Abstract
Aim
To explore whether killer cell immunoglobulin-like receptors (KIR) gene polymorphisms are associated with susceptibility to persistent hepatitis B virus (HBV) infection or HBV clearance.
Methods
Fifteen known KIR genes were determined in 150 chronic hepatitis B patients, 251 spontaneously recovered controls, and 412 healthy controls by the sequence specific primer polymerase chain reaction (SSP-PCR) method. KIR genotype frequency (gf) differences were tested for significance by two-tailed Fisher exact test or χ2 test. Multifactorial analysis was also performed by logistic analysis (the SAS system).
Results
Framework genes KIR2DL4, KIR3DL2, KIR3DL3, and KIRZ were present in all individuals. The frequencies of KIR2DS2 and KIR2DS3 were higher in chronic hepatitis B patients, than in both healthy and spontaneously recovered controls. The frequencies of activating KIR2DS1, KIR3DS1, and the inhibitory KIR2DL5 were higher in spontaneously recovered controls than in chronic hepatitis B patients and healthy controls.
Conclusion
KIR polymorphisms may be associated with susceptibility to HBV infection or HBV clearance. It could be suggested that KIR2DS2 and KIR2DS3 were HBV-susceptive genes, which induced a persistent yet weak inflammatory reaction that resulted in continuous injury of live tissues and chronic hepatitis. KIR2DS1, KIR3DS1, and KIR2DL5, on the other hand, may be protective genes that facilitated the clearance of HBV.
Hepatitis B virus (HBV) infection, one of the major viral diseases in the word, especially in China, causes a broad spectrum of liver diseases (1). It has been documented that there are currently about 350 million people with persistent HBV infection worldwide. Persistent HBV infection has been regarded as a multi-factorial disorder relevant to virus, host age, sex, environment, and concurrent infections with the hepatitis C and D virus, and human immunodeficiency virus (HIV) (2). Segregation analysis and twin studies strongly support the role of host genetic background in determining the course of HBV infection (3-5). Actually, the susceptibility to infectious diseases is governed by a number of different factors, such as cytokine production, antigen presentation, and receptor recognition. Of note, genetic susceptibility to persistent HBV infection or HBV clearance are likely polygenic, pertaining to genes such as the genes of human leukocyte antigen (HLA) and class cytokine receptor.
It has been reported that killer cell immunoglobulin-like receptor (KIR) genes present diversity in the Chinese population (6,7). However, it is unknown whether KIR genes participate in the regulation of HBV infectious process. KIR gene family, located on human chromosome 19q13.4, encodes HLA class I receptors expressed by natural killer (NK) cells and subsets of T-cells. KIR genes are organized in a highly polymorphic, multi-gene family with considerable allelic polymorphism. The genes have been divided into distinct groups, depending on the number of external immunoglobulin domains (2D or 3D). The presence of a long cytoplasmatic tail with two immune tyrosine-based inhibitory motifs (ITIM) allows the transduction of inhibitory signals and characterizes the inhibitory KIRs (2DL, 3DL), whereas the presence of short cytoplasmatic tails corresponds to the activating KIR receptors (2DS, 3DS) (8-11). Theoretically, NK cells and T cells activation may be regulated by one of the two following mechanisms: the presence of and signaling through activating receptors on a large proportion of effector cells (ie, KIR haplotypes containing many activating receptors) or the presence of inhibitory receptor-ligand combinations that send relatively poor inhibitory signals. Upon interaction with HLA class I molecules expressed on the surface of target cells, KIR genes provide activating or suppressing signals to regulate the activation of NK cells and T cells, thereby playing an important role in antiviral and anti-tumor immunity (12).
Previous studies have demonstrated that KIR genes are involved in the pathogenesis of a variety of diseases, including rheumatoid arthritis, vasculitis (13,14), psoriatic arthritis (15), type 1 diabetes mellitus (16), and hepatitis C virus (17). However up to now, the role of KIR polymorphisms in patients with HBV infection has not been investigated. Therefore, the present study was designed to investigate the KIR gene polymorphisms in a large cohort of 150 chronic hepatitis B patients, 251 spontaneously recovered cases, and 412 healthy controls by means of sequence specific primer polymerase chain reaction (SSP-PCR), with special attention given to the relationship between KIRs and the HBV infection or HBV clearance.
