Abstract
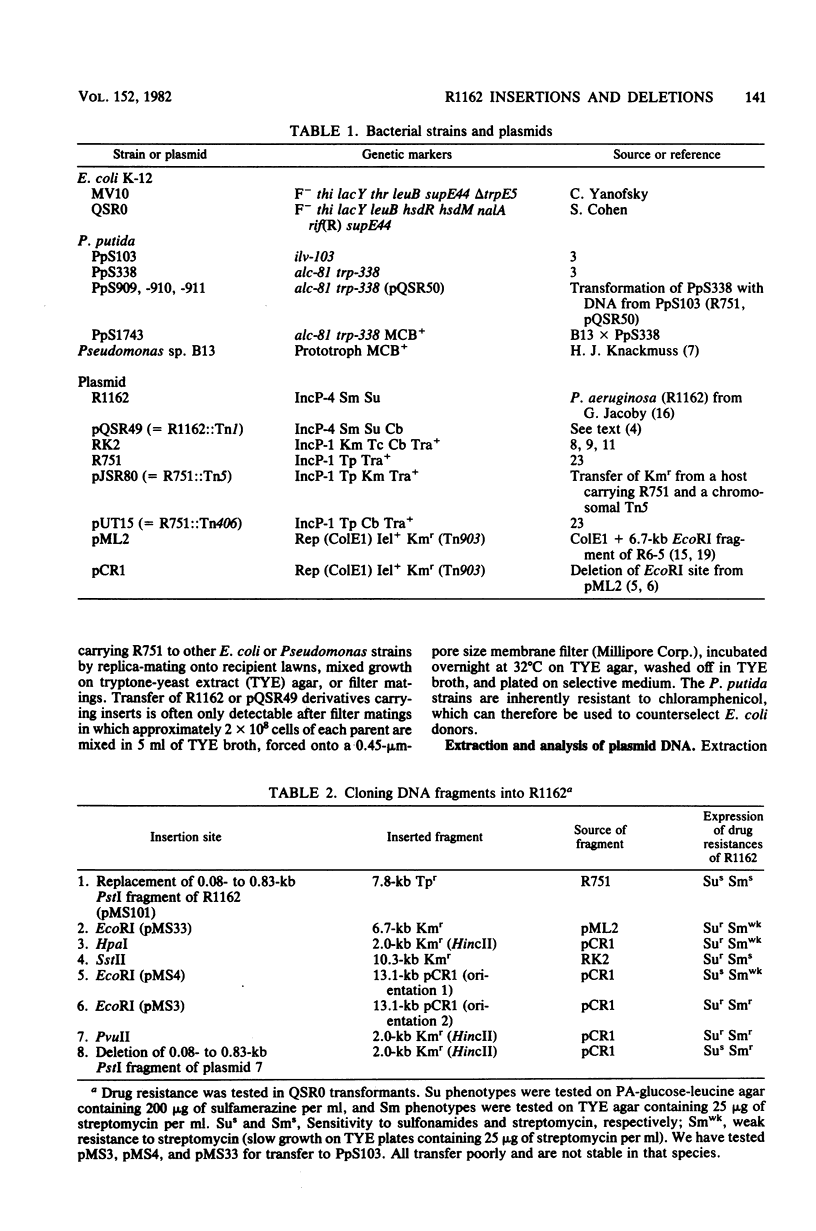
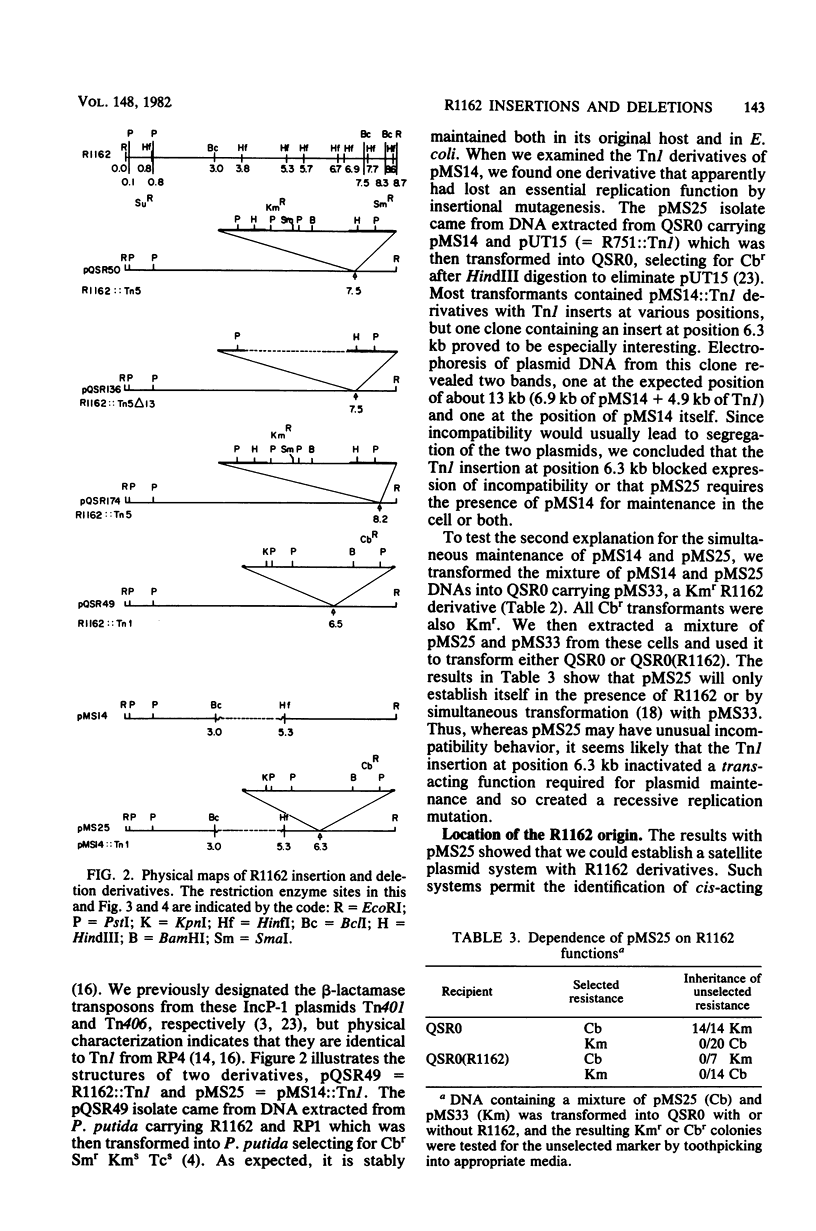
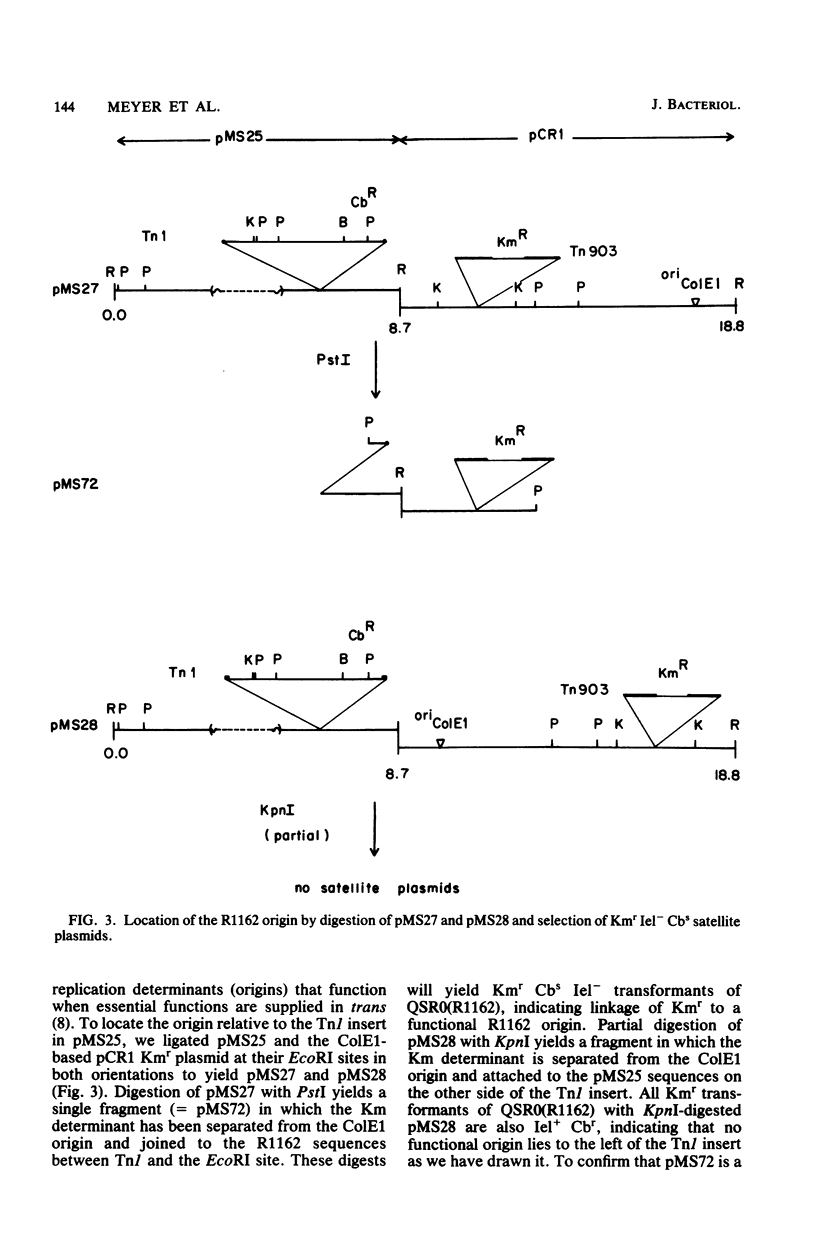
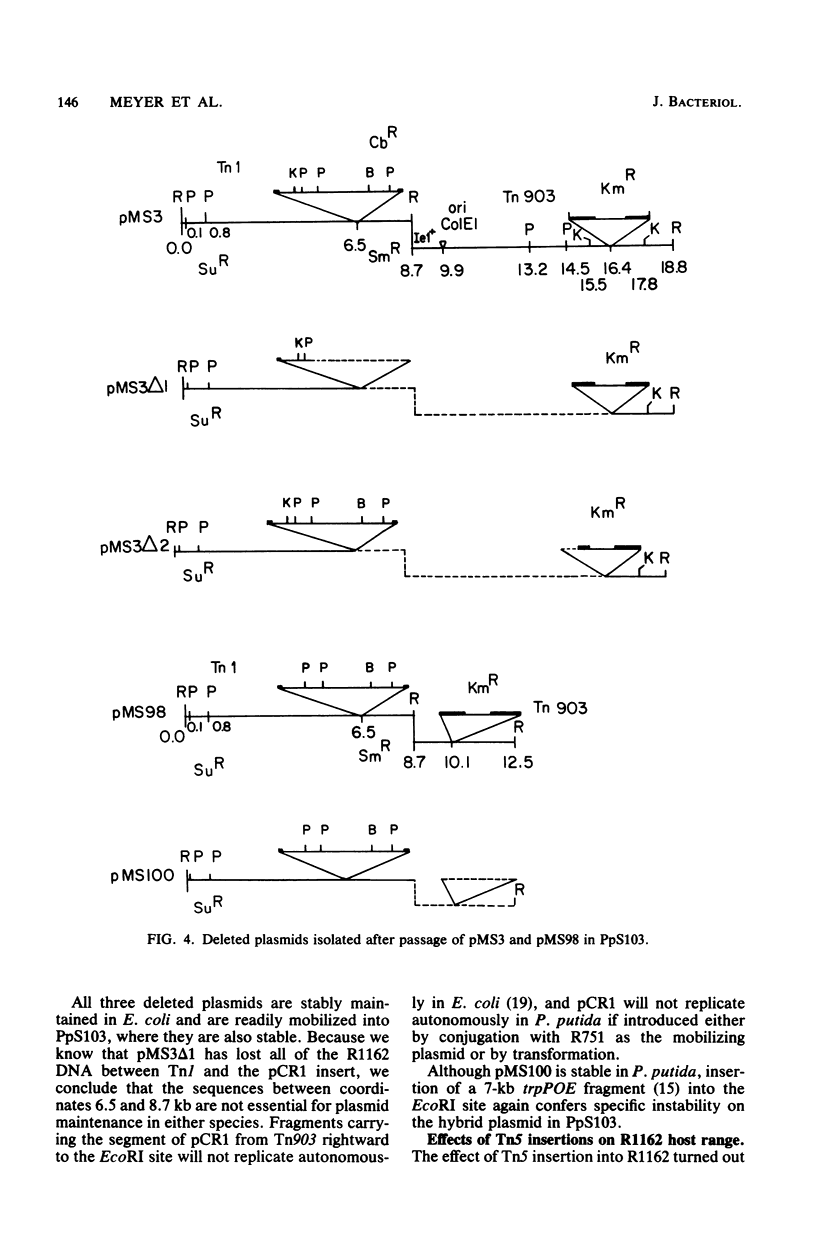
R1162 is an 8.7-kilobase (kb) broad-host-range replicon encoding resistance to streptomycin and sulfa drugs. In vitro deletion of 1.8-kb DNA between coordinates 3.0 and 5.3 kb did not affect plasmid maintenance, but a Tn1 insertion at coordinate 6.3 kb led to a recessive defect in plasmid maintenance. The only cis-acting region necessary for plasmