Abstract
Werner and Bloom syndromes are human diseases characterized by premature age-related defects including elevated cancer incidence. Using a novel Saccharomyces cerevisiae model system for aging and cancer, we show that cells lacking the RecQ helicase SGS1 (WRN and BLM homologue) undergo premature age-related changes, including reduced life span under stress and calorie restriction (CR), G1 arrest defects, dedifferentiation, elevated recombination errors, and age-dependent increase in DNA mutations. Lack of SGS1 results in a 110-fold increase in gross chromosomal rearrangement frequency during aging of nondividing cells compared with that generated during the initial population expansion. This underscores the central role of aging in genomic instability. The deletion of SCH9 (homologous to AKT and S6K), but not CR, protects against the age-dependent defects in sgs1Δ by inhibiting error-prone recombination and preventing DNA damage and dedifferentiation. The conserved function of Akt/S6k homologues in lifespan regulation raises the possibility that modulation of the IGF-I–Akt–56K pathway can protect against premature aging syndromes in mammals.
INTRODUCTION
Sgs1 is the only known Saccharomyces cerevisiae member of the RecQ helicase family of DNA unwinding proteins, which is involved in the maintenance of genomic stability in organisms ranging from Escherichia coli to humans. Mutations in at least three of the five human genes encoding RecQ helicases (BLM, WRN, and RECQ4) promote genomic instability and cancer. Specific mutations in BLM cause Bloom's Syndrome (BS), which is characterized by increased sister chromatid exchanges and a wide variety of tumors, as well as diabetes and immunodeficiency (Hickson et al., 2001; Mohaghegh and Hickson, 2002; Thompson and Schild, 2002; Kaneko and Kondo, 2004). WRN mutations, which cause Werner's syndrome (WS), promote cancer, cardiovascular diseases, diabetes, and immunodeficiency and accelerate other age-related changes, including graying of the hair, wrinkling of the skin, and osteoporosis (Epstein et al., 1966; Ozgenc and Loeb, 2005). The first connection between the yeast WRN/BLM homologue and premature aging was demonstrated by the fact that mutants lacking SGS1 showed a shortened replicative life span (Sinclair et al., 1997). Furthermore, expression of either human BLM or WRN gene in yeast was able to suppress certain recombination phenotypes of yeast lacking SGS1 (Yamagata et al., 1998), indicating that these human genes are, at least in part, functional homologues of SGS1. Sgs1 is required to prevent defective DNA recombination, which causes premature G2/M arrest and senescence (Johnson et al., 2001; McVey et al., 2001). RecQ helicases catalyze the DNA unwinding through the conserved RecQ domain, which was originally described for the E. Coli RecQ protein (Gangloff et al., 1994; Sun et al., 1999). In the S. cerevisiae Sgs1 (1,447 aa), the RecQ domain is followed, as in most of the RecQ proteins, by a RecQ C-terminal domain (a C-terminal extension from the helicase domain, which is involved in protein–protein interactions) and the HRCD domain (helicase/RNase D C terminal, which is involved in DNA binding). Moreover, Sgs1 contains key domains, including one required for S-phase checkpoint function and others that interact with Top3, Rad51, or checkpoint kinase Rad53 (Miyajima et al., 2000b; Mullen et al., 2000; Onodera et al., 2002; Ui et al., 2005). The structural complexity of this protein may explain the multiplicity of cellular processes in which Sgs1 is involved (S-phase checkpoint, replication, double-strand break repair, recombination, telomere maintenance, etc.), as well as the complexity of phenotypes associated with its deficiency (Bennett and Keck, 2004; Cobb and Bjergbaek, 2006; Killoran and Keck, 2006).
The helicase and Top3 interaction domains, but not the S-phase checkpoint function domain, were shown to be required to prevent rapid clonal senescence in S. cerevisiae telomerase mutants. This was measured by monitoring population doublings (Azam et al., 2006). The Sgs1–Top3 complex has also been implicated in interactions with mismatch repair proteins (Myung et al., 2001). Because mismatch proteins act to limit recombination between similar, but not identical, DNA sequences, it is possible that RecQ proteins also act to limit recombination by reversing inappropriate heteroduplex intermediates. In fact, hMSH2/6 forms a complex with human BLM–p53–RAD51 in response to the damaged DNA forks during double-stranded–break repair (Pedrazzi et al., 2003; Yang et al., 2004). The hyperrecombination and erroneous (homeologous) recombination seen in sgs1Δ cells appear to play a central role in the high level of gross chromosomal rearrangements (GCRs) observed in these mutant strains (Onoda et al., 2000; Myung et al., 2001; Myung and Kolodner, 2002; Schmidt et al., 2006). Sister chromatid exchange and exchange of genetic material between different chromosomes may be important features of RecQ helicase deficiency because they are commonly observed in human BS cells (Ellis et al., 1995; German, 1995). WS cells do not show an elevated sister chromatid–exchange frequency, but they do display increased illegitimate recombination and a high frequency of large chromosomal deletions (Shen and Loeb, 2000).
In this paper, we develop a simple yeast system modeling BS and WS, based on studies of the chronological life span (CLS) and on the monitoring of cell-cycle profile, DNA mutations, chromosomal rearrangements, and dedifferentiation of sgs1Δ mutants during chronological aging. Using this model system, we describe a marked age-dependent increase in various categories of mutations in cells lacking SGS1, which can be prevented by the deletion of SCH9, a homologue of mammalian AKT and S6 kinase (Toda et al., 1988; Urban et al., 2007). We provide evidence that points to enhanced cellular protection against stress, tighter G1 arrest, and reduced recombination errors as mechanisms by which lack of Sch9 activity protects against genomic instability and dedifferentiation associated with accelerated aging in sgs1Δ mutants.
RESULTS
Aging in sgs1Δ mutants
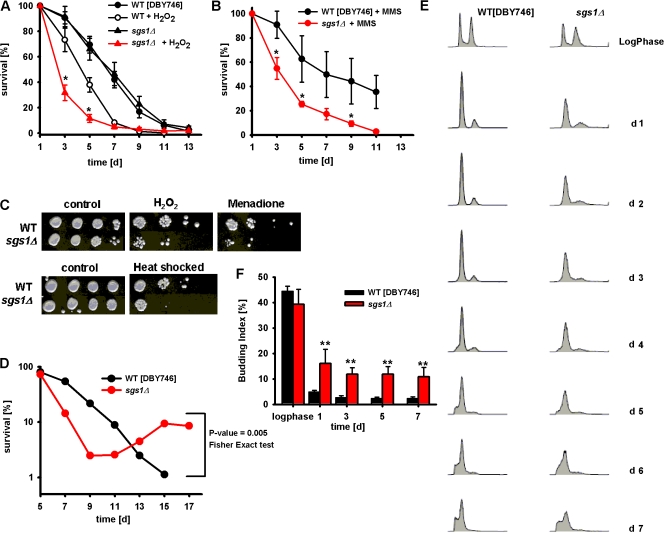
Yeast sgs1Δ of three different genetic backgrounds (DBY746, BY4741, and SP1) did not have a shortened CLS when maintained in standard glucose/ethanol-containing medium (Fig. 1 A and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200707154/DC1). This observation was confirmed by measuring CLS using the phloxin B live/dead staining (Kucsera et al., 2000), which ruled out the possibility that wild-type cells may live longer than sgs1Δ mutants but may be unable to form a colony. However, premature death was observed after chronic exposure of sgs1Δ mutants to either hydrogen peroxide or the alkylating agent methyl methane sulfonate (MMS; Fig. 1, A and B). Sgs1-deficient cells aging chronologically were also hypersensitive to acute heat shock or acute treatment with hydrogen peroxide or the superoxide/H2O2-generating agent menadione (Fig. 1 C).
Figure 1.
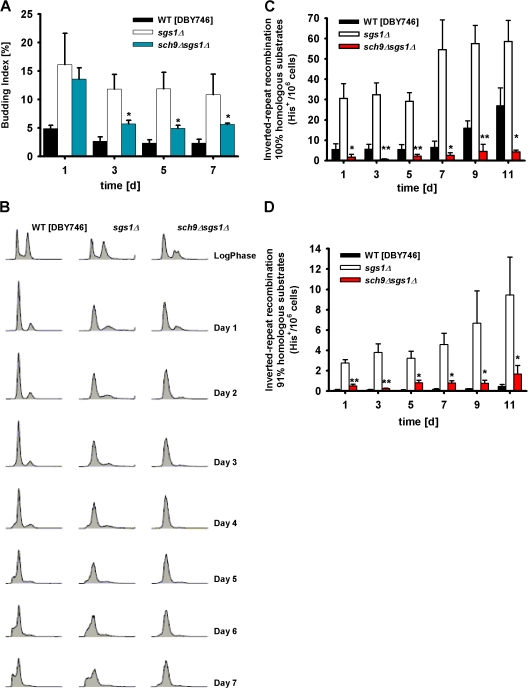
Chronological survival of sgs1Δ mutants. (A and B) Chronological survival in synthetic complete medium (SDC) of wild-type (DBY746) and sgs1Δ mutant cells in standard conditions or after chronic exposure (days 1 and 3) to either 1 mM H2O2 (A) or 0.001% MMS (B). Data represent the mean ± SEM (n = 12–17). *, P < 0.05 (sgs1Δ treated cells vs. wild-type treated cells). (C) Oxidative stress and heat shock resistance of wild-type and sgs1Δ mutant. Day-3 cells were exposed to 100 mM H2O2 for 30 min, 250 μM menadione for 60 min (top), or heat shock treatment at 55°C for 90 min (bottom). These experiments were repeated three times with similar results. (D) Survival and regrowth/dedifferentiation during the high mortality phase (day 7–17) of wild-type and sgs1Δ mutant. A representative experiment is shown (see Table I). (E) Cell-cycle profiles of wild-type (DBY746) and sgs1Δ mutant cells during log phase and from day 1–7. (F) Budding index of wild-type and sgs1Δ mutant cells. Data represent the mean ± SEM (n = 7–8). **, P < 0.01 (sgs1Δ cells vs. wild-type cells).
Increased chronological age-dependent dedifferentiation and G1 arrest defects in yeast sgs1Δ mutants
We have previously shown that yeast mutants reminiscent of cancer cells, which dedifferentiate and grow by using the nutrients released by dead cells, can emerge within aging S. cerevisiae cultures (Fabrizio et al., 2004). The frequency of this phenotype was doubled in cells lacking cytosolic superoxide dismutase 1 and greatly reduced in long-lived mutants or mutants overexpressing superoxide dismutases (Fabrizio et al., 2004). Because both WS and BS are associated with elevated age-dependent cancer incidence, we tested whether mutations in SGS1 affected this dedifferentiation phenotype. Subpopulations of cells were considered to be dedifferentiating/regrowing if the viability counts during the high mortality phase (days 7–17) either increased by at least 50% or did not decrease in three consecutive counts (6 d) as expected, based on the normal age-dependent exponential increase in mortality rates (Finch, 1990). Early dedifferentiation occurred in 3 out of 38 wild-type DBY746 cultures but in 18 out of 38 sgs1Δ cultures (48%; P = 0.005 [Fisher's exact test]; Fig. 1 D and Table I). The frequency of dedifferentiation in wild-type cells was lower than that reported in our previous study (Fabrizio et al., 2004) because dedifferentiation after day 17 was not considered. Similarly, in the SP1 genetic background, early dedifferentiation occurred in two out of three wild-type cultures (67%) and three out of three sgs1Δ cultures (100%), and in the BY4741 background in one out of six wild-types cultures (17%) but three out of six sgs1Δ cultures (50%; Table I). These results suggest that Sgs1 contributes to the prevention of age-dependent dedifferentiation.
Table I. Percentage of age-dependent early dedifferentiation (days 7–17).
| Strains
|
|||
|---|---|---|---|
| Background | Wild type % (n) |
sgs1Δ
% (n) |
sch9Δsgs1Δ
% (n) |
| DBY746 | 8.0 (38) | 47.4 (38)a | 12.0 (25)b |
| BY4741 | 16.7 (6) | 50.0 (6) | 0.0 (6) |
| SP1 | 66.7 (3) | 100.0 (3) | 0.0 (3) |
P = 0.005 (vs. wild type; Fisher's exact test).
P = 0.036 (vs. sgs1Δ; Fisher's exact test).
To further test the hypothesis that Sgs1 might exert a role in blocking cellular dedifferentiation, we determined cell-cycle and budding index profiles during aging. FACS analysis showed a similar profile for both log-phase wild-type and sgs1Δ mutant cells (Fig. 1 E and Table II). Starting on day 1, wild-type cells were mostly G1 arrested with only 5% of cells with small buds (Fig. 1 F), and a lower percentage of cells were in S phase (Table II). The efficiency of G1 arrest is indicated by the increased height and narrowness of the G1-DNA content peak and the smaller amount of cells with an S-phase or G2/M content of DNA. In accordance with the dedifferentiation phenotype showed by the Sgs1-deficient strain (Fig. 1 D), sgs1Δ mutant cultures displayed a slightly lower G1 arrest than wild-type cells but a threefold increase in budding index (15%) and a 2–10-fold higher percentage of cells in S phase (Fig. 1, E and F; and Table II).
Table II. Percentage of cells in each cell-cycle compartment at early time points.
| Strain
|
||||
|---|---|---|---|---|
| Time point | Cell-cycle phase |
Wild type (DBY746) |
sgs1Δ | sch9Δsgs1Δ |
| Log phase | sub G1 | 0.0 | 0.6 | 0.4 |
| G1 | 31.5 | 35.2 | 59.4 | |
| S | 16.6 | 11.8 | 3.0 | |
| G2/M | 51.9 | 52.9 | 37.6 | |
| Day 1 | sub G1 | 0.3 | 0.3 | 0.0 |
| G1 | 82.0 | 69.5 | 71.7 | |
| S | 0.0 | 4.0 | 12.3 | |
| G2/M | 18.0 | 26.4 | 15.6 | |
| Day 2 | sub G1 | 0.4 | 0.2 | 0.0 |
| G1 | 82.5 | 68.8 | 86.9 | |
| S | 1.5 | 12.6 | 7.5 | |
| G2/M | 16.0 | 18.5 | 5.7 | |
| Day 3 | sub G1 | 2.9 | 2.0 | 0.0 |
| G1 | 79.4 | 72.9 | 86.6 | |
| S | 7.8 | 14.0 | 9.3 | |
| G2/M | 12.7 | 13.1 | 4.1 | |
Data are from FACS analysis measurements presented in Figs. 1 E and 5 B and calculated using MODFIT.
Premature genomic instability in sgs1Δ mutants
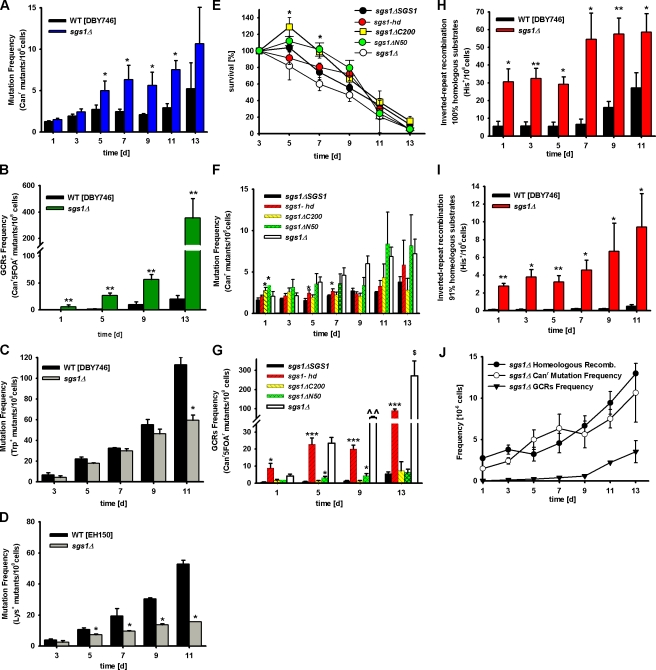
To characterize the type of DNA damage occurring during normal and premature aging (Lombard et al., 2005), we monitored the frequency of Canr mutations, GCRs, base substitutions (trp1-289 reversions, trp−→TRP1+), and small DNA insertions/deletions (lys2-Bgl→LYS+; Madia et al., 2007) in both wild-type and Sgs1-deficient S. cerevisiae aging chronologically (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200707154/DC1). Canr mutations impart cellular resistance to the arginine analogue canavanine (l-canavanine sulfate) and are easily detectable. During growth, they are mostly caused by a combination of frameshifts and point mutations in the CAN1 gene (Chen et al., 1998). In our previous papers, we showed that in the wild-type DBY746 genetic background, spontaneous Canr mutations increased progressively with age and reached a frequency of approximately three to five per one million cells by day 11–13 (Fig. 2 A; Fabrizio et al., 2004, 2005). Similar to what had been shown in growing yeast cells (Myung et al., 2001), mutations in SGS1 did not induce an increase of Canr frequency on day 1 of a chronological aging study compared with the wild-type strain (Fig. 2 A). However, the frequency increased rapidly in sgs1Δ mutants and reached a threefold higher level than in wild-type cells by days 7 and 9 (Fig. 2 A). An analogous, but delayed, effect on Canr mutations was observed in the longer-lived BY4741 genetic background (Fig. S3 A).
Figure 2.
Premature genomic instability in sgs1Δ mutants. (A) Age-dependent mutation frequency in CAN1 gene, measured as Canr mutants/106 cells of wild-type (DBY746) and sgs1Δ cells. The mean ± SEM is shown (n = 12–16). (B) Age-dependent GCR frequency measured as Canr5FOAr mutants/108 cells, in sgs1Δ mutants compared with wild-type cells. The mean ± SEM is shown (n = 7–11). (C) Age-dependent spontaneous base-substitution mutations measured as tryptophan reversions in wild-type and sgs1Δ mutant. Data are presented as mean ± SEM. (D) Age-dependent spontaneous small insertion/deletion mutations measured as Lys+ revertants in the EH150 genetic background. Mutation frequencies over time were measured as cumulative appearance of Trp+ or Lys+/108 cells. The means ± SEM are presented (n = 6–9). (E–G) Survival (E) of mutants carrying sgs1Δ, sgs1-ΔC200, sgs1-ΔN50, sgs1-hd alleles, and sgs1ΔSGS1 in DBY746 background and age-dependent mutation frequency of Canr mutants (F) and GCRs (G). The means ± SEM are shown (n = 7–8). (H and I) Age-dependent homologous (H) and homeologous (I) recombination frequency measured as His+/106 cells of wild-type and sgs1Δ mutant cells. Data are shown as mean ± SEM (n = 4). (J) Age-dependent homeologous recombination events, Canr mutation, and GCR frequencies per million cells in the DBY746 strain lacking SGS1. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (vs. wild type). ^, P < 0.05; ^^, P < 0.01 (sgs1Δ vs. sgs1-hd).
To gain further insight into the type of age-dependent mutations, we analyzed the spectra of mutations in 10 Canr mutant colonies from 7-d-old sgs1Δ mutant cultures (Table III). We were only able to PCR amplify and sequence the entire CAN1 gene in 5 out of 10 Canr mutants from 7-d-old sgs1Δ cultures (Table III), which suggests that half of the CAN1 genes contain GCRs that prevent sequencing. Sequencing of the CAN1 gene revealed that four mutants carried a single point mutation (three with base substitution and one with deletion) and one a double mutation (Table III). The four-base substitutions were C→A transversions (two of four) and T→C transitions (two of four), whereas the deletions were ΔA and ΔT (Table III). Thus, in addition to suppressing GCRs during growth (Myung et al., 2001; Myung and Kolodner, 2002), Sgs1 protects against GCRs and, possibly, other mutations during chronological aging.
Table III. Spectrum of mutations observed in Canr colonies from day 7.
| Clone | Mutation type | Position from ATG | Sequence | ||
|---|---|---|---|---|---|
| Wild typea | |||||
| 1 | Base substitution | C→T | Proline-leucine | 656 | TGTTCCCTGTC |
| 2 | Base substitution | T→G | Asparagine-lysine | 1173 | AAATTCAAATA |
| Insertion | G | 1710 | GAGGC-G-AATTGT | ||
| 3 | Duplication | 248bp | 184–431 | ||
| 4 | Base substitution | G→C | Alanine-proline | 709 | TTTTAGCCATTA |
| Deletion | TG | Isoleucine-arginineframeshift | 1098–1099 | TTGCTATTGAGAA | |
| 5 | Base substitution | G→T | Glycine-valine | 353 | ACGCCGGCCCAG |
| Insertion | A | 1341 | CAAA-A-GTTTTCG | ||
| 6 | Base substitution | C→T | Proline-serine | 937 | GCTGCAAACCCCA |
| 7 | no PCR | ||||
| 8 | Deletion | AT | Frameshift | 1129–1130 | TGTCACATATCTT |
| Insertion | T | Frameshift (T3-T4) | 1086 | CCCTTT-T-ATTATT | |
| 9 | Base substitution | G→T | Tryptophan-cysteine | 531 | GGTTTTCTTGGCA |
| Deletion | A | Frame-shift (A3-A2) | 663 | CAAATATTACGGT | |
| Insertion | T | Frameshift (T3-T4) | 1086 | CCCTTT-T-ATTATT | |
| 10 | Base substitution | G→A | Glutamic acid-lysine | 679 | GAATTCGAGTTCT |
| Base substitution | G→T | Valine-phenylalanine | 907 | GAACTATTTGGTA | |
| Deletion | A | Frameshift | 1217 | TATCAAAGAACAC | |
| sgs1Δ b | |||||
| 1 | Base substitution | C→A | Tyrosine-STOP | 591 | GGACGTACAAAG |
| Deletion | A | Frameshift (A6-A5) | 969 | TCAAAAAAGTTGC | |
| 2 | Deletion | T | Frameshift | 1756 | GAATGTTGTAGC |
| 3 | partially sequenced | ||||
| 4 | no PCR | ||||
| 5 | Base substitution | T→C | Valine-alanine | 659 | GTTCCCTGTCAAA |
| 6 | no PCR | ||||
| 7 | partially sequenced | ||||
| 8 | no PCR | ||||
| 9 | Base substitution | T→C | Tryptophan-arginine | 529 | GGTTTTCTTGGGC |
| 10 | Base substitution | C→A | Alanine-aspartic acid | 1166 | ATTTCTGCCGCAA |
| sch9Δsgs1Δ c | |||||
| 1 | Insertion | CT | Frameshift | 1006–1007 | GCTCTCT-CT-ATT |
| 2 | Base substitution | G→A | Glycine-serine | 670 | ATTACGGTGAATT |
| 3 | Base substitution | G→T | Glutamic acid-STOP | 184 | TGGCGAGGATAC |
| 4 | partially sequenced | ||||
| 5 | no PCR | ||||
| 6 | Base substitution | A→C | Tyrosine-serine | 1184 | TATTTACGTTGGT |
| 7 | Base substitution | G→T | Glycine-valine | 911 | TAGTTGGTATCAC |
| 8 | Base substitution | G→A | Glycine-serine | 922 | ACTGCTGGTGAAG |
| 9 | Base substitution | C→A | Phenylalan-leucine | 1193 | ACGTTGGTTCCTA |
| 10 | Deletion | T | Frameshift leucine-phenyalan | 543 | AATCACTTTTGCC |
| Base substitution | G→C | No change | 544 | AATCACTTTTGCC | |
Base substitutions: bold, insertions; underline, deletions.
Total clones: 10 (no PCR, 1/10; sequenced, 9/10).
Total clones: 10 (no PCR, 3/10; partially sequenced, 2/10; sequenced, 5/10).
Total clones: 10 (no PCR, 1/10; partially sequenced, 1/10; sequenced, 8/10).
Kolodner and co-workers had shown, during the growth phase, a 20-fold increase in the rate of GCRs in sgs1Δ mutants when compared with wild-type yeast (Myung et al., 2001;Myung and Kolodner, 2002). We measured the GCR frequency in log-phase (OD600 of ∼1) wild-type and Sgs1-deficient cultures and obtained similar results (wild type, 0.51 ± 0.03 × 10−8 cells; sgs1Δ, 8.20 ± 0.45 × 10−8 cells). During chronological aging, the frequency of GCRs underwent a 40-fold age-dependent increase in wild-type cells and a 110-fold increase in the sgs1Δ mutant between days 1 and 13 (Fig. 2 B), underscoring the central role of aging in genomic instability. It should be noted that this increase in GCR frequency was observed on day 13 when <5% of the initial population was still alive. Thus, it represents a very high genomic instability in the subpopulation that survives until day 13. The GCR results are in agreement with the inability to totally or partially PCR amplify the CAN1 gene in half of the Canr mutants that emerged from sgs1Δ cultures (Table III).
Point and insertion/deletion mutations in sgs1Δ mutants
As an additional method to monitor the occurrence of point mutations in cells aging chronologically, we measured the frequency of age-dependent C→T base substitutions, which cause reversion of the trp1-289 amber mutation present in the DBY746 genetic background and allow growth in the absence of tryptophan (Fig. S2). The frequency of trp1-289 reversion mutation was similar between wild-type and sgs1Δ mutants between days 3 and 9 and was significantly lower in sgs1Δ mutants compared with that in wild-type cells at day 11 (Fig. 2 C). These results were in agreement with the absence of C→T base substitutions in the sequenced CAN1 genes (Table III).
To further characterize the link between Sgs1 and the age-dependent mutations and as an indirect measure of nonhomologous end-joining (NHEJ) repair (Heidenreich et al., 2003), we examined how lack of Sgs1 affected the frequency of small insertions of 2 or 5 bp or deletions of 1 or 4 bp, which cause a frameshift and allow the expression of Lys2 (Fig. S2). We monitored the frequency of reversion of the lysine auxotrophy phenotype of a lys2-Bgl mutant aging chronologically (Boulton and Jackson, 1996; Heidenreich et al., 2003). Although the frequency of LYS+ mutations was similar in young wild-type and sgs1Δ mutants, it increased by 13-fold in wild-type cells but only sixfold in sgs1Δ mutants by day 11. The sequencing of the mutated CAN1 gene from sgs1Δ mutants at day 7 confirmed that the frequency of small insertions/deletions was reduced (Table III). Collectively, these data indicate that C→T substitutions and NHEJ-dependent insertions/deletions occur at a normal or reduced rate in SGS1-deficient yeast (Fig. 2, C and D; and Table III).
Role of Sgs1 functional domains in age-dependent genomic instability
The characterization of the yeast SGS1 gene indicated that different SGS1 domains have distinct functions (Miyajima et al., 2000a,b; Mullen et al., 2000; Lo et al., 2006). As a first step toward mapping the different Sgs1 domains required for age-dependent mutation and dedifferentiation avoidance, we generated yeast strains lacking either the region encoding for the C-terminal 200 aa (sgs1-ΔC200) or the N-terminal 50 aa (sgs1-ΔN50) or bearing a mutation that disrupts the helicase activity (sgs1-hd) by a single amino acid change within the ATP binding domain of Sgs1 (K706A; Mullen et al., 2000; Azam et al., 2006). The mutated, as well as the wild-type, SGS1 alleles (sgs1Δ-SGS1) were integrated at the LEU2 locus of sgs1Δ mutant. Chronological survival assay, Canr mutation frequency, and GCR frequency assays were performed with these strains.
Yeast strains lacking the helicase domain (sgs1-hd) or the N-terminal 50-aa region (sgs1-ΔN50) showed no difference in survival when compared with the strain lacking the entire protein (sgs1Δ) or the wild type (Fig. 2 E). Notably, the cell density of yeast lacking the S-phase checkpoint domain (sgs1-ΔC200) continued to increase slightly until day 5 (Fig. 2 E), which is in agreement with a helicase-independent role for the Sgs1 S-phase domain in regulating G1 arrest and dedifferentiation during aging. Because this growth effect was not observed in sgs1Δ, it appears that the lack of the checkpoint domain in cells that otherwise maintain other functions is more permissive to the observed additional 20% growth. Lack of the S-phase checkpoint function did not affect the age-dependent Canr mutation frequency (Fig. 2 F). However, at day 1, sgs1-ΔC200 cells displayed a significantly higher mutation frequency compared with the wild type (sgs1-ΔC200, 2.75 ± 0.38 × 10−6; sgs1ΔSGS1, 1.62 ± 0.26 × 10−6; P < 0.05), indicating that the sgs1-ΔC200 cells have a slightly higher mutation rate during growth (Fig. 2 F). The mutated Sgs1 bearing a deletion in the N-terminal 50 aa (sgs1-ΔN50) lacks residues that are essential for the interaction with Top3, a type I topoisomerase that cooperates with Sgs1p during homologous recombination (Bennett and Wang, 2001; Fabre et al., 2002; Onodera et al., 2002). The age-dependent mutation frequency (Canr) of the sgs1-ΔN50 mutant was also elevated on day 1 (Fig. 2 F; sgs1-ΔN50, 3.40 ± 0.74 × 10−6; sgs1ΔSGS1, 1.62 ± 0.26 × 10−6; P < 0.05) but did not display a significant age-dependent increase when compared with sgs1ΔSGS1 cells (Fig. 2 F). Similar Canr frequency results were obtained with the sgs1-hd allele, which carries a single point mutation in the helicase domain and has no detectable DNA helicase activity (Mullen et al., 2000; Fig. 2 F).
Our data indicate that only the combination of various activities of the Sgs1 protein completely protects against age-dependent Canr mutations. These activities may include some that we have not investigated or remain unknown. In contrast, the DNA helicase activity appears to be involved in the avoidance of a major portion of GCRs during aging (Fig. 2 G). Until day 5 of a standard chronological aging study, the sgs1-hd allele showed a similar GCR frequency compared with that of sgs1Δ mutants. At days 9 and 13, the Sgs1-deficient strain showed a two- to threefold higher GCR frequency compared with sgs1-hd. These results suggest that the helicase activity of Sgs1 plays a central role in the avoidance of age-dependent GCRs but that other domains of Sgs1 also protect against gross chromosomal damage.
Hyperhomologous and homeologous recombination in aging sgs1Δ mutants
To determine whether erroneous recombination, previously shown to play a major role in GCR occurrence during growth in sgs1Δ mutants (Myung et al., 2001), might be responsible for both Canr mutations and GCRs during aging, we measured the frequency of homologous and homeologous recombinational events in wild-type and Sgs1-deficient strains. We generated wild-type DBY746 and sgs1Δ mutants in which we could measure the frequency of recombination between inverted-repeat substrates with 100 or 91% homology, as described previously (Datta et al., 1996). Both homologous and homeologous recombination were much higher in sgs1Δ mutants compared with wild-type cells on day 1 (end of the growth phase; Fig. 2, H and I). During chronological aging, the frequency of homologous recombinational events remained stable up to day 5 and then increased by two to threefold in both wild-type and sgs1Δ cells. Homeologous recombination frequency increased steadily between days 5 and 11 in both wild-type cells and sgs1Δ mutants (Fig. 2, H and I). The maximum difference between sgs1Δ mutants and wild-type cells was observed by day 5–7 with the sgs1Δ/wild-type ratio reaching 8- and 30-fold for homologous and homeologous recombination, respectively. Based on our data and those of Myung et al. (2001), this high level of homeologous recombination is likely to contribute to the significant increase of Canr mutations and GCRs observed in the sgs1Δ mutants during aging (Fig. 2 J).
Age-dependent mutations in SGS1 deletion mutants originate both in nonquiescent and quiescent cells
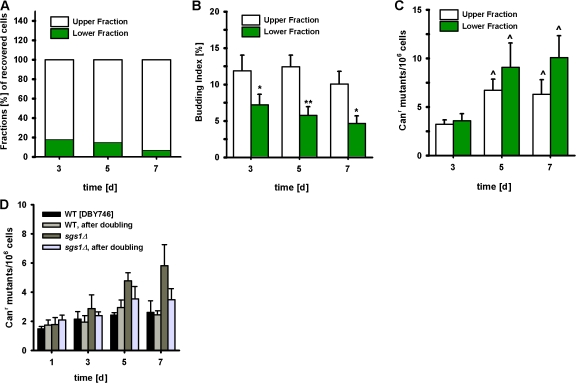
The results presented indicate a significant age-dependent increase in mutation accumulation, GCRs, and recombinational events in the Sgs1-deficient strain when compared with the wild type. To understand whether the initial age-dependent DNA damage occurs in nondividing cells and to investigate the association between cell-cycle arrest and mutation frequency, we made use of the fractionation method recently described by Allen et al. (2006). The cultures subjected to the Percoll density gradient separated into two distinct fractions: the upper fraction, mainly composed of a heterogeneous population of nonquiescent cells that includes dividing, nondividing, apoptotic, and necrotic cells and the lower fraction of quiescent cells (Allen et al., 2006). The Canr mutation frequency of both fractions derived from day-3, -5, and -7 cultures were measured.
In the DBY746 background, quiescent cells accounted for 40% of the wild-type population by day 3 (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200707154/DC1). This percentage gradually decreased to 13% by day 7. In contrast, on days 3 and 7, respectively, only 20 and 7% of the sgs1Δ cells were observed in the lower quiescent fraction (Fig. 3 A). Moreover, sgs1Δ cells displayed significantly higher percentages of budding indices (Fig. 3 B) over time in both fractions compared with wild type (Fig. S4 B), which suggests a defect in cell-cycle/G1 arrest and inability to enter into a true “quiescent” state. As expected, the deletion of SGS1 caused a significant age-dependent increase of mutation frequency in both the upper and lower fractions when compared with the wild type (Fig. 3 C and Fig. S4 C). The mutation frequency levels detected in Sgs1-deficient cells were of the same order of magnitude before and after treatment with Percoll gradient, indicating that the cell separation procedure itself (including high Percoll concentrations) was not responsible for the increase of mutations.
Figure 3.
Premature genomic instability in sgs1Δ mutants originates from both nonquiescent and quiescent cells. (A) Fractions (%) of recovered cells from sgs1Δ chronological aging cultures subjected to a Percoll density-gradient separation. (B and C) Budding index (%; B) and CAN1 mutation frequency (C) over time measured as Canr mutants/106 cells are measured in upper and lower fractions. Data are shown as mean ± SEM (n = 4–5). *, P < 0.05; **, P < 0.01 (lower fraction vs. upper fraction). ^, P < 0.05 (vs. day 3). (D) CAN1 mutation frequency over time measured as Canr mutants/106 cells of wild-type and sgs1Δ mutant cells before and after doubling. On days 1, 3, 5, and 7 of a chronological aging study, cells were allowed to undergo one population doubling before plating on canavanine selective medium. The mean ± SEM is shown (n = 4).
To determine whether the initial DNA damage might form during cell division and not during aging, we allowed cells from days 1, 3, 5, and 7 of a standard CLS study to undergo one population doubling before treatment with the toxic canavanine. If the initial DNA damage did not occur during aging but during the reentry of aging cells into the cell cycle, we would expect the population doubling of all the 2 × 107 cells to cause a major increase in mutation frequency. Instead, we observed that the frequency of mutations normalized for population size remained the same for cells exposed to canavanine before or after one population doubling (Fig. 3 D), which suggests that at least the initial DNA damage in the CAN1 gene occurs in nondividing cells during aging and becomes a mutation, deletion, or GCR during the first round of replication.
Deletion of oncogene homologue SCH9, but not severe calorie restriction (CR), blocks premature genomic instability in sgs1Δ mutants
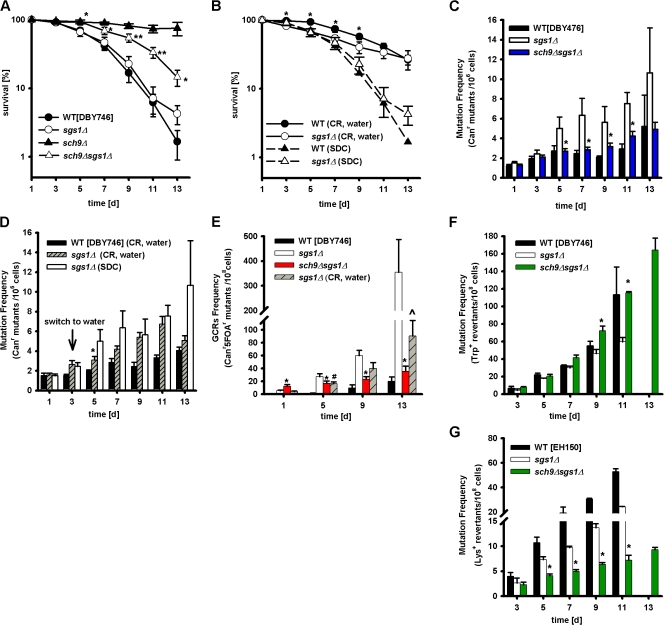
We previously showed that yeast lacking SCH9 or components of the Ras–PKA pathway are long-lived (Fabrizio et al., 2001, 2005; Longo, 2003). We, and others, have proposed that the down-regulation of the Sch9 and Ras–cAMP–PKA pathway delays aging and partially mediates the life-span extension caused by CR (Longo and Finch, 2003; Kaeberlein et al., 2005). CR is a well studied intervention that consistently extends the life span of organisms ranging from yeast to mice (Weindruch and and Walford, 1988; Weindruch, 1996; Lin et al., 2000; Fabrizio and Longo, 2003). Furthermore, recent results from our laboratory showed that genomic instability was reduced at all ages in long-lived mutants lacking SCH9 (Fabrizio et al., 2004, 2005). In the present study, we set out to determine whether the deletion of this oncogene homologue could also exert a protective effect against premature chronological age-dependent genomic instability in the sgs1Δ-null mutants.
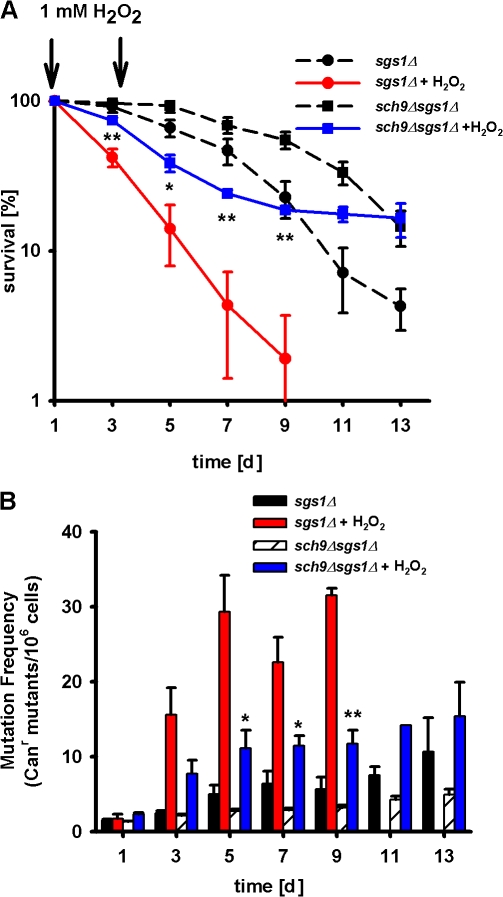
Deletion of SCH9 led to a significant CLS increase in sgs1Δ mutants, although the sch9Δsgs1Δ double mutant did not survive as long as the single sch9Δ mutant (Fig. 4 A and Fig. S3, B and C; Fabrizio et al., 2001, 2005). Notably, CR sgs1Δ mutants did not have a significantly longer mean life span compared with sgs1Δ mutants in glucose/ethanol medium, although they survived better at advanced ages (Fig. 4 B and Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200707154/DC1).
Figure 4.
Effect of deletion of oncogene homologue SCH9 and starvation/severe CR on premature aging and genomic instability of sgs1Δ mutants. (A) Survival of wild type and mutants lacking SGS1, SCH9, or both. The mean ± SEM is shown (n = 12–17). *, P < 0.05; **, P < 0.01 (sch9Δsgs1Δ vs. sgs1Δ). (B) Survival of wild-type strain and sgs1Δ mutant under starvation/CR. Data represent the mean ± SEM (n = 10–16). *, P < 0.05 (between calorie-restricted sgs1Δ and wild-type cells). (C) Age-dependent mutation frequency measured as Canr mutants/106 cells of wild type and mutants lacking SGS1 or both SCH9 and SGS1. The mean ± SEM is shown (n = 12–17). *, P < 0.05 (sch9Δsgs1Δ vs. sgs1Δ). (D) Age-dependent mutation frequency measured as Canr mutants/106 cells of calorie-restricted wild type and mutants lacking SGS1. Data represent the mean ± SEM (n = 10–16). *, P < 0.05 (calorie-restricted sgs1Δ vs. sgs1Δ in SDC). (E) GCR frequency measured as Canr5FOAr mutants/108 cells of wild type and mutants lacking SGS1 or both SCH9 and SGS1 and calorie-restricted sgs1Δ mutant cells. Data represent the mean ± SEM (n = 7–11). *, P < 0.05; ^, P < 0.05 (sch9Δsgs1Δ vs. sgs1Δ in SDC or CR sgs1Δ, respectively); #, P < 0.05 (CR sgs1Δ vs. SDC sgs1Δ). (F and G) Effect of the SCH9 deletion on the age-dependent trp1-289 reversion frequency (F) and the rate of small insertion/deletion mutations in the lysΔBgl strain (EH150) of sgs1Δ mutants (G). The means ± SEM are shown (n = 7–14). *, P < 0.05 (sch9Δsgs1Δ vs. sgs1Δ).
