test. Bars, 5 μm. (C) Still images (a–p) from time series (0–5 h) of targeting cells showing transcription at the lamina. At 0 h, in Dox (−) condition, targeted locus (a and d, arrows) showed no accumulation of MS2-YFP (b and d, arrows) and no CFP-SKL protein in peroxisomes (c and d). In Dox (+) condition, transcription was observed at the targeted nuclear lamina as observed by the accumulation of MS2-YFP (f, j, and n, arrows) and CFP-SKL localization (k, l, o, and p) over time. Images are projections of deconvolved z stacks taken every 20 min. Arrows: (e, i, and m) decondensed targeted locus; (h, l, and p) accumulation of MS2-YFP. Bar, 5 μm. See Video 4 (available at http://www.jcb.org/cgi/content/full/jcb.200706060/DC1) for the entire time series.
To directly visualize transcription at the nuclear lamina in living mammalian cells, targeting cells were transiently transfected with pVitro2–Tet-On + MS2-YFP and incubated with cumate for 12 h to induce targeting to the nuclear lamina. Fig. 9 C shows still images (a–p) from a time series (0–5 h) of targeting cells during transcriptional activation at the nuclear lamina. Cells that exhibited the locus targeted and plastered onto the nuclear lamina and expressed Tet-On and MS2-YFP (based on YFP signal) were selected for imaging. At 0 h, in the absence of Dox, the locus was condensed (Fig. 9 C, a and d, arrows), there was no transcription, and no MS2-YFP accumulation was visualized at the locus (Fig. 9 C, b and d, green, arrows). At 20 min after the addition of Dox, MS2-YFP was first observed at the locus with no visible decondensation of the locus or accumulation of CFP-SKL protein in the peroxisomes (Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200706060/DC1). By 1 h in Dox (+) condition, the targeted locus started to decondense (Fig. 9 C, e and h, arrows) and an increase of MS2-YFP was observed at the locus (Fig. 9 C, f, arrow). CFP-SKL protein was first observed at 1 h 20 min after induction (Video 4), although it was more apparent at later time points, and by 2 h peroxisome labeling was very apparent (Fig. 9 C, k and l; and Video 4). By 5 h in the presence of Dox, the targeted locus continued to be transcribed at the nuclear lamina (Fig. 9 C, n, arrow) and an increased CFP-SKL protein signal marked the peroxisomes (Fig. 9 C, o and p). Thus, our live-cell data shows that transcription can be induced and RNA synthesis visualized at the nuclear periphery in mammalian cells.
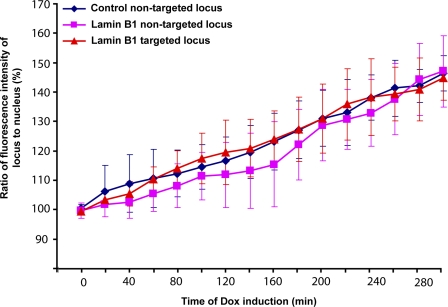
Having visualized transcription at the nuclear periphery, we were next interested in determining whether there was a significant difference in the kinetics of transcription when the locus was located at an internal nuclear site versus at the periphery. To do so, control or targeting cells were transiently transfected with pVitro2–Tet-on + MS2-YFP and induced for 12 h with cumate for control or targeting fusion expression and cells were imaged in the YFP and mCherry channels. The ratio of MS2-YFP fluorescence intensity of the locus to the entire nucleoplasm (mean values) was expressed as a percentage and was plotted against time of Dox induction. Such analysis indicated that transcription of the lamin B1–targeted loci at the nuclear periphery as well as at internal nuclear regions paralleled that of the control nontargeted loci at the nuclear interior (Fig. 10). mRNA synthesis was first detected at 20 min after Dox addition in both control and targeting cells. The rates of mRNA synthesis increased at similar levels over the 5-h period of examination (Fig. 10). The ability to induce transcription of this locus at the nuclear periphery is consistent with the dynamic and freely diffusible nature of most nuclear proteins (for review see Misteli, 2001).
Figure 10.
The kinetics of transcriptional induction of the targeted locus parallels that of the nontargeted locus. Control or targeting cells were transiently transfected with pVitro2–Tet-on + MS2-YFP and induced for 12 h with cumate for targeting fusion expression. The transfected cells (control nontargeted [n = 7], lamin B1–bound nontargeted [n = 8], and lamin B1–targeted [n = 10] cells) were then initially imaged in the mCherry and YFP channels in Dox (−) condition (0 h). Subsequently, imaging was continued in Dox (+) condition every 20 min for a total of 5 h. The ratio of fluorescence intensity of the locus to the entire nucleus (mean values) was expressed as a percentage and plotted against time of Dox induction (each time point is represented by the mean and standard deviation).
Discussion
At the nuclear envelope, lamins bind to chromatin either directly or indirectly through LAPs (for reviews see Goldman et al., 2002; Schirmer and Foisner, 2007) and hence are thought to regulate higher order chromatin structure. In addition to the lamins and various lamina-associated proteins, a multitude of nuclear envelope transmembrane proteins of unknown function have recently been identified (for review see Schirmer and Gerace, 2005), indicating an increased complexity of the nuclear envelope. In vitro studies showed that lamin B1 binds to native core histones (Taniura et al., 1995) and to specific DNA sequences called matrix attachment regions (Luderus et al., 1992). Analysis of D. melanogaster B-type lamin (Dmo) revealed TRAT amino acid and NLS sequences in the tail domain that directly interact with histone H2A (Mattout et al., 2007). Recently, a genome-wide screen in D. melanogaster Kc cells using DNA adenine methyltransferase identification technology showed that ∼500 genes interact with Dmo (Pickersgill et al., 2006). However, these genes show no consensus DNA-binding sequence for B-type lamin that would enable anchoring of perinuclear chromatin to the lamina. In the absence of such a DNA-consensus sequence, it becomes difficult to study the anchoring process. However, the targeting system that we developed has allowed us to address important issues relating to the spatial and temporal aspects of targeting as well as its functional consequences.
Live-cell studies from different organisms in interphase nuclei showed that chromatin, in general, exhibits constrained diffusional random walk motion, with a mean radius of confinement of ∼0.5 μm in the mammalian nucleus (for reviews see Spector, 2003; Lanctot et al., 2007). These studies also indicated that unlike in yeast and D. melanogaster, wherein the dynamic nature of chromosomes allows their probing of significant portions of the nuclear volume, the mammalian chromosome visits a limited nuclear volume during interphase. Furthermore, the diffusion coefficient of human chromatin is fourfold lower than that of yeast (Chubb et al., 2002). This constraint on mammalian chromatin could be a reflection of the complexity of the genome and the constraint posed by the physical attachment of chromatin to nuclear compartments (Chubb et al., 2002). Although chromosomes are organized as distinct territories in interphase nuclei, several studies have shown that they are dynamic in early G1 and can be repositioned with respect to each other or other nuclear compartments (for reviews see Spector, 2003; Thomson et al., 2004; Lanctot et al., 2007). Furthermore, chromatin has been shown to frequently exhibit long-range movements of >2 μm during this phase of the cell cycle (Walter et al., 2003). This is also the time window when lamin B1 is incorporated into the polymerizing lamina (Moir et al., 2000). It is very likely that the constrained diffusion of chromatin results from interaction of chromatin to structural elements such as the nuclear lamina or the nuclear pore complex (Marshall, 2002). It has been suggested that the chromatin–nuclear lamina interactions, which are disrupted during mitosis, are reformed during early G1 phase of the cell cycle and, indeed, this temporal window of increased chromatin mobility is in fact when many aspects of nuclear organization are established (Parada et al., 2003; for review see Spector, 2003). We observed targeting of the locus to the nuclear lamina during the same temporal window, correlating with the dynamics of the lamin B1 protein and bulk chromatin. Hence, we propose, based on our targeting study, that the contribution by the lamina in establishing nuclear architecture and chromatin organization occurs during the early G1 phase of the cell cycle.
An important question regarding the role of spatial positioning to gene function is whether subnuclear position of a gene, relative to nuclear landmarks, influences gene activity (for reviews see Spector, 2003; Misteli, 2005). Although a classical case involving the phenomenon of position-effect variegation of the brown locus in D. melanogaster indicates that repositioning to a nearby heterochromatic region results in trans-silencing (Csink and Henikoff, 1996; Dernburg et al., 1996), λ5 transgene insertion into heterochromatin in mouse pre-B cells does not result in silencing, and repositioning away from heterochromatin was not required for gene activation (Lundgren et al., 2000). Together, these studies indicate that absolute gene position may not be critical for gene activation or that position may be critical for some genes and not for others. The accessibility of chromosome territories (Verschure et al., 1999; for review see Cremer and Cremer, 2001) and the dynamic nature of chromatin and nucleoplasmic proteins (Becker et al., 2002; Dundr et al., 2002; Kimura et al., 2002; Phair et al., 2004; Chen et al., 2005) provides the opportunity for genes anywhere within a chromosome territory to be easily accessible to both gene activators and repressors (Meaburn and Misteli, 2007). Our findings are consistent with the dynamic accessibility by nuclear proteins, wherein upon transcriptional activation the transcription machinery, as well as the pre-mRNA processing machinery, was recruited to the locus irrespective of its nuclear location, resulting in location-independent gene expression. However, we are not excluding the possibility that there may be other microenvironments along the nuclear periphery, or for that matter at internal nuclear regions, that may be more attuned to transcriptional silencing. It is also interesting to consider that the large size of the 200-copy gene array could generate a microenvironment. Nonetheless, our study shows that there is not a discernible effect on expression of the peripheral versus internal nuclear compartments on this array.
Perhaps the best cases that correlate changes in gene position to gene activation/expression in relation to nuclear periphery in mammals occur during differentiation of specialized cell types, such as erythroid cells, lymphocytes, or embryonic stem cells (for review see Lanctot et al., 2007). For example, the repositioning of genes, such as immunoglobulin heavy chain in B lymphocytes, C-maf in T cells, Mash1 in neuronal cells, Cftr in adenocarcinoma cells, and MyoD in myoblasts, from the nuclear periphery to the nuclear interior upon activation has been well documented (Kosak et al., 2002; Hewitt et al., 2004; Zink et al., 2004; Lee et al., 2006; Williams et al., 2006; Lanctot et al., 2007). However, gene activation is not always associated with movement away from the nuclear periphery, as was observed in other studies based on interferon γ, β-globin, and the CD8 locus during differentiation or microarray gene expression analysis on lamin B1−/− mouse fibroblasts (Hewitt et al., 2004; Kim et al., 2004; Ragoczy et al., 2006; Malhas et al., 2007). This is consistent with our observation that a lamina-targeted genetic locus can be activated and transcribed at the lamina. Most importantly, although we observed a similar kinetics of transcriptional induction between the targeted and nontargeted loci, we did observe some differences in the percentage of cells that showed recruitment of the gene expression machinery. However, the complexities of the mammalian nuclear periphery and the fact that we are comparing data points from two different cell lines (although with the same genomic integration site of lac operator) makes it difficult to determine the absolute significance of the observed differences. Therefore, as is the case in yeast and D. melanogaster, it is likely that there will not be a single rule that dictates the relationship between the nuclear periphery and transcription.
In summary, we have developed an inducible live-cell system in which the position of a genetic locus can be targeted from a more internal nuclear region to the nuclear periphery in mammalian cells. Interestingly, the mechanism of targeting of the locus to the nuclear periphery requires one round of cell division and nuclear reassembly. Transcriptional analysis indicated that the targeted locus retained its transcriptional activity, which is similar to the locus at an internal nuclear region. This new cell system is a powerful tool to study the dynamics of gene function at the nuclear periphery during normal physiology and in disease states, such as envelopathies, in living cells.
Materials and methods
Plasmid constructs
pTet-On was purchased from Clontech Laboratories, Inc., pVitro2 was purchased from Invivogen, pCMV5-CymR and pCMV-CuO were purchased from Qbiogene, and pCMV–lamin B1 and pCMV–lamin A were purchased from Open Biosystems. pCMV-CuO–lacI-mCherry was constructed by cloning lacI-mCherry fragment (mCherry was a gift from R. Tsien [University of California, San Diego, La Jolla, CA] and was initially PCR amplifed) into pCMV-CuO. pCMV-CuO–lacI-mCherry–lamin B1 was constructed by cloning lamin B1 into pCMV-CuO–lacI-mCherry. pSV2-YFP–lamin A was obtained by cloning lamin A into pSV2-YFP-C1 (Janicki et al., 2004). pVitro2–Tet-On + MS2-YFP was made by sequential cloning of Tet-On and MS2-YFP into pVitro2 dual promoter vector. pCMV–MS2-YFP, pSV2-YFP–RNA poly II, pSV2-YFP-SF2/ASF, and pYFP-rtTA-N1 plasmids were previously described (Janicki et al., 2004). pCMV–H2A-YFP and pSV2-YFP–LAP2β were subcloned by S. Janicki (The Wistar Institute, Philadelphia, PA).
Cell culture and transfection
Cells were grown in DME supplemented with penicillin-streptomycin (Invitrogen) and 10% tet system–approved FBS (Clontech Laboratories, Inc.). Transient transfection was performed on cells growing on glass coverslips using Fugene 6 reagent (Roche) as per the manufacturer's protocol or by electroporation (Janicki et al., 2004) using a Gene Pulser II (Bio-Rad Laboratories). Because immediate addition of cumate (Qbiogene) drug seemed to decrease transfection efficiency, 200 μg/ml cumate was added 2 h after transfection and cells were left for 12 h for expression before processing for imaging.
Development of stable cell lines
Parental U2OS-2-6-3 cells (Janicki et al., 2004) grown in DME supplemented with 100 μg/ml each of penicillin-streptomycin and hygromycin B (Invitrogen) and 10% tet system–approved FBS were transfected with 2 μg pCMV5-CymR using Fugene 6. Stable clones were selected in 400 μg/ml G418 drug (Invitrogen) for 10 d. 100 individual colonies were picked and expanded. Although the clones were being frozen, each clone was seeded on coverslips, transfected with pCMV-CuO–lacI-mCherry, treated in the absence or presence of cumate (control), and screened by visual examination for absence of fluorescence, respectively, with a fluorescence microscope (Axioplan 2i; Carl Zeiss, Inc.) using a 40×/1.3 NA oil immersion objective lens (planApo-Neofluar). Images were captured using a charge-coupled device camera (Orca; Hammamatsu) and OpenLab Software (Improvision). By this visual screen, the best clone, which expressed the highest CymR repressor and hence negative fluorescence in the absence of cumate, was labeled as the U2OS-2-6-3–CymR repressor clone. The repressor clone was then subjected to a second round of transfection as before, but in parallel with pCMV-CuO–lacI-mCherry and pCMV-CuO–lacI-mCherry–lamin B1, and then selected as before but in the presence of 1 μg/ml puromycin (Sigma-Aldrich) for 7 d. 100 individual colonies for each transfection were picked, expanded, and screened by visual examination as before. The best clones, which gave no expression in the absence of cumate but optimum expression in the presence of cumate, were considered to be tightly regulated and were labeled as U2OS-2-6-3–CymR–lacI-mCherry (control cells) and U2OS-2-6-3–CymR–lacI-mCherry–lamin B1 (targeting cells). To make a targeting line stably expressing histone H2A-YFP, targeting cells were transfected with pCMV–H2A-YFP as before. 2 d after transfection, H2A-YFP–expressing fluorescent cells were sorted using a FACSVantage SE cell sorter with DiVa option (Becton Dickinson), using a helium-neon laser at 488 nm to generate the stable targeting line H2A-YFP.
Cell synchronization
On day 0, targeting cells were subjected to serum starvation in 0.5% FBS for a period of 48 h in 10-cm dishes to synchronize cells at G0 phase of cell cycle. On day 2, after 48 h of serum starvation, culture medium was replaced with fresh medium containing 10% FBS (to release from G0 phase) and 2 mM hydroxyurea (commonly used for G1/S-phase block) for a period of 20 h. To prepare asynchronous cells, targeting cells were treated as previously described but in the absence of hydroxyurea. At the end of 20 h, dishes containing G1/S-blocked cells or asynchronous cells were trypsinized and plated separately on glass coverslips with or without hydroxyurea, respectively. The plated cells were then allowed to attach to coverslips for a period of 4 h, and then induced with cumate to express the targeting fusion. After 12 h of cumate induction, both G1/S-blocked and asynchronous cells were fixed and DNA was stained with HOECHST 33342 (Invitrogen).
Immunofluorescence
Cells were rinsed once briefly with PBS and fixed for 15 min in 2% formaldehyde in PBS, pH 7.4, at room temperature. Fixed cells were permeabilized in PBS containing 0.5% Triton X-100 and 1% normal goat serum for 5 min on ice. After permeabilization, rabbit polyclonal anti–lamin B1 (N. Chaudhary and G. Blobel, Rockefeller University, New York, NY) primary antibody was added (1:500) for 1 h at room temperature. Cells were rinsed briefly with PBS containing 1% normal goat serum, and then incubated with anti–rabbit IgG Alexa Fluor 486 (Invitrogen) secondary antibody (1:1,000) for 1 h at room temperature. When required, DNA was stained with HOECHST 33342 for 5 min. Immunolabeled cells or formaldehyde-fixed cells expressing fluorescent proteins in cumate (+) and Dox (−/+) conditions were mounted in medium containing 10% PBS, pH 8.0, plus 1 mg/ml paraphenylenediamine. Cells were observed on a DeltaVision RT system (Applied Precision) as in Quantitation of RNA synthesis at the locus. Z stacks at 0.2-μm intervals were collected and deconvolved using SoftWoRx 2.50 software (Applied Precision).
Live-cell deconvolution microscopy
For time-lapse imaging of transcriptional activation, targeting cells grown on coverslips were transiently transfected with pVitro2–Tet-On + MS2-YFP and induced with cumate as before. Cells were then placed in an FCS2 chamber (Bioptechs), and phenol red–free Leibovitz's L15 (live cell) medium (Invitrogen) containing cumate was perfused into the chamber from a syringe. A second syringe with live-cell medium containing 200 μg/ml cumate and1 μg/ml Dox was also attached to the FCS2 chamber through an additional inlet (initially closed). The whole setup was mounted on a restoration microscope system (DeltaVision RT; Applied Precision) equipped with an inverted microscope (1X-70; Olympus), rapid shutters, and a 60×/1.4 NA oil immersion objective lens (planApo; Olympus) using CFP/YFP/mCherry filters (Chroma Technology Corp.), a cooled charge-coupled device camera (Photometrics Cool SNAP HQ; Roper Scientific) in a thermally insulated temperature-controlled chamber at 37°C. To minimize photosensitivity/phototoxicity from blue light during live-cell imaging, our DeltaVision RT has a sharp cut-off (long-pass) filter at ∼420 nm. Z stacks (0.4 μm, 1×1 binning) of transcriptionally inactive cells with a plastered locus (mCherry channel), a uniform diffuse nucleoplasmic signal (YFP channel) with no peroxisomal signal (CFP channel), were collected at 0 min. Immediately after imaging, live-cell medium with cumate and Dox from the second syringe was perfused into the FCS2 chamber and corrected for any z drift before the next time frame of imaging began at 20 min, and imaging was continued with an occasional perfusion of fresh medium. For time-lapse imaging during mitosis, targeting cells H2A-YFP were grown on coverslips and preinduced with 70 μg/ml cumate for 2–3 h and then placed in an FCS2 chamber, and live-cell medium containing 70 μg/ml cumate was perfused into the chamber from a syringe. To minimize the exposure of cells to fluorescent light, cells showing a nontargeted spherical locus at around G2/M phase were chosen by visualization of the H2A-YFP chromatin pattern and nuclear size and imaged through mitosis. Z stacks (0.4 μm, 2×2 binning) were collected (mCherry and YFP channel) every 10 min with occasional perfusion of fresh medium. Z stacks were deconvolved using SoftWoRx 2.50. In-focus sections of deconvolved images were projected, saved in tif format, and processed using Photoshop 7.0 (Adobe) and Illustrator 10.0 (Adobe). tif images from the time series were then combined using QuickTime Pro (Apple) to make videos.
Quantitation of RNA synthesis at the locus
Control cells or targeting cells grown on live-cell coverslips were transiently transfected with pVitro2–Tet-on + MS2-YFP and induced for 12 h with cumate for targeting fusion expression. The transfected cells were then imaged in the mCherry and YFP channels in Dox (−) condition (0 h), followed by imaging in Dox (+) condition every 20 min for a total period of 5 h. Raw images were then projected and the mean fluorescence intensity of the locus and of the whole nuclei were extracted using edit polygon and edit 2D polygon finder programs in SoftWoRx 2.50. The ratio of fluorescence intensity of the locus to the entire nucleus (mean values) was expressed as a percentage and was plotted against time of Dox induction.
Online supplemental material
Fig. S1 shows that the expressed targeting fusion is associated with the lac operator DNA at the targeted locus at the nuclear periphery. Fig. S2 shows serial z sections, which show targeting of the locus to the nuclear periphery. Video 1 shows that targeting of the locus to the lamina occurs at the end of mitosis. Video 2 shows serial z sections at a single time point, which show that the locus is targeted to the nuclear periphery. Video 3 shows that the targeted locus at the nuclear lamina remains targeted upon passage through mitosis. Video 4 shows transcription from the targeted locus at the lamina. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200706060/DC1.
Supplemental Material
Acknowledgments
We would like to thank Edith Heard, Prasanth Kannanganattu, and Veena Parnaik for invaluable suggestions, help, and critical review of the manuscript. We would also like to thank all members of the Spector laboratory for critical review of the manuscript and helpful discussions, as well as Jim Duffy and Catherine Eberstark for artistic services.
D.L. Spector is supported by grant 71407 from the National Institutes of Health/National Institute of General Medical Sciences.
Abbreviations used in this paper: CuO, cumate operator; LAP, lamina-associated polypeptide; pCMV, cytomegalovirus promoter; TRE, tetracycline response element.
References
- Andrulis, E.D., A.M. Neiman, D.C. Zappulla, and R. Sternglanz. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 394:592–595. [DOI] [PubMed] [Google Scholar]
- Becker, M., C. Baumann, S. John, D.A. Walker, M. Vigneron, J.G. McNally, and G.L. Hager. 2002. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3:1188–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont, A.S. 2001. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 11:250–257. [DOI] [PubMed] [Google Scholar]
- Belmont, A.S., Y. Zhai, and A. Thilenius. 1993. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J. Cell Biol. 123:1671–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner, J.H., and P. Walter. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.R., and P.A. Silver. 2007. Transcriptional regulation at the nuclear pore complex. Curr. Opin. Genet. Dev. 17:100–106. [DOI] [PubMed] [Google Scholar]
- Casolari, J.M., C.R. Brown, S. Komili, J. West, H. Hieronymus, and P.A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439. [DOI] [PubMed] [Google Scholar]
- Chen, D., M. Dundr, C. Wang, A. Leung, A. Lamond, T. Misteli, and S. Huang. 2005. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 168:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, C.H., A.E. Carpenter, B. Fuchsova, T. Johnson, P. de Lanerolle, and A.S. Belmont. 2006. Long-range directional movement of an interphase chromosome site. Curr. Biol. 16:825–831. [DOI] [PubMed] [Google Scholar]
- Chubb, J.R., S. Boyle, P. Perry, and W.A. Bickmore. 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12:439–445. [DOI] [PubMed] [Google Scholar]
- Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301. [DOI] [PubMed] [Google Scholar]
- Cremer, M., J. von Hase, T. Volm, A. Brero, G. Kreth, J. Walter, C. Fischer, I. Solovei, C. Cremer, and T. Cremer. 2001. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 9:541–567. [DOI] [PubMed] [Google Scholar]
