Abstract
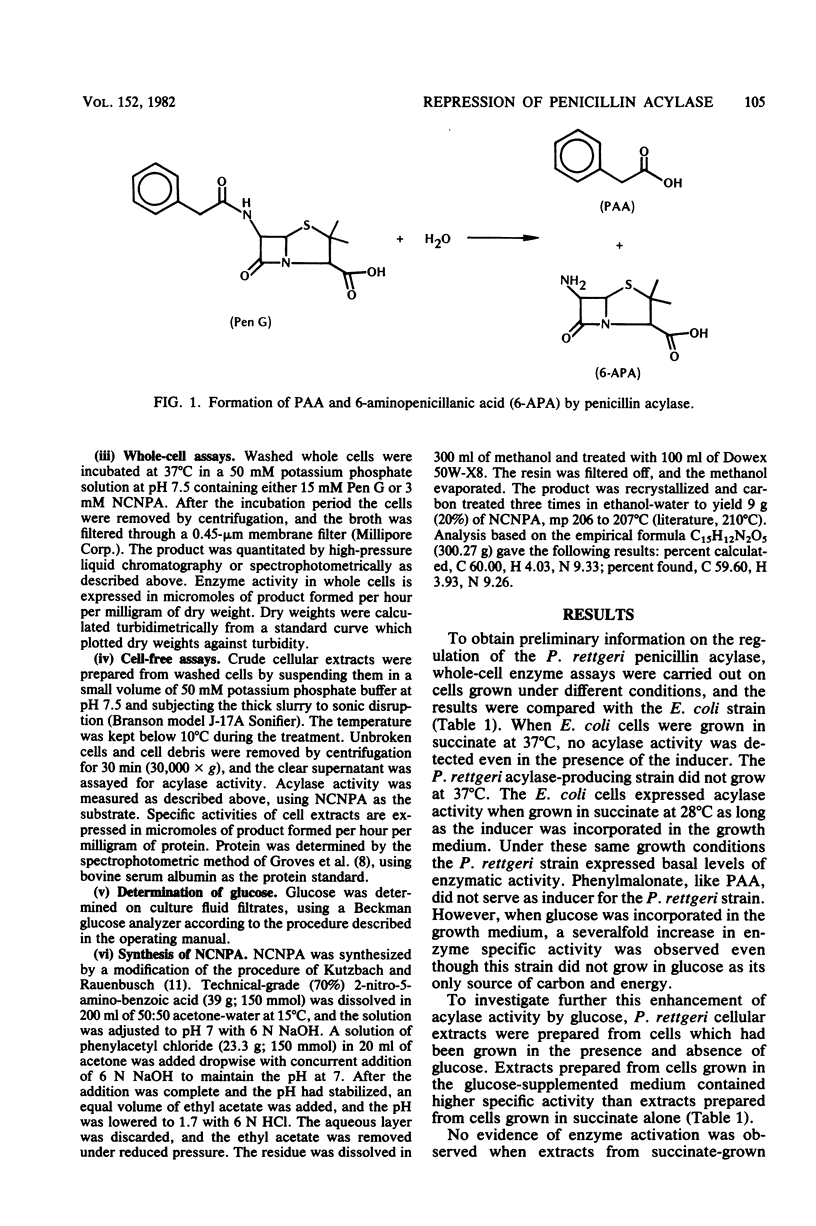
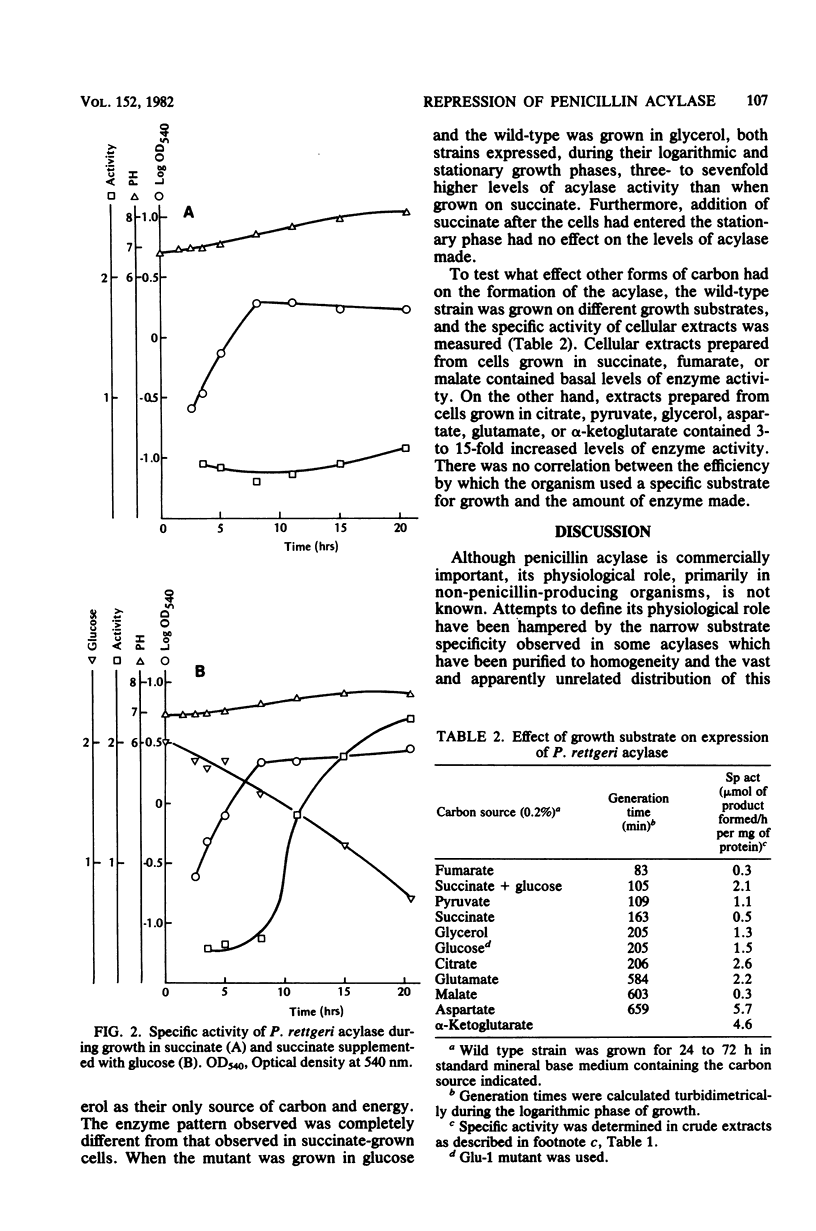
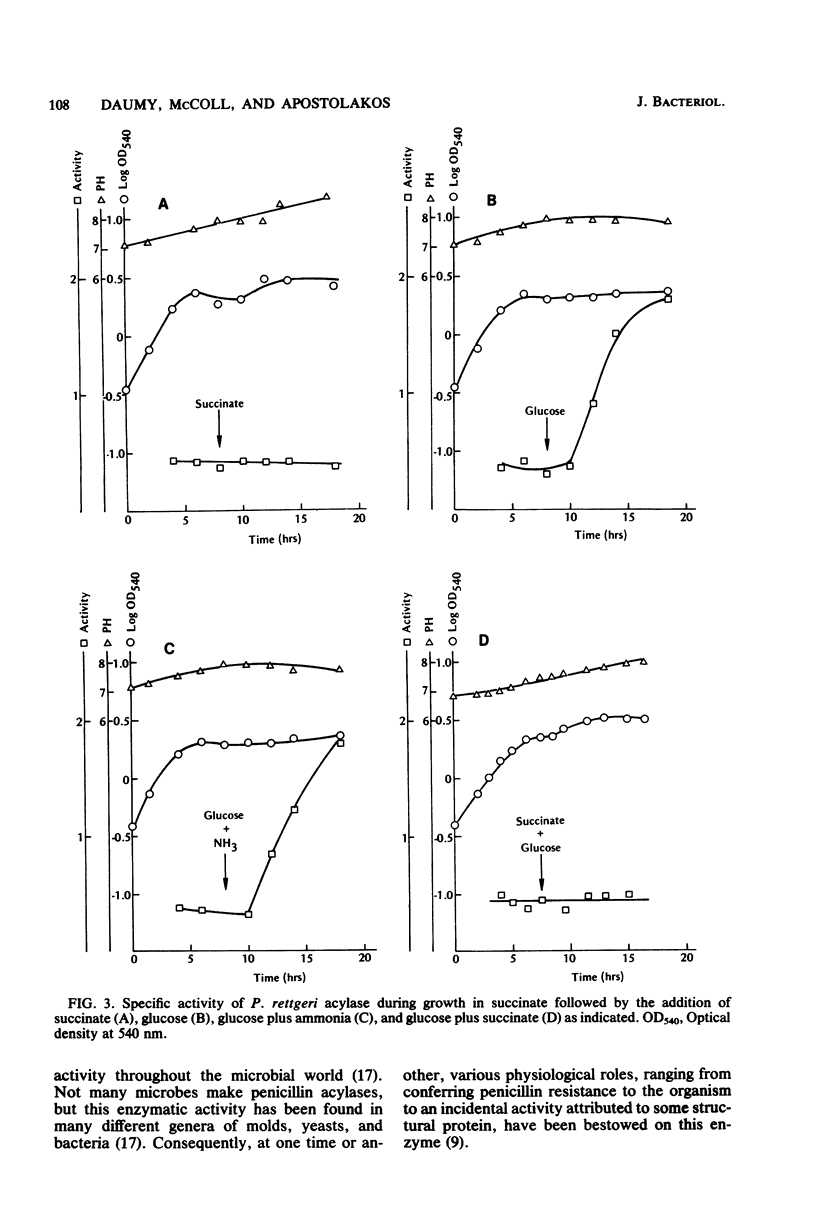
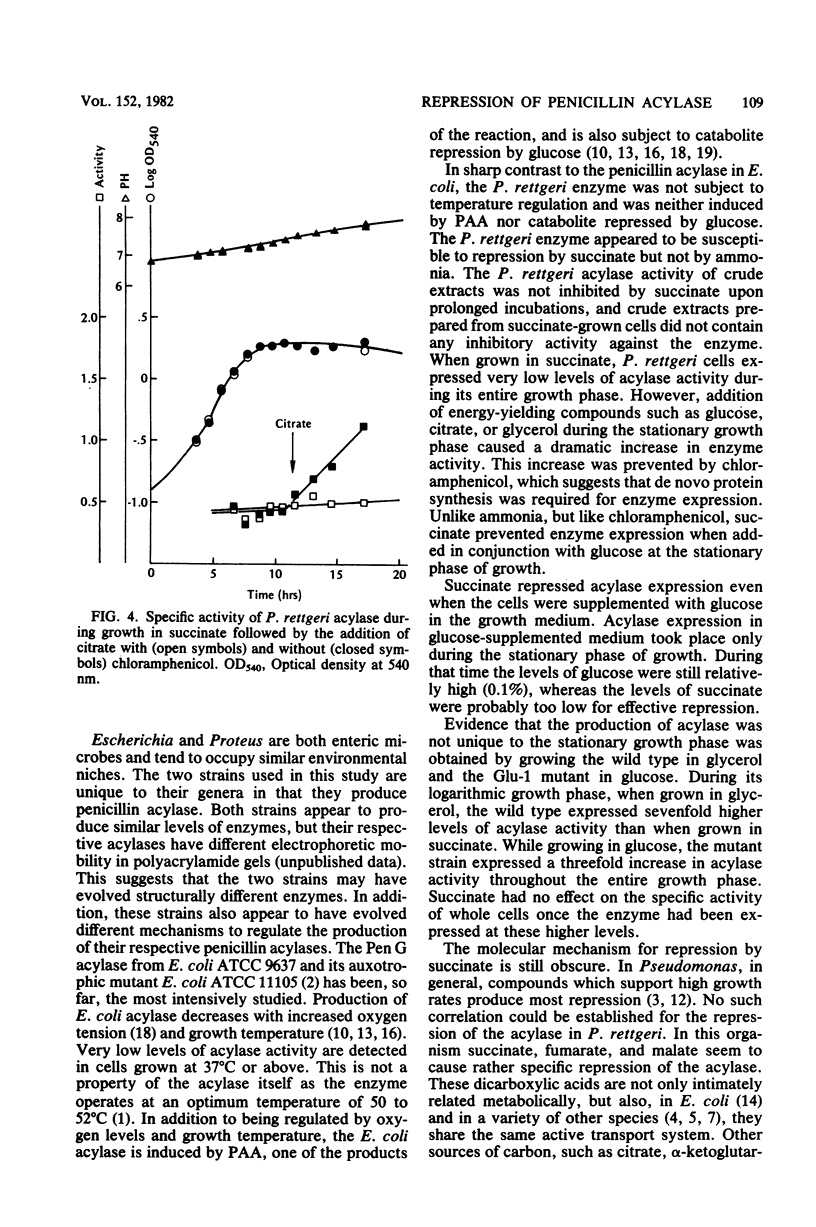
The regulation of the penicillin acylase in proteus rettgeri ATCC 31052 was compared with that of the enzyme in Escherichia coli ATCC 9637. Unlike the E. coli acylase, the P. rettgeri enzyme was not induced by phenylacetic acid, nor was it subject to catabolite repression by glucose. The P. rettgeri acylase appears to be expressed constitutively but is subject to repression by the C4-dicarboxylic acids of the tricarboxylic acid cycle, succinate, fumarate, and malate.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURKHOLDER P. R. Determination of vitamin B12 with a mutant strain of Escherichia coli. Science. 1951 Nov 2;114(2966):459–460. doi: 10.1126/science.114.2966.459. [DOI] [PubMed] [Google Scholar]
- Finan T. M., Wood J. M., Jordan D. C. Succinate transport in Rhizobium leguminosarum. J Bacteriol. 1981 Oct;148(1):193–202. doi: 10.1128/jb.148.1.193-202.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang D. M., Shaikh K. Regulation of penicillin acylase in Escherichia coli by cyclic AMP. Biochim Biophys Acta. 1976 Feb 18;425(1):110–114. doi: 10.1016/0005-2787(76)90220-3. [DOI] [PubMed] [Google Scholar]
- Ghei O. K., Kay W. W. Properties of an inducible C 4 -dicarboxylic acid transport system in Bacillus subtilis. J Bacteriol. 1973 Apr;114(1):65–79. doi: 10.1128/jb.114.1.65-79.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves W. E., Davis F. C., Jr, Sells B. H. Spectrophotometric determination of microgram quantities of protein without nucleic acid interference. Anal Biochem. 1968 Feb;22(2):195–210. doi: 10.1016/0003-2697(68)90307-2. [DOI] [PubMed] [Google Scholar]
- Hamilton-Miller J. M. Penicillinacylase. Bacteriol Rev. 1966 Dec;30(4):761–771. doi: 10.1128/br.30.4.761-771.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessie T. G., Neidhardt F. C. Formation and operation of the histidine-degrading pathway in Pseudomonas aeruginosa. J Bacteriol. 1967 Jun;93(6):1800–1810. doi: 10.1128/jb.93.6.1800-1810.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitov M. M., Klapovskaia K. I., Kleiner G. I. Indutsirovannyi biosintez atsilazy Escherichia coli. Mikrobiologiia. 1967 Sep-Oct;36(5):912–917. [PubMed] [Google Scholar]
- Lo T. C., Bewick M. A. The molecular mechanisms of dicarboxylic acid transport in Escherichia coli K12. The role and orientation of the two membrane-bound dicarboxylate binding proteins. J Biol Chem. 1978 Nov 10;253(21):7826–7831. [PubMed] [Google Scholar]
- SZENTIRMAI A. PRODUCTION OF PENICILLIN ACYLASE. Appl Microbiol. 1964 May;12:185–187. doi: 10.1128/am.12.3.185-187.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Vandamme E. J., Voets J. P. Microbial penicillin acylases. Adv Appl Microbiol. 1974;17(0):311–369. doi: 10.1016/s0065-2164(08)70563-x. [DOI] [PubMed] [Google Scholar]
- Vojtísek V., Slezák J. Penicillinamidohydrolase in Escherichia coli. II. Synthesis of the enzyme, kinetics and specificity of its induction and the effect of O2. Folia Microbiol (Praha) 1975;20(4):289–297. doi: 10.1007/BF02878110. [DOI] [PubMed] [Google Scholar]
- Vojtísek V., Slezák J. Penicillinamidohydrolase in Escherichia coli. III. Catabolite repression, diauxie, effect of cAMP and nature of the enzyme induction. Folia Microbiol (Praha) 1975;20(4):298–306. doi: 10.1007/BF02878111. [DOI] [PubMed] [Google Scholar]