Methods
Study subjects
The 813 samples analyzed for this study comprised 150 samples from patients with chronic hepatitis B (CHB), 251 spontaneously recovered (SR) controls, and 412 healthy unrelated adult Chinese recruited from Shandong Provincial Hospital and Jinan Infectious Disease Hospital, both in Jinan, between October 2004 and August 2006. CHB patients were required to have a history of hepatitis B viral infection for more than one year and elevated levels of alanine aminotransferase/aspartate aminotransferase or total bilirubin when recruited. Those who were negative for hepatitis B surface antigen (HBsAg) and positive for both hepatitis B surface antibody (HBsAb) and hepatitis B core antibody (HBcAb) were defined as SR controls. All the recruited subjects had no serological evidence of hepatitis C virus, hepatitis D virus, and HIV coinfection; they had no diabetes, malignant tumor, or any autoimmune diseases (18). All studies were performed after informed consent was obtained from the subjects.
Research approach
Genome DNA extraction. Genomic DNA sample was extracted from 5mL EDTA (EDTA) anticoagulated peripheral blood with a standard salting-out procedure and stored at -20°C before use.
Genotyping with SSP-PCR method. KIR genotyping was performed by the sequence-specific primer polymerase chain reaction (SSP-PCR) method in all the recruited subjects. KIR locus typing was performed to detect the presence or absence of a total of 14 KIR loci and one pseudogene KIRZ. Among them, 8 KIR genes (2DLI, 2DL2, 2DL3, 2DL4, 2DL5, 3DL1,3DL2, and 3DL3) were responsible for inhibitory functions and 6 KIR genes (2DS1, 2DS2, 2DS3, 2DS4, 2DS5, and 3DS1) for conveying activating signals. The SSP-PCR primers used for the detection of KIR loci were based on primer sites that have been previously described (15). Among the 29 formatted couple primers (Shanghai BoYa biotechnology Co.Ltd, Shangai, China), 2DS5 gene uses one couple primer and each of the 14 surplus genes uses two couple primers, so as to ensure a detectable rate of positive gene (KIR genes primer in Table 1). The framework genes (2DL4, 3DL2, and 3DL3) served as a positive marker of PCR. PCR was conducted by the Gene Amp PCR system 9700-R (Applied Biosystems, Foster City, CA, USA). Briefly, 1µL of genomic DNA was amplified in a volume of approximately 20 µL system including 6µL primers, 2 µL 10 × PCR buffer,1.6 µL MgCL2 (25 000 µM), 0.4 µL dNTP (10 000 µM), 0.125 µL Taq polymerase (5 U/µL), and 8.875 µL dH2O. After the initial denaturation for 1 minute at 96°C, the samples were amplified in the following way: 5 cycles of 25 seconds at 96°C, 45 seconds at 65°C, and 30 seconds at 72°C; 21 cycles of 25 seconds at 96°C, 45 seconds at 60°C, and 30 seconds at 72°C; 5 cycles of 25 seconds at 96°C, 1 minute at 55°C, 2 minutes at 72°C; and a prolongation of 10-minute at 72°C.
Table 1.