Mutations in SCH9, but not CR, prevented the age-dependent Canr mutations (Fig. 4, C and D) and GCRs (Fig. 4 E) caused by Sgs1 deficiency. In general, CR showed a positive protective trend, which was significant only at day 5. In agreement with the GCR frequency results, we were able to PCR amplify and sequence the entire CAN1 gene in 80% of Canr mutant colonies from a day-7 sch9Δsgs1Δ culture but only 50% of the CAN1 genes obtained from sgs1Δ colonies (Table III). The most likely explanation is that GCRs caused a deletion of the region containing the primer sequence for the PCR amplification.
Notably, lack of SCH9 actually increased trp1-289 reversions in sgs1Δ mutants back to the wild-type level (Fig. 4 F), suggesting that the sgs deletion causes genomic instability but also prevents the activity of an error-prone repair system that causes C→T base substitutions, possibly by promoting recombination or death instead. Although C→T (G→A) base substitution was absent in sgs1Δ, it was present in sch9Δsgs1Δ (Table III). In contrast, lack of SCH9 further decreased the small DNA insertions/deletions that cause Lys+ reversions (Fig. 4 G), indicating that NHEJ repair may be reduced in these mutants.
To investigate the mechanisms responsible for the protective effect of lack of SCH9 against sgs1Δ defects, we monitored age-dependent changes in cell-cycle profiles. The deletion of SCH9 in sgs1Δ mutants increased the G1 arrest, as shown by the FACS analysis data, and caused a major reduction in budding index (Fig. 5, A and B; and Table II). Consistently, the frequency of chronological age-dependent dedifferentiation was reduced from 50% in sgs1Δ cultures to 12% in sch9Δsgs1Δ cultures (2 out of 21; P = 0.036 [Fisher's exact test]; Table I). Similarly, dedifferentiation was not observed in any of the sch9Δsgs1Δ cultures in the SP1 or BY4741 genetic backgrounds, although the number of samples was small (Table I). Thus, the deletion of SCH9 reversed the dedifferentiation defects of sgs1Δ mutant cells by significantly reducing the budding index and tightening the G1 arrest (Fig. S4, A and B). Furthermore, it abolished the age-dependent increase in Canr mutation frequency originated by both nonquiescent and quiescent sgs1Δ cells to levels similar to those of wild-type cells (Fig. S4 C).
Figure 5.
Effect of lack of SCH9 on the cell-cycle profiles and recombinational events of sgs1Δ mutants. (A) Age-dependent variation of the percentage of budded cells measured as described in Materials and methods. Data represent the mean ± SEM (n = 3). (B) Representative cell-cycle profile (FACS) of wild type and mutants lacking SGS1 or both SCH9 and SGS1. (C and D) Homologous (C) and homeologous (D) recombination frequency measured as His+ colonies/106 cells during chronological aging of wild-type, sgs1Δ, and sch9Δsgs1Δ mutants. The mean ± SEM is shown (n = 4). *, P < 0.05; **, P < 0.01 (sch9Δsgs1Δ vs. sgs1Δ).
In the previous section, we showed that the deletion of SGS1 caused a major increase in homologous and homeologous recombination during chronological aging. To determine whether the protective effect of sch9Δ might be caused by attenuating recombination, we monitored age-dependent homologous and homeologous recombination frequency. Both homologous and homeologous recombination were greatly reduced in sch9Δsgs1Δ compared with sgs1Δ cells (Fig. 5, C and D). Together with the previous studies by Kolodner and co-workers, our results suggest that the lack of SCH9 prevents the age-dependent genomic instability defects in sgs1Δ mutants in part through inhibition of erroneous recombination between sister chromatids (Myung et al., 2001;Myung and Kolodner, 2002).
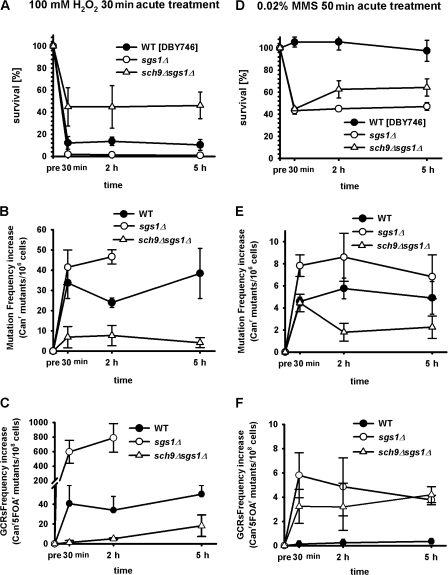
To determine whether the lack of Sch9 might protect against genomic instability in sgs1Δ mutants by either protecting against the initial damage of DNA or enhancing DNA repair, we monitored the changes in Canr mutations and GCR frequencies upon treatment of oxidant (100 mM hydrogen peroxide) or alkylating agent (0.02% MMS). After treatment, 3-d-old cells were washed four times and allowed to recover in a toxin-free medium for up to 5 h. Canr mutations and GCRs were measured 30 min and 2 and 5 h after treatment. The lack of Sch9 activity protected sgs1Δ against oxidation-dependent viability loss as well as Canr mutations and GCRs (Fig. 6, A–C). The deletion of SCH9 also partially reversed the hypersensitivity of sgs1Δ to chronic treatment with H2O2 (Fig. 7, A and B).
Figure 6.
Effect of lack of SCH9 on the time-dependent repair of oxidation or alkylation-induced mutations in sgs1Δ mutants. (A–C) Survival of wild-type (DBY746), sgs1Δ, or sch9Δsgs1Δ mutants after 30 min of exposure to 100 mM H2O2 (A). After treatment, cells were centrifuged, washed four times, and resuspended in H2O2-free medium obtained from day-3 wild-type culture. Mutation frequency, measured as Canr mutants/106 cells (B) and GCRs (Canr5FOAr mutants/108cells; C), was monitored at the indicated times (30 min and 2 and 5 h). (D) Survival of wild-type, sgs1Δ, or sch9Δsgs1Δ mutants after exposure to 0.02% MMS for 50 min. After treatment, cells were centrifuged, washed four times, and resuspended in MMS-free medium obtained from day-3 wild-type culture. Mutation frequency, measured as Canr mutants/106 cells (E) and GCRs (F; Canr5FOAr mutants/108cells), was monitored at the indicated times (30 min and 2 and 5 h). In both experiments, ∼30 min was the minimum required time from the end of treatment to centrifuge, wash, and resuspend the cells before plating. The mean ± SEM is shown (n = 3).
Figure 7.
Protective effect of lack of SCH9 against oxidative stress in sgs1Δ mutants. (A and B) Chronological survival (A) and mutation frequency (B) over time in the CAN1 gene, measured as Canr mutants/106 cells of sgs1Δ and sch9Δsgs1Δ cells, either untreated or chronically (days 1 and 3) exposed to 1 mM H2O2. Data represent the mean ± SEM (n = 4). *, P < 0.05; **, P < 0.01 (sch9Δsgs1Δ-treated vs. sgs1Δ-treated cells).
Although the majority of sgs1Δ cells died after 2-h treatment with hydrogen peroxide, the survivors showed a 20-fold increase of Canr mutation frequency and a 65-fold increase in GCR frequency (Fig. 6, B and C). To exclude the possibility that this increase in DNA damage in both wild-type and sgs1Δ cells was not representative of the damage caused by oxidative treatment but rather a characteristic of very sick cells, both strains were exposed to a reduced dose of hydrogen peroxide (50 mM). The lower dose of hydrogen peroxide had a smaller effect on cell death but caused DNA damage comparable to that at the higher dose (unpublished data).
In contrast to the results with oxidative stress, cells lacking SGS1 died after treatment with MMS, even in combination with sch9Δ mutations (Fig. 6 D). Although the lack of SCH9 protected against MMS-induced Canr mutations, it did not protect against GCRs (Fig. 6, D and F). These results indicate that the SCH9 deletion protects against DNA damage in sgs1Δ mutants in part by preventing the initial oxidation-dependent DNA damage, which is in agreement with our previous results indicating that superoxide is a major contributor to age-dependent mutations (Fabrizio et al., 2003, 2004).
DISCUSSION
Mutations in the human WRN and BLM RecQ helicases cause accelerated aging (WS) and premature age-dependent cancer (BS), respectively. Because S. cerevisiae is known to express only one RecQ helicase, the combination of the deletion of the SGS1 gene and CLS analysis provides a simple system to study the fundamental role of RecQ helicases in age-dependent genomic instability. Whereas Sgs1-deficient strains either had a normal or slightly decreased CLS when maintained in standard glucose/ethanol medium, they showed increased mortality under starvation/extreme CR (Fig. 4 B, water) or when treated with hydrogen peroxide or the mutagen MMS (Fig. 1, A and B). sgs1Δ mutants showed early dedifferentiation and regrowth phenotype associated with G1 arrest deficiencies and a three- to fourfold increase of budding index (Fig. 1, E and F; and Table I) reminiscent of the early cancer phenotype of BS. This early/increased dedifferentiation phenotype was observed in three different genetic backgrounds.