- Croft, J.A., J.M. Bridger, S. Boyle, P. Perry, P. Teague, and W.A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink, A.K., and S. Henikoff. 1996. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature. 381:529–531. [DOI] [PubMed] [Google Scholar]
- Dernburg, A.F., K.W. Broman, J.C. Fung, W.F. Marshall, J. Philips, D.A. Agard, and J.W. Sedat. 1996. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 85:745–759. [DOI] [PubMed] [Google Scholar]
- Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L.I. Rothblum, R.D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 298:1623–1626. [DOI] [PubMed] [Google Scholar]
- Gartenberg, M.R., F.R. Neumann, T. Laroche, M. Blaszczyk, and S.M. Gasser. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 119:955–967. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T.I., K. Byrd, and V.G. Corces. 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 6:1025–1035. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., S. Boyle, H. Fiegler, K. Woodfine, N.P. Carter, and W.A. Bickmore. 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 118:555–566. [DOI] [PubMed] [Google Scholar]
- Goldman, R.D., Y. Gruenbaum, R.D. Moir, D.K. Shumaker, and T.P. Spann. 2002. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16:533–547. [DOI] [PubMed] [Google Scholar]
- Hetzer, M.W., T.C. Walther, and I.W. Mattaj. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21:347–380. [DOI] [PubMed] [Google Scholar]
- Hewitt, S.L., F.A. High, S.L. Reiner, A.G. Fisher, and M. Merkenschlager. 2004. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur. J. Immunol. 34:3604–3613. [DOI] [PubMed] [Google Scholar]
- Ishii, K., G. Arib, C. Lin, G. Van Houwe, and U.K. Laemmli. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 109:551–562. [DOI] [PubMed] [Google Scholar]
- Janicki, S.M., T. Tsukamoto, S.E. Salghetti, W.P. Tansey, R. Sachidanandam, K.V. Prasanth, T. Ried, Y. Shav-Tal, E. Bertrand, R.H. Singer, and D.L. Spector. 2004. From silencing to gene expression: real-time analysis in single cells. Cell. 116:683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., P.G. McQueen, M.K. Lichtman, E.M. Shevach, L.A. Parada, and T. Misteli. 2004. Spatial genome organization during T-cell differentiation. Cytogenet. Genome Res. 105:292–301. [DOI] [PubMed] [Google Scholar]
- Kimura, H., K. Sugaya, and P.R. Cook. 2002. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak, S.T., J.A. Skok, K.L. Medina, R. Riblet, M.M. Le Beau, A.G. Fisher, and H. Singh. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 296:158–162. [DOI] [PubMed] [Google Scholar]
- Lanctot, C., T. Cheutin, M. Cremer, G. Cavalli, and T. Cremer. 2007. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 8:104–115. [DOI] [PubMed] [Google Scholar]
- Lee, H., J.C. Quinn, K.V. Prasanth, V.A. Swiss, K.D. Economides, M.M. Camacho, D.L. Spector, and C. Abate-Shen. 2006. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 20:784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderus, M.E., A. de Graaf, E. Mattia, J.L. den Blaauwen, M.A. Grande, L. de Jong, and R. van Driel. 1992. Binding of matrix attachment regions to lamin B1. Cell. 70:949–959. [DOI] [PubMed] [Google Scholar]
- Lundgren, M., C.M. Chow, P. Sabbattini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell. 103:733–743. [DOI] [PubMed] [Google Scholar]
- Malhas, A., C.F. Lee, R. Sanders, N.J. Saunders, and D.J. Vaux. 2007. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J. Cell Biol. 176:593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W.F. 2002. Order and disorder in the nucleus. Curr. Biol. 12:R185–R192. [DOI] [PubMed] [Google Scholar]
- Marshall, W.F., A.F. Dernburg, B. Harmon, D.A. Agard, and J.W. Sedat. 1996. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol. Biol. Cell. 7:825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout, A., M. Goldberg, Y. Tzur, A. Margalit, and Y. Gruenbaum. 2007. Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J. Cell Sci. 120:77–85. [DOI] [PubMed] [Google Scholar]
- Meaburn, K.J., and T. Misteli. 2007. Cell biology: chromosome territories. Nature. 445:379–781. [DOI] [PubMed] [Google Scholar]