Sequence specific polymerase chain reaction (PCR) primers of killer cell immunoglobulin-like receptors (KIR) genes
| Primer |
|||
|---|---|---|---|
| KIR gene | forward (5′-3′) | reverse (5′- 3′) | Base pair |
| 2DL1 |
GTT GGT CAG ATG TCA TGT TTG AA |
GGT CCC TGC CAG GTC TTG CG |
127 |
| TGG ACC AAG AGT CTG CAG GA |
TGT TGT CTC CCT AGA AGA CG |
330 |
|
| 2DL2 |
CTG GCC CAC CCA GGT CG |
GGA CCG ATG GAG AAG TTG GCT |
173 |
| GAG GGG GAG GCC CAT GAA T |
TCG AGT TTG ACC ACT CGT AT |
150 |
|
| 2DL3 |
CTT CAT CGC TGG TGC TG |
AGG CTC TTG GTC CAT TAC AA |
550 |
| TCC TTC ATC GCT GGT GCT G |
GGC AGG AGA CAA CTT TGG ATC A |
800 |
|
| 2DL4 |
CAG GAC AAG CCC TTC TGC |
CTG GGT GCC GAC CAC T |
254 |
| ACC TTC GCT TAC AGC CCG |
GGG TTT CCT GTG ACA GAA ACA G |
288 |
|
| 2DL5 |
GCG CTG TGG TGC CTC G |
GAC CAC TCA ATG GGG GAG C |
214 |
| TGC AGC TCC AGG AGC TCA |
GGG TCT GAC CAC TCA TAG GGT |
194 |
|
| 3DL1 |
CGC TGT GGT GCC TCG A |
GGT GTG AAC CCC GAC ATG |
197 |
| CCC TGG TGA AAT CAG GAG AGA G |
TGT AGG TCC CTG CAA GGG CAA |
181 |
|
| 3DL2 |
CAA ACC CTT CCT GTC TGC CC |
GTG CCG ACC ACC CAG TGA |
245 |
| CCC ATG AAC GTA GGC TCC G |
CAC ACG CAG GGC AGG G |
130 |
|
| 3DL3 |
GTC AGA TGT CAG GTT TGA GCG |
CAT GGA ATA GTT GAC CTG GGA AC |
112 |
| GCA GCT CCC GGA GCT TG |
GGG TCT GAC CAC GCG TG |
190 |
|
| 2DS1 |
CTTCTCCATCAGTCGCATGAA |
CTTCTCCATCAGTCGCATGAG |
102 |
| CTTCTCCATCAGTCGCATGAA |
AGAGGGTCACTGGGAGCTGAC |
102 |
|
| 2DS2 |
TTC TGC ACA GAG AGG GGA AGT A |
AGG TCA CTG GGA GCT GAC AA |
173 |
| CGG GCC CCA CGG TTT |
GGT CAC TCG AGT TTG ACC ACT CA |
240 |
|
| 2DS3 |
TGG CCC ACC CAG GTC G |
TGA AAA CTG ATA GGG GGA GTG AGG |
242 |
| CTA TGA CAT GTA CCA TCT ATC CAC |
AAG CAG TGG GTC ACT TGA C |
190 |
|
| 2DS4 |
CTG GCC CTC CCA GGT CA |
TCT GTA GGT TCC TGC AAG GAC AG |
204 |
| CTG GCC CTC CCA GGT CA |
GGA ATG TTC CGT TGA TGC |
2000 |
|
| 2DS5 |
TGA TGG GGT CTC CAA GGG |
TCC AGA GGG TCA CTG GGC |
125 |
| 3DS1 |
AGC CTG CAG GGA ACA GAA G |
GCC TGA CTG TGG TGC TCG |
300 |
| CCT GGT GAA ATC AGG AGA GAG |
GTC CCT GCA AGG GCA C |
177 |
|
| 2DP1 | GTC TGC CTG GCC CAG CT |
GTG TGA ACC CCG ACA TCT GTA C |
205 |
| CCA TCG GTC CCA TGA TGG | CAC TGG GAG CTG ACA ACT GAT G | 90 | |
Agarose gel electrophoreses and image scanning. The PCR products, together with approximately 3µL 100 base pairs (bp) DNA ladder as molecular weight marker (MBI, San Francisco, CA, USA), were electrophoresed on 1.5% agarose gels with bromophenol blue, keeping voltage at 160 V for 30 minutes. After electrophoresis, the agarose gel was scanned and imaged by Alphaimager TM 2200 instrument (Alpha Innotech Corporation, San Leandro, CA, USA) and each sample was genotyped. A result of electrophoresis of the KIR genes PCR products is shown in Figure 1. All primers were validated to be gene-specific by PCR product sequencing.