In agreement with the studies performed in dividing S. cerevisiae (Myung et al., 2001), young (day 1) Sgs1-deficient cultures displayed a frequency of CAN1 mutations similar to that of wild type in two different genetic backgrounds (DBY746 and BY4741). However, a higher increase in age-dependent CAN1 mutation frequency and GCR formation occurred in sgs1Δ mutants compared with wild-type cells (Fig. 2, A and B). Notably, specific base substitutions (C→T) and small DNA insertion/deletion mutations were actually reduced in aging sgs1Δ mutants, which raises the possibility that the higher rate of recombination between sister chromatids performed in these mutants replaces other types of error-prone repair systems that cause base substitutions or small insertions/deletions. Interestingly, the loss of multiple Sgs1-dependent functions might be at the base of part of the age-dependent DNA damage in sgs1Δ cells because cells lacking only the helicase domain, the C-terminal checkpoint domain, or the Top3-binding domain of Sgs1 did not show similar levels of Canr mutations when compared with cells lacking the entire protein (sgs1Δ). An alternative explanation is that additional domains that are important for genomic stability remain to be identified. In contrast, the lack of the DNA helicase domain of Sgs1 was sufficient to cause a major age-dependent increase in the formation of GCRs, whereas at advanced ages this increase was lower than that caused by the deletion of the entire SGS1 gene (Fig. 2 G, sgs1Δ).
It is remarkable that the age-dependent frequency of GCRs that occurred in nondividing cells reached a level that was 40-fold higher than that obtained after many cycles of cell divisions required to reach a population density of ∼100 × 106 cells on day 1. This underscores the importance of performing age-dependent studies in addition to studies in the growth phase.
The following results suggest that reduced recombination is a key component of the protective effect of the lack of SCH9 against the premature aging phenotypes caused by the deletion of SGS1: The protection against age-dependent Canr mutations and GCR, but not trp, reversions (Fig. 4, C, E, and F); the reversion in sch9Δsgs1Δ of the major increase in the rate of homologous and homeologous recombination caused by the deletion of SGS1 (Fig. 5, C and D); the attenuation of GCRs confirmed by the frequency results and the sequencing data (a lack of major portions of the CAN1 gene observed in only 20% of sch9Δsgs1Δ but 50% of sgs1Δ; Fig. 4 E and Table III); and previous studies conclude that homeologous recombination plays a central role in GCRs in growing sgs1Δ cells (Myung et al., 2001).
The other major protective effect caused by the deletion of SCH9 appears to be the prevention of the initial DNA damage. In fact, sch9Δsgs1Δ cells were protected against both acute and chronic treatment with hydrogen peroxide (Figs. 6 and Figs.7). The acute oxidative stress experiments suggest that sch9Δ is blocking the initial DNA damage or mutations rather than increasing the rate of repair. This protection is likely to involve antioxidant enzymes as well as additional protective systems (Longo et al., 1996, 1999; Fabrizio et al., 2003).
The G1-arrest defects of sgs1Δ could explain the sixfold increase in the percentage of age-dependent dedifferentiation (Fig. 1 and Table I). These defects, which may derive from the lack of the cell-cycle arrest C200 domain of Sgs1, are reversed by the deletion of SCH9 (Fig. 5 and Table I). In contrast, the finding that the frequency of age-dependent mutations is higher in the quiescent fraction of sgs1Δ cells compared with the nonquiescent fraction suggests that the cell-cycle defects are not a major promoter of mutation (Fig. 3). One possibility is that a major portion of the sgs1Δ cells that have DNA damage but fail to arrest, die, possibly because of double-strand breaks. Starvation/extreme CR showed a promising protective trend but not a significant effect against genomic instability during most of the CLS of sgs1Δ mutants. Although further experiments in yeast and other model systems should be performed to test further the role of CR against age-dependent DNA damage, these results are consistent with our previous conclusion that even extreme CR is not as effective as mutations in SCH9 in protecting against age-dependent damage and aging (Fig. 4 and Fig. S5; Longo, 1997; Longo et al., 1997; Fabrizio et al., 2005).
We previously showed that superoxide plays a primary role in chronological age-dependent DNA damage and mutations. Our model is that the DNA damage caused by oxidative and other types of stress accumulated during aging in nondividing cells generates double-strand breaks during the first round of replication after the exit from G0. Cells lacking SGS1 attempt to repair this damage by homologous recombination between sister chromatids but generate a large number of GCRs, especially at advanced age.
This study raises the possibility that down-regulation of Akt/mTOR/S6kinase or upstream proteins implicated in life-span regulation may protect against age-dependent DNA damage and cancer, including the premature defects observed in WS and BS (Hasty and Vijg, 2002). Naturally, the connection between oncogenes or protooncogenes and age-dependent genomic instability demonstrated in the S. cerevisiae model must first be established in higher eukaryotes. However, our findings are consistent with the recent observation that in the Ercc1 −/− mouse, a model of the human XPF progeroid syndrome, the growth hormone/IGF1 hormonal axis is suppressed in response to cytotoxic DNA damage (Niedernhofer et al., 2006; Vijg and Suh, 2006), which raises the possibility that reduced IGF-I–Akt–mTOR–S6kinase signaling might protect against the accelerated rate of DNA damage in this model. Furthermore, these mice showed similar patterns of metabolic and growth changes to those already observed under CR conditions (Spindler, 2005). Thus, it will be important to determine whether the down-regulation of the IGF-I–Akt–mTOR–S6kinase pathway may have a similar effect in protection of mouse model systems for WS and BS.
MATERIALS AND METHODS
Yeast strains and growth conditions
The majority of the experiments were performed in DBY746 (MATα,leu2-3,112,his3Δ1,trp1-289,ura3-52,GAL+; provided by D. Botstein, Massachusetts Institute of Technology, Cambridge, MA). Strains BY4741 (MATa,his3Δ1leu2Δ0,met15Δ0,ura3Δ0; Open Biosystems) and SP1 (MATα,his,leu2,ura3,trp1,ade8 can1; provided by J. Valentine, University of California, Los Angeles, Los Angeles, CA) were used to confirm the results obtained with DBY746. Strain EH150 (MATa,lys2ΔBglII,trp1-Δ,his3-Δ200,ura3-52,ade2-1o) was used for the small insertion/deletion mutation assay (provided by E. Heidenreich, Institute of Cancer Research, Medical University of Vienna, Vienna, Austria).The sch9Δ mutant has been described previously (Fabrizio et al., 2001).
All the mutant strains were originated in the different backgrounds by one-step gene replacement according to Brachmann et al. (1998). To generate the sgs1 mutated derivatives, we used the following plasmids: pJM526 (SGS1), pJM511 (sgs1-hd), pJM531 (sgs1-DN50), and pJM512 (sgs1-DC200; provided by B. Johnson, University of Pennsylvania School of Medicine, Philadelphia, PA; Mullen et al., 2000). Each plasmid was digested with BStEII and transformed in DBY746. Proper integration of a single copy of each sgs1 mutant at the LEU2 locus was confirmed by PCR. Strains for intron-based inverted-repeat recombination assay were constructed using plasmids provided by S. Jinks-Robertson (Emory University, Atlanta, GA; Datta et al., 1996). Plasmids pRS406 (carrying two 100% homologous inverted repeats [IRs]) and pRS407 (carrying two 91% homologous IRs) were linearized with StuI before transformation. Ura+ transformants with a single copy of integrated HIS3∷intron-IR cassette derived from wild-type (DBY746), sgs1Δ, or sch9Δ sgs1Δ strains were used for recombination assay. CLS of cells incubated in either minimal medium containing glucose (synthetic dextrose complete [SDC]) or water was measured as described previously (Fabrizio and Longo, 2003). In brief, yeast were grown in synthetic medium (SDC) containing 2% glucose and supplemented with amino acids, adenine, and uracil (as previously described in Kaiser et al. [1994]), as well as a fourfold excess of the supplements tryptophan, leucine, uracil, and histidine. CLS was monitored in expired SDC medium by measuring colony-forming units (CFUs) every 48 h. The number of CFUs on day 1 was considered as the initial survival (100%) and was used to determine the age-dependent mortality. CLS was also monitored by counting yeast cells stained with 0.01 mg/ml phloxin B using a standard hemocytometer chamber (Kucsera et al., 2000). For life-span experiments in water (CR), yeast were grown in SDC for 3 d, washed with sterile distilled water, and resuspended in water. Yeast cells were washed with water every 48 h to avoid the accumulation of nutrient release from dead cells.
Stress-resistance assays
Heat-shock resistance was measured by spotting serial dilutions of cells removed from day-3 postdiauxic-phase cultures onto yeast extract peptone dextrose medium (YPD) plates and incubating at 55 (heat shocked) and 30°C (control) for 90 min. After heat shock, plates were transferred at 30°C and incubated for 2–3 d.
For oxidative stress–resistance assays, acute treatment exponentially growing (OD600 = 1) or day-3 postdiauxic-phase cells were diluted to an OD600 of 0.1 in potassium phosphate buffer, pH 7.4, and treated with 250 μM menadione for 60 min. Serial dilutions of untreated and menadione-treated cells were spotted onto YPD plates and incubated at 30°C for 2–3 d. Alternatively, cells were diluted to an OD of 1 in potassium phosphate buffer, pH 6, and treated with 100 mM hydrogen peroxide for 30 min.
SDC liquid medium cell cultures were also chronically treated with either 1 mM hydrogen peroxide or 0.001% MMS on days 1 and 3 of a typical chronological aging study.
Mutation-frequency measurements
Spontaneous mutation frequency was evaluated by measuring the frequency of mutations of the CAN1 (YEL063) gene. In brief, overnight inoculations were diluted in liquid SDC medium and incubated at 30°C. Cell viability was measured every 2 d starting at day 1 by plating appropriate dilutions onto YPD plates and counting the CFUs. To identify the canavanine-resistant mutants (Canr) in the liquid culture, an appropriate number of cells (starting amount, 2 × 107 cells) was harvested by centrifugation, washed once with sterile water, and plated on selective medium (SDC-ARG supplemented with 60 μg/ml l-canavanine sulfate). Mutant colonies were counted after 3–4 d. The mutation frequency was expressed as the ratio of Canr over total viable cells.
The Canr mutator phenotype can be conferred by any mutations that block the expression of the CAN1 gene (Chen and Kolodner, 1999). Thus, we measured base substitutions, small insertions/deletions, or GCRs to specifically characterize the age-dependent type of mutations occurring in wild-type and mutant strains (Madia et al., 2007).
Large-scale measurement of GCRs
To detect GCRs, we generated a DBY476 background strain in which we replaced HXT13 (YEL069), encoding for a highly redundant hexose transporter, with a URA3 cassette as described by Chen and Kolodner (1999). HXT13 is located 7.5 kb telomeric to CAN1 on chromosome V. The experiment was conducted similarly to that described for the can1 mutations but the detection for the loss of both CAN1 and URA3 was performed on SDC-ARG plates containing 1 mg/ml 5-fluoroorotic acid (5FOA) and60 μg/ml l-canavanine. Mutant colonies were counted after 3–4 d.