- Mendjan, S., M. Taipale, J. Kind, H. Holz, P. Gebhardt, M. Schelder, M. Vermeulen, A. Buscaino, K. Duncan, J. Mueller, et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 21:811–823. [DOI] [PubMed] [Google Scholar]
- Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science. 291:843–847. [DOI] [PubMed] [Google Scholar]
- Misteli, T. 2005. Concepts in nuclear architecture. Bioessays. 27:477–487. [DOI] [PubMed] [Google Scholar]
- Moir, R.D., M. Yoon, S. Khuon, and R.D. Goldman. 2000. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 151:1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada, L.A., J.J. Roix, and T. Misteli. 2003. An uncertainty principle in chromosome positioning. Trends Cell Biol. 13:393–396. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D.T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill, H., B. Kalverda, E. de Wit, W. Talhout, M. Fornerod, and B. van Steensel. 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38:1005–1014. [DOI] [PubMed] [Google Scholar]
- Ragoczy, T., M.A. Bender, A. Telling, R. Byron, and M. Groudine. 2006. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 20:1447–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, E.C., and L. Gerace. 2005. The nuclear membrane proteome: extending the envelope. Trends Biochem. Sci. 30:551–558. [DOI] [PubMed] [Google Scholar]
- Schirmer, E.C., and R. Foisner. 2007. Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 313:2167–2179. [DOI] [PubMed] [Google Scholar]
- Shav-Tal, Y., X. Darzacq, S.M. Shenoy, D. Fusco, S.M. Janicki, D.L. Spector, and R.H. Singer. 2004. Dynamics of single mRNPs in nuclei of living cells. Science. 304:1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker, D.K., T. Dechat, A. Kohlmaier, S.A. Adam, M.R. Bozovsky, M.R. Erdos, M. Eriksson, A.E. Goldman, S. Khuon, F.S. Collins, et al. 2006. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA. 103:8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, D.L. 1993. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9:265–315. [DOI] [PubMed] [Google Scholar]
- Spector, D.L. 2003. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72:573–608. [DOI] [PubMed] [Google Scholar]
- Spector, D.L. 2006. SnapShot: cellular bodies. Cell. 127:1071. [DOI] [PubMed] [Google Scholar]
- Taddei, A., G. Van Houwe, F. Hediger, V. Kalck, F. Cubizolles, H. Schober, and S.M. Gasser. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 441:774–778. [DOI] [PubMed] [Google Scholar]
- Taniura, H., C. Glass, and L. Gerace. 1995. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol. 131:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, I., S. Gilchrist, W.A. Bickmore, and J.R. Chubb. 2004. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr. Biol. 14:166–172. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, T., N. Hashiguchi, S.M. Janicki, T. Tumbar, A.S. Belmont, and D.L. Spector. 2000. Visualization of gene activity in living cells. Nat. Cell Biol. 2:871–878. [DOI] [PubMed] [Google Scholar]
- Tumbar, T., G. Sudlow, and A.S. Belmont. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschure, P.J., I. van Der Kraan, E.M. Manders, and R. van Driel. 1999. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 147:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, J., L. Schermelleh, M. Cremer, S. Tashiro, and T. Cremer. 2003. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 160:685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R.R., V. Azuara, P. Perry, S. Sauer, M. Dvorkina, H. Jorgensen, J. Roix, P. McQueen, T. Misteli, M. Merkenschlager, and A.G. Fisher. 2006. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 119:132–140. [DOI] [PubMed] [Google Scholar]
- Zhao, K., A. Harel, N. Stuurman, D. Guedalia, and Y. Gruenbaum. 1996. Binding of matrix attachment regions to nuclear lamin is mediated by the rod domain and depends on the lamin polymerization state. FEBS Lett. 380:161–164. [DOI] [PubMed] [Google Scholar]
- Zink, D., M.D. Amaral, A. Englmann, S. Lang, L.A. Clarke, C. Rudolph, F. Alt, K. Luther, C. Braz, N. Sadoni, et al. 2004. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 166:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.