Figure 1.
Amplification production electrophoresis picture of a chronic hepatitis B patient. The corresponding digits represent: 1,2 – KIR3DL1; 3,4 – KIR3DL2; 5,6 – KIR3DS1; 7,8 – KIR3DL3; 9,10 – KIR2DL5; 11,12 – KIRZ; 13,14 – KIR2DL1; 15,16 – KIR2DL2; 17,18 – KIR2DL3; 19,20 – KIR2DL4; 21,22 – KIR2DS2; 23,24 – KIR2DS3; 25,26 – KIR2DS4; 27 – KIR2DS5; 28,29 – KIR2DS1; M – M-DNA marker. All KIR phenotypes were positive except KIR2DS3 (19,20).
Statistical analysis
The phenotype frequency (pf, %) of each KIR was calculated as the percentage of positive numbers among all specimens. Genotype frequency (gf) was calculated with the formula gf = 1-. KIR genotype frequency (gf) differences were tested for significance by two-tailed Fisher exact test or χ2 test. Analyses were performed by the SAS 9.0 statistical package (SAS, Cary, NC, USA).
Results
In this study, all the tested KIR genes were present in both patient groups and control group in different frequencies. Framework genes KIR2DL4, KIR3DL2, KIR3DL3, and KIRZ were present in all individuals (Table 2).
Table 2.
Killer cell immunoglobulin-like receptors (KIR) phenotype frequency (pf) and genotype frequencies (gf) in patients and control subjects*
| Group |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KIR | healthy controls (N = 412) | chronic hepatitis B (N = 150) | spontaneously recovered controls (N = 251) | |||||||||
| + | pf (%) | gf (%) | + | pf (%) | gf | PC | + | pf (%) | gf (%) | PS | P | |
| Inhibitory | ||||||||||||
| 2DL1 | 407 | 98.79 | 89.00 | 150 | 100 | 100 | 1 | 248 | 98.80 | 89.05 | 1 | 1 |
| 2DL2 | 79 | 19.17 | 10.10 | 21 | 11.00 | 7.263 | 0.172 | 39 | 15.54 | 0.081 | 0.251 | 0.773 |
| 2DL3 | 407 | 98.79 | 89.00 | 150 | 100 | 100 | 1 | 237 | 94.42 | 76.38 | 0.045 | 0.062 |
| 2DL4 | 412 | 100 | 100 | 150 | 100 | 100 | 1 | 251 | 100 | 100 | 1 | 1 |
| 2DL5 | 216 | 52.43 | 31.03 | 102 | 68.00 | 43.43 | 0.001 | 200 | 79.68 | 54.92 | 0 | 0.001 |
| 3DL1 | 407 | 98.79 | 89.00 | 150 | 100 | 100 | 1 | 247 | 98.41 | 87.39 | 0.736 | 0.654 |
| 3DL2 | 412 | 100 | 100 | 150 | 100 | 100 | 1 | 251 | 100 | 100 | 1 | 1 |
| 3DL3 | 412 | 100 | 100 | 150 | 100 | 100 | 1 | 251 | 100 | 100 | 1 | 1 |
| Activating | ||||||||||||
| 2DS1 | 205 | 49.76 | 29.16 | 84 | 56.00 | 33.67 | 0.215 | 180 | 71.71 | 46.81 | 0 | 0.001 |
| 2DS2 | 114 | 27.67 | 14.95 | 64 | 42.67 | 24.28 | 0.001 | 63 | 25.10 | 13.46 | 0.526 | 0.001 |
| 2DS3 | 79 | 19.17 | 10.09 | 44 | 29.33 | 15.93 | 0.011 | 47 | 18.73 | 9.85 | 0.919 | 0.014 |
| 2DS4 | 399 | 96.84 | 82.22 | 148 | 98.67 | 88.47 | 0.375 | 251 | 100 | 100 | 0.022 | 0.059 |
| 2DS5 | 133 | 32.20 | 17.66 | 64 | 42.67 | 24.28 | 0.028 | 114 | 45.42 | 26.12 | 0.001 | 0.605 |
| 3DS1 | 176 | 42.72 | 24.32 | 70 | 46.67 | 26.97 | 0.442 | 164 | 65.34 | 41.13 | 0 | 0.001 |
| Pseudogene | ||||||||||||
| KIRZ | 412 | 100 | 100 | 150 | 100 | 100 | NS | 251 | 100 | 1 | 1 | 1 |
*Abbreviations: + – positive case numbers; PC – P value for the comparison between chronic hepatitis group and healthy control group; PS – P value for the comparison between spontaneously recovered group and healthy control group; P – P-value for the comparison between spontaneously recovered group and chronic hepatitis group.