Measurement of age-dependent small insertion/deletion mutations
Based on the experimental design proposed by Heidenreich et al. (2003) and Heidenreich and Wintersberger (1998), we generated sgs1Δ and sch9Δsgs1Δ mutants in a Lys− strain (EH150) in which a lys2ΔBglII mutation was constructed by filling in a BglII restriction site of the LYS2 gene. The resulting +4 shift in the ORF causes an auxotrophy for lysine that can be reversed by small age-dependent insertion/deletion mutations. Using these strains, we have monitored the age-dependent insertion/deletion events after plating 108 cells on selective SDC-LYS plates. The experiments were performed similarly to that described for the can1 mutations.
Measurement of age-dependent single-base substitution mutations.
To monitor the frequency of reversion of a base substitution, we used the strain DBY746, which carries a trp1-289 amber mutation (C→T at residue 403 of the coding sequence), and measured the frequency of trp1-289 to Trp+ reversions (Capizzi and Jameson, 1973). The experimental protocol was similar to the one described for the small insertion/deletion mutations detection. 108 cells were plated onto selective medium (SDC-TRP). The experiments were performed similarly to that described for the can1 mutations.
Recombination assay
To monitor the level of homologous (100%) and homeologous (91%) recombination during chronological aging, we generated mutants in which we integrated linearized plasmids carrying either 100% homologous IRs (pRS406) or 91% homeologous IRs (pRS407; Datta et al., 1996). Ura + transformants with a single copy of integrated HIS3∷intron-IR cassette derived from wild-type (DBY746), sgs1Δ, or sch9Δ sgs1Δ strains were used. Starting from day 1 of a standard chronological aging study, homologous or homeologous His+ recombinants were measured by plating 5 × 107 viable cells (washed twice with sterile water) every 2 d on selective medium (synthetic complete medium lacking HIS and supplemented with galactose according to Datta et al. [1996]). The number of His + colonies was counted 4 d after plating, and the frequency of homologous or homeologous recombination was calculated based on the number of viable cells.
FACS analysis and budding index
Cells from exponentially proliferating cultures of wild-type, sgs1Δ, and sch9Δsgs1Δ strains were inoculated into SDC medium at an initial density of 5 × 105/ml and continuously cultured for 7 d at 30°C with rotary shaking. Aliquots of cells removed from each culture at the indicated times were pelleted by centrifugation and resuspended in water (for determining the fraction of budding cells) or 70% ethanol. To measure budding index, cells were sonicated using a Sonic Dismembrator sonicator (model 60; Thermo Fisher Scientific) for 10 s at power setting 5. The fraction of budding cells (budding index) of at least 500 cells from each aliquot was visually determined using a microscope (E600 Eclipse; Nikon) with a 40× phase-contrast objective. To measure DNA content by flow cytometry, cells suspended in 70% ethanol were pelleted by centrifugation, washed with 50 mM sodium citrate, pH 7.5, and resuspended in 0.5 ml of this same buffer containing 0.5 mg/ml RNase (Invitrogen). After overnight incubation at 37°C, 0.5 ml sodium citrate buffer containing 2 μM SYTOX green (Invitrogen) was added to each sample. Stained cells were briefly sonicated as described in this section, and DNA content was measured using a FacsCaliber flow cytometer (BD Biosciences) at a maximum flow rate of 500 cells per second. Flow cytometry data were processed using CellQuest (BD Biosciences) and Flojo (Tree Star, Inc.) software. Data were calculated using MODFIT (Verity Software House). Data from FACS measurements presented in Figs. 1 E and 5 B are reported in Table II.
Separation of cells through a Percoll density gradient
A Percoll density gradient (GE Healthcare) was prepared using the protocol described by Allen et al. (2006). In brief, ∼2 × 109 cells from sgs1Δ and sch9Δsgs1Δ SDC cultures were collected at days 3, 5, and 7 of a chronological aging study. Cells were pelleted and resuspended in 1 ml Tris buffer, overlaid on the preformed Percoll gradient, and centrifuged at 400 g av for 60 min at 20°C. Fractions were collected, washed once with 40 ml Tris buffer, pelleted, and resuspended in ddH2O. Immediately after the separation, cells were plated on YPD and on SDC-ARG supplemented with 60 μg/ml l-canavanine sulfate solid media to measure viability (CFUs) and determine mutation frequency, respectively. Three independent experiments were performed.
Lower and upper fraction cells were examined microscopically, without previous sonication, for the presence of new buds. Five fields of 50–60 cells were examined per time point and the budding percentage was calculated by comparing the number of cells with new buds to the total number of cells counted.
CAN1 sequencing
Canavanine-resistant clones from wild-type, sgs1Δ, and sch9Δsgs1Δ strains were collected on day 7 of a CLS study. Genomic DNA was isolated using standard glass beads/chloroform-phenol procedure. The two primer sets used for PCR amplification to cover the CAN1 open reading frame were the following: v-534 (5′-GAAGAGTGGTTGCGAACAGAG-3′) and v-555 (5′-CCTAAGAACCTCCCTTCGTTTT-3′) to amplify a 1080-bp region (−226 to approximately +854); and v-557 (5′-ATCACTTTTGCCCTGGAACTTA-3′) and v-270 (5′-CGTGGAAATGTGATCAAAGGTA-3′) to amplify a 1275-bp region (+535 to approximately +1809). 30 ng/ml of genomic DNA in a 50-μl reaction was subject to 30–35 cycles of amplification (94°C, 30 s; 52°C, 45 s, 72°C, 2 min 15 s). PCR products were gel purified and sequenced using the amplification primers (both directions). The additional primers used to confirm the sequencing results when necessary were the following: v-279 (5′-CTTAACTCCTGTAAAAAC-3′), v-280 (5′-TATTGGTATGATTGCCCTTG-3′), v-281 (5′-GAGTTCTGGGTCGCTTCCATC-3′), v-282 (5′-CAATCTACTTCCTACGTTTC-3′), and v-283 (5′-CGGTAACCCCAGCACCAC-3′). All primers were synthesized by Integrated DNA Technologies, Inc. Sequencing was performed by Laragen, Inc. Identification of mutation was performed using Mutation Surveyor v3.00 (Softgenetics).
Statistical analysis
Longevity curves and mutation-frequency curves were analyzed by eithert test (P < 0.001, P < 0.01, and P < 0.05) or Mann-Whitney test on the data for each pair of strains at each day of the chronological aging study. Fisher's exact test was used to compare the percentage of dedifferentiation between wild-type, sgs1Δ, and sch9Δsgs1Δ mutant cultures.
Online supplemental material
Fig. S1 shows chronological survival of sgs1Δ in BY4741 and SP1 backgrounds. Fig. S2 shows the methods of monitoring age-dependent mutation frequency using CLS. Fig. S3 shows premature genomic instability in the sgs1Δ mutant in the BY4741 background and the effect of lack of SCH9 on premature aging in the sgs1Δ mutant in the BY4741 and SP1 backgrounds. Fig. S4 shows the separation of quiescent and nonquiescent cells, through a Percoll density gradient, from chronologically aging wild-type and sch9Δsgs1Δ cultures. Fig. S5 shows the effect of lack of SCH9 and CR on survival and mutation frequency of the sgs1Δ mutant. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200707154/DC1.
Acknowledgments
We thank Dr. Sue Jinks-Robertson for providing us plasmids pRS406 and pS407 and Dr. E. Heindereich for providing strain EH150. We thank Myron Goodman and Michael Lieber for very helpful comments.
This work was supported in part by an American Federation for Aging Research grant and by National Institutes of Health grants AG20642, AG025135, and GM075308.
F. Madia and C. Gattazzo contributed equally to this paper.
Abbreviations used in this paper: 5FOA, 5-fluoroorotic acid; BS, Bloom's Syndrome; CFU, colony-forming unit; CLS, chronological life span; CR, calorie restriction; GCR, gross chromosomal rearrangement; IR, inverted repeat; MMS, methyl methane sulfonate; NHEJ, nonhomologous end joining; SDC, synthetic dextrose complete; WS, Werner's Syndrome; YPD, yeast extract peptone dextrose medium.
References
- Allen, C., S. Buttner, A.D. Aragon, J.A. Thomas, O. Meirelles, J.E. Jaetao, D. Benn, S.W. Ruby, M. Veenhuis, F. Madeo, and M. Werner-Washburne. 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam, M., J.Y. Lee, V. Abraham, R. Chanoux, K.A. Schoenly, and F.B. Johnson. 2006. Evidence that the S. cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res. 34:506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, R.J., and J.C. Wang. 2001. Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: studies of fusion proteins. Proc. Natl. Acad. Sci. USA. 98:11108–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, R.J., and J.L. Keck. 2004. Structure and function of RecQ DNA helicases. Crit. Rev. Biochem. Mol. Biol. 39:79–97. [DOI] [PubMed] [Google Scholar]
- Boulton, S.J., and S.P. Jackson. 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C.B., A. Davies, G.J. Cost, E. Caputo, J. Li, P. Hieter, and J.D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Capizzi, R.L., and J.W. Jameson. 1973. A table for the estimation of the spontaneous mutation rate of cells in culture. Mutat. Res. 17:147–148. [DOI] [PubMed] [Google Scholar]
- Chen, C., and R.D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81–85. [DOI] [PubMed] [Google Scholar]
- Chen, C., K. Umezu, and R.D. Kolodner. 1998. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell. 2:9–22. [DOI] [PubMed] [Google Scholar]
- Cobb, J.A., and L. Bjergbaek. 2006. RecQ helicases: lessons from model organisms. Nucleic Acids Res. 34:4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, A., A. Adjiri, L. New, G.F. Crouse, and S. Jinks Robertson. 1996. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccaromyces cerevisiae. Mol. Cell. Biol. 16:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, N.A., J. Groden, T.Z. Ye, J. Straughen, D.J. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 83:655–666. [DOI] [PubMed] [Google Scholar]
- Epstein, C.J., G.M. Martin, A.L. Schultz, and A.G. Motulsky. 1966. Werner's syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore). 45:177–221. [DOI] [PubMed] [Google Scholar]
- Fabre, F., A. Chan, W.D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA. 99:16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., and V.D. Longo. 2003. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2:73–81. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., F. Pozza, S.D. Pletcher, C.M. Gendron, and V.D. Longo. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 292:288–290. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., L.L. Liou, V.N. Moy, A. Diaspro, J.S. Valentine, E.B. Gralla, and V.D. Longo. 2003. SOD2 Functions downstream of Sch9 to extend longevity in yeast. Genetics. 163:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., L. Battistella, R. Vardavas, C. Gattazzo, L.L. Liou, A. Diaspro, J.W. Dossen, E.B. Gralla, and V.D. Longo. 2004. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 166:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., C. Gattazzo, L. Battistella, M. Wei, C. Cheng, K. McGrew, and V.D. Longo. 2005. Sir2 blocks extreme life-span extension. Cell. 123:655–667. [DOI] [PubMed] [Google Scholar]
- Finch, C.E. 1990. Longevity, Senescence, and the Genome. University of Chicago Press, Chicago. 948 pp.