KIR phenotype and genotype frequency in patients and control subjects
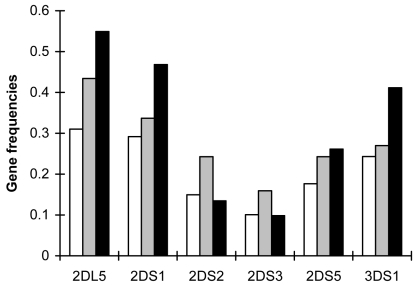
The total carriage frequency of KIR2DL5, KIR2DS2, KIR2DS3, and KIR2DS5 was higher in CHB patients than in health control subjects (P = 0.001, P = 0.001, P = 0.011, and P = 0.028, respectively).
The total carriage frequency of KIR2DS2 and KIR2DS3 was higher (P = 0.001, P = 0.014, respectively), while the frequency of KIR2DL5, KIR2DS1, and KIR3DS1 was lower (P = 0.001, P = 0.001, and P = 0.001, respectively) in CHB patients than in SR controls. As for KIR2DS5, there was no significant difference between these two groups (P = 0.605).
The total carriage frequency of KIR2DL5, KIR2DS1, KIR2DS5, and KIR3DS1 in SR controls was significantly higher than in healthy controls (P<0.001, P<0.001, P<0.001, and P<0.001, respectively). There was no significant difference in the frequency of KIR2DS2 and KIR2DS3 between the two groups (P = 0.526, P = 0.919, respectively) (Table 2, Figure 2).
Figure 2.
Comparison of killer cell immunoglobulin-like receptors gene frequencies. Closed bars – healthy controls; open bars – chronic hepatitis B group; gray bars – spontaneously recovered controls.
Activating KIRs in healthy controls and SR controls
We divided SR and healthy controls into 2 groups as follows: group one with a single activating KIR gene and group two with 2 or more activating KIR genes. Out of 251 SR controls, only 56 were in the group one and 195 in the group two (22.31% and 77.69%, respectively). On the contrary, the percentage in healthy controls was 66.26% and 33.74%, respectively.
Discussion
Our data clearly showed that there was a difference in the frequencies of these KIR genes in three different sets of subjects. To date, most of the studies on human genes relevant to HBV infection have focused on HLA genetic alteration. For example, genetic association analyses based on Gambian, European, and Asian cohorts have implicated the role of HLA allele DRB1*1302 in the clearance of HBV infection (21-23). As KIR molecules modulate cell function upon the recognition of HLA class I, it can be inferred that KIR gene may also exert a crucial role in the pathogenesis of HBV infection. In the present study, we analyzed 15 known KIR genes in CHB, SR control, and healthy control group. We found that the frequencies of KIR2DS2 and KIR2DS3 in CHB patients were significantly higher than in normal controls, but not in SR controls, indicating that the two genes may serve as HBV infectious susceptibility genes. Recent evidence has suggested that a subset of T cells expressing KIR2DS2 can mediate vascular damage in patients with rheumatoid arthritis, implicating a role of activating KIR in rheumatoid arthritis (16) and other autoimmune diseases (14). Although the samples used in this study were different, our results were, at least in part, in agreement with these findings. This suggests that KIR2DS2 is involved in inflammatory reaction and also that the excessive inflammatory reaction leads to liver tissue damage.
Clinically, HBV infection does not invariably result in chronic hepatitis since the host possesses the ability to eliminate the virus spontaneously in most cases. It is believed that the antibody response to viral envelope antigens contributes to clearance of the virus and that cytotoxic T cells mediate viral clearance by killing the infected cells. In addition, one study has shown that cytotoxic T lymphocytes inhibit HBV gene expression through the secretion of antiviral cytokines and that the expression of these cytokines may be the principal mechanism of viral clearance during HBV infection (24). It is hypothesized that chronic infection is related to a weak T-cell response to viral antigens. Recent research has demonstrated that KIRs expressed on the cell surface of NK and T cells play a role in the regulation of innate and acquired anti-virus immune responses through the transduction of inhibitory or activating signals (11,19,20). In CHB, the excessive expression of KIR2DS2 and KIR2DS3 may weakly activate NK or T cells' cytotoxicity and regulate it, leading to deferrable and persistent destruction of the hepatocytes. KIR2DS2 and KIR2DS3 are observed in high positive linkage disequilibrium (25), which might be a reason for the simultaneous increase of both of them.
To further analyze our data according to the number of activating KIRs and inhibitory KIRs, we found that there was a significant difference in the two kinds of KIRs between SR controls and healthy control group. This may mean that, in SR patients, more activating KIRs send more activating signals to NK cells or to T cells, resulting in HBV clearance or recruitment of other cells of the immune system.
In this study, both the activating KIR2DS1, KIR3DS1 and the inhibitory KIR2DL5 were higher in SR patients than in healthy controls and CHB patients. So, we suggested that these KIR genes may play a role in the clearance of HBV.
In conclusion, our results implicate that KIR2DS2 and KIR2DS3 may act as HBV susceptive genes to induce a persistent weak inflammatory reaction that results in continuous injury of live tissues and chronic hepatitis; whereas, KIR2DS1, KIR3DS1, and KIR2DL5 may be protective genes that facilitate the clearance of HBV.
Acknowledgment
We are very grateful to every one involved in the collection of samples from Jinan Infectious Hospital and Shandong Provincial Hospital, especially Chen Shi-jun (Jinan Infectious Hospital, Jinan, China) and Li Gui-qin and Song Zhen (Shandong Provincial Hospital). We also thank Wen Xin and Li Jing (Shandong Provincial Hospital) for assistance in sample collection and DNA isolation.
This study was supported by Chinese National Natural Science Foundation grants No. 30371304 and Shandong Provincial Natural Science Foundation grants yz2006c72.
Reference
- 1.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–45. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Thursz M. Genetic susceptibility in chronic viral hepatitis. Antiviral Res. 2001;52:113–6. doi: 10.1016/S0166-3542(01)00175-9. [DOI] [PubMed] [Google Scholar]
- 3.Hann HW, Kim CY, London WT, Whitford P, Blumberg BS. Hepatitis B virus and primary hepatocellular carcinoma: family studies in Korea. Int J Cancer. 1982;30:47–51. doi: 10.1002/ijc.2910300109. [DOI] [PubMed] [Google Scholar]
- 4.Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, et al. Hepatitis B virus markers in Chinese twins. Anticancer Res. 1989;9:737–41. [PubMed] [Google Scholar]
- 5.Shimbo S, Zhang ZW, Qu JB, Wang JJ, Zhang CL, Song LH, et al. Urban-rural comparison of HBV and HCV infection prevalence among adult women in Shandong Province, China. Southeast Asian J Trop Med Public Health. 1997;28:500–6. [PubMed] [Google Scholar]
- 6.Yan LX, Zhu FM, Jiang K, He J. Diversity of the killer cell immunoglobulin-like receptor gene KIR2DS4 in the Chinese population. Tissue Antigens. 2007;69:133–8. doi: 10.1111/j.1399-0039.2006.00746.x. [DOI] [PubMed] [Google Scholar]