- Gangloff, S., J.P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German, J. 1995. Bloom's syndrome. Dermatol. Clin. 13:7–18. [PubMed] [Google Scholar]
- Hasty, P., and J. Vijg. 2002. Aging. Genomic priorities in aging. Science. 296:1250–1251. [DOI] [PubMed] [Google Scholar]
- Heidenreich, E., and U. Wintersberger. 1998. Replication-dependent and selection-induced mutations in respiration-competent and respiration-deficient strains of Saccharomyces cerevisiae. Mol. Gen. Genet. 260:395–400. [DOI] [PubMed] [Google Scholar]
- Heidenreich, E., R. Novotny, B. Kneidinger, V. Holzmann, and U. Wintersberger. 2003. Non-homologous end joining as an important mutagenic process in cell cycle-arrested cells. EMBO J. 22:2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson, I.D., S.L. Davies, J.L. Li, N.C. Levitt, P. Mohaghegh, P.S. North, and L. Wu. 2001. Role of the Bloom's syndrome helicase in maintenance of genome stability. Biochem. Soc. Trans. 29:201–204. [DOI] [PubMed] [Google Scholar]
- Johnson, F.B., R.A. Marciniak, M. McVey, S.A. Stewart, W.C. Hahn, and L. Guarente. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20:905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M., R.W. Powers III, K.K. Steffen, E.A. Westman, D. Hu, N. Dang, E.O. Kerr, K.T. Kirkland, S. Fields, and B.K. Kennedy. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 310:1193–1196. [DOI] [PubMed] [Google Scholar]
- Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 202 pp.
- Kaneko, H., and N. Kondo. 2004. Clinical features of Bloom syndrome and function of the causative gene, BLM helicase. Expert Rev. Mol. Diagn. 4:393–401. [DOI] [PubMed] [Google Scholar]
- Killoran, M.P., and J.L. Keck. 2006. Sit down, relax and unwind: structural insights into RecQ helicase mechanisms. Nucleic Acids Res. 34:4098–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucsera, J., K. Yarita, and K. Takeo. 2000. Simple detection method for distinguishing dead and living yeast colonies. J. Microbiol. Methods. 41:19–21. [DOI] [PubMed] [Google Scholar]
- Lin, S.J., P.A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 289:2126–2128. [DOI] [PubMed] [Google Scholar]
- Lo, Y.C., K.S. Paffett, O. Amit, J.A. Clikeman, R. Sterk, M.A. Brenneman, and J.A. Nickoloff. 2006. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol. Cell. Biol. 26:4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard, D.B., K.F. Chua, R. Mostoslavsky, S. Franco, M. Gostissa, and F.W. Alt. 2005. DNA repair, genome stability, and aging. Cell. 120:497–512. [DOI] [PubMed] [Google Scholar]
- Longo, V.D. 1997. The pro-senescence role of Ras2 in the chronological life span of yeast. Ph.D. thesis. University of California, Los Angeles, Los Angeles. 41 pp.
- Longo, V.D. 2003. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp. Gerontol. 38:807–811. [DOI] [PubMed] [Google Scholar]
- Longo, V.D., and C.E. Finch. 2003. Evolutionary medicine: from dwarf model systems to healthy centenarians. Science. 299:1342–1346. [DOI] [PubMed] [Google Scholar]
- Longo, V.D., E.B. Gralla, and J.S. Valentine. 1996. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 271:12275–12280. [DOI] [PubMed] [Google Scholar]
- Longo, V.D., L.M. Ellerby, D.E. Bredesen, J.S. Valentine, and E.B. Gralla. 1997. Human Bcl-2 reverses survival defects in yeast lacking superoxide dismutase and delays death of wild-type yeast. J. Cell Biol. 137:1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo, V.D., L.L. Liou, J.S. Valentine, and E.B. Gralla. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys. 365:131–142. [DOI] [PubMed] [Google Scholar]
- Madia, F., C. Gattazzo, P. Fabrizio, and V.D. Longo. 2007. A simple model system for age-dependent DNA damage and cancer. Mech. Ageing Dev. 128:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey, M., M. Kaeberlein, H.A. Tissenbaum, and L. Guarente. 2001. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics. 157:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima, A., M. Seki, F. Onoda, M. Shiratori, N. Odagiri, K. Ohta, Y. Kikuchi, Y. Ohno, and T. Enomoto. 2000. a. Sgs1 helicase activity is required for mitotic but apparently not for meiotic functions. Mol. Cell. Biol. 20:6399–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima, A., M. Seki, F. Onoda, A. Ui, Y. Satoh, Y. Ohno, and T. Enomoto. 2000. b. Different domains of Sgs1 are required for mitotic and meiotic functions. Genes Genet. Syst. 75:319–326. [DOI] [PubMed] [Google Scholar]
- Mohaghegh, P., and I.D. Hickson. 2002. Premature aging in RecQ helicase-deficient human syndromes. Int. J. Biochem. Cell Biol. 34:1496–1501. [DOI] [PubMed] [Google Scholar]
- Mullen, J.R., V. Kaliraman, and S.J. Brill. 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 154:1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung, K., and R.D. Kolodner. 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 99:4500–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung, K., A. Datta, C. Chen, and R.D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113–116. [DOI] [PubMed] [Google Scholar]
- Niedernhofer, L.J., G.A. Garinis, A. Raams, A.S. Lalai, A.R. Robinson, E. Appeldoorn, H. Odijk, R. Oostendorp, A. Ahmad, W. van Leeuwen, et al. 2006. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 444:1038–1043. [DOI] [PubMed] [Google Scholar]
- Onoda, F., M. Seki, A. Miyajima, and T. Enomoto. 2000. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom's syndrome gene. Mutat. Res. 459:203–209. [DOI] [PubMed] [Google Scholar]
- Onodera, R., M. Seki, A. Ui, Y. Satoh, A. Miyajima, F. Onoda, and T. Enomoto. 2002. Functional and physical interaction between Sgs1 and Top3 and Sgs1-independent function of Top3 in DNA recombination repair. Genes Genet. Syst. 77:11–21. [DOI] [PubMed] [Google Scholar]
- Ozgenc, A., and L.A. Loeb. 2005. Current advances in unraveling the function of the Werner syndrome protein. Mutat. Res. 577:237–251. [DOI] [PubMed] [Google Scholar]
- Pedrazzi, G., C.Z. Bachrati, N. Selak, I. Studer, M. Petkovic, I.D. Hickson, J. Jiricny, and I. Stagljar. 2003. The Bloom's syndrome helicase interacts directly with the human DNA mismatch repair protein hMSH6. Biol. Chem. 384:1155–1164. [DOI] [PubMed] [Google Scholar]
- Schmidt, K.H., V. Pennaneach, C.D. Putnam, and R.D. Kolodner. 2006. Analysis of gross-chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol. 409:462–476. [DOI] [PubMed] [Google Scholar]
- Shen, J.C., and L.A. Loeb. 2000. The Werner syndrome gene: the molecular basis of RecQ helicase-deficiency diseases. Trends Genet. 16:213–220. [DOI] [PubMed] [Google Scholar]
- Sinclair, D.A., K. Mills, and L. Guarente. 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 277:1313–1316. [DOI] [PubMed] [Google Scholar]
- Spindler, S.R. 2005. Rapid and reversible induction of the longevity, anticancer and genomic effects of caloric restriction. Mech. Ageing Dev. 126:960–966. [DOI] [PubMed] [Google Scholar]
- Sun, H., R.J. Bennett, and N. Maizels. 1999. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 27:1978–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, L.H., and D. Schild. 2002. Recombinational DNA repair and human disease. Mutat. Res. 509:49–78. [DOI] [PubMed] [Google Scholar]
- Toda, T., S. Cameron, P. Sass, and M. Wigler. 1988. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP- dependent protein kinase catalytic subunits. Genes Dev. 2:517–527. [DOI] [PubMed] [Google Scholar]
- Ui, A., M. Seki, H. Ogiwara, R. Onodera, S. Fukushige, F. Onoda, and T. Enomoto. 2005. The ability of Sgs1 to interact with DNA topoisomerase III is essential for damage-induced recombination. DNA Repair (Amst.). 4:191–201. [DOI] [PubMed] [Google Scholar]
- Urban, J., A. Soulard, A. Huber, S. Lippman, D. Mukhopadhyay, O. Deloche, V. Wanke, D. Anrather, G. Ammerer, H. Riezman, et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 26:663–674. [DOI] [PubMed] [Google Scholar]
- Vijg, J., and Y. Suh. 2006. Ageing: chromatin unbound. Nature. 440:874–875. [DOI] [PubMed] [Google Scholar]
- Weindruch, R. 1996. The retardation of aging by caloric restriction: studies in rodents and primates. Toxicol. Pathol. 24:742–745. [DOI] [PubMed] [Google Scholar]
- Weindruch, R., and R. Walford. 1988. The Retardation of Aging and Disease by Dietary Restriction. Charles C. Thomas, Springfield, IL. 436 pp.
- Yamagata, K., J. Kato, A. Shimamoto, M. Goto, Y. Furuichi, and H. Ikeda. 1998. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA. 95:8733–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., R. Zhang, X.W. Wang, S.P. Linke, S. Sengupta, I.D. Hickson, G. Pedrazzi, C. Perrera, I. Stagljar, S.J. Littman, et al. 2004. The mismatch DNA repair heterodimer, hMSH2/6, regulates BLM helicase. Oncogene. 23:3749–3756. [DOI] [PubMed] [Google Scholar]