- 7.Jiang K, Zhu FM, Lv QF, Yan LX. Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens. 2005;65:556–63. doi: 10.1111/j.1399-0039.2005.00412.x. [DOI] [PubMed] [Google Scholar]
- 8.Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, et al. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. 1995;182:875–84. doi: 10.1084/jem.182.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. 2000;51:268–80. doi: 10.1007/s002510050620. [DOI] [PubMed] [Google Scholar]
- 10.Selvakumar A, Steffens U, Dupont B. Polymorphism and domain variability of human killer cell inhibitory receptors. Immunol Rev. 1997;155:183–96. doi: 10.1111/j.1600-065X.1997.tb00951.x. [DOI] [PubMed] [Google Scholar]
- 11.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–63. doi: 10.1016/S1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 12.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–50. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 13.Yen JH, Moore BE, Nakajima T, Scholl D, Schaid DJ, Weyand CM, et al. Major histocompatibility complex class I-recognizing receptors are disease risk genes in rheumatoid arthritis. J Exp Med. 2001;193:1159–67. doi: 10.1084/jem.193.10.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yen JH, Lin CH, Tsai WC, Wu CC, Ou TT, Hu CJ, et al. Killer cell immunoglobulin-like receptor gene's repertoire in rheumatoid arthritis. Scand J Rheumatol. 2006;35:124–7. doi: 10.1080/03009740500381252. [DOI] [PubMed] [Google Scholar]
- 15.Martin MP, Nelson G, Lee JH, Pellett F, Gao X, Wade J, et al. Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J Immunol. 2002;169:2818–22. doi: 10.4049/jimmunol.169.6.2818. [DOI] [PubMed] [Google Scholar]
- 16.van der Slik AR, Koeleman BP, Verduijn W, Bruining GJ, Roep BO, Giphart MJ. KIR in type 1 diabetes: disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes. 2003;52:2639–42. doi: 10.2337/diabetes.52.10.2639. [DOI] [PubMed] [Google Scholar]
- 17.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–4. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 18.Deng G, Zhou G, Zhai Y, Li S, Li X, Li Y, et al. Association of estrogen receptor alpha polymorphisms with susceptibility to chronic hepatitis B virus infection. Hepatology. 2004;40:318–26. doi: 10.1002/hep.20318. [DOI] [PubMed] [Google Scholar]
- 19.Selvakumar A, Steffens U, Dupont B. Polymorphism and domain variability of human killer cell inhibitory receptors. Immunol Rev. 1997;155:183–96. doi: 10.1111/j.1600-065X.1997.tb00951.x. [DOI] [PubMed] [Google Scholar]
- 20.Lowe EJ, Turner V, Handgretinger R, Horwitz EM, Benaim E, Hale G, et al. T-cell alloreactivity dominates natural killer cell alloreactivity in minimally T-cell-depleted HLA-non-identical paediatric bone marrow transplantation. Br J Haematol. 2003;123:323–6. doi: 10.1046/j.1365-2141.2003.04604.x. [DOI] [PubMed] [Google Scholar]
- 21.Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065–9. doi: 10.1056/NEJM199504203321604. [DOI] [PubMed] [Google Scholar]
- 22.Höhler T, Gerken G, Notghi A, Lubjuhn R, Taheri H, Protzer U, et al. HLA-DRB1*1301 and *1302 protect against chronic hepatitis B. J Hepatol. 1997;26:503–7. doi: 10.1016/S0168-8278(97)80414-X. [DOI] [PubMed] [Google Scholar]
- 23.Ahn SH, Han KH, Park JY, Lee CK, Kang SW, Chon CY, et al. Association between hepatitis B virus infection and HLA-DR type in Korea. Hepatology. 2000;31:1371–3. doi: 10.1053/jhep.2000.7988. [DOI] [PubMed] [Google Scholar]
- 24.Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. 2000;51:268–80. doi: 10.1007/s002510050620. [DOI] [PubMed] [Google Scholar]
- 25.Bontadini A, Testi M, Cuccia MC, Martinetti M, Carcassi C, Chiesa A, et al. Distribution of killer cell immunoglobulin-like receptor genes in the Italian Caucasian population. J Transl Med. 2006;4:44. doi: 10.1186/1479-5876-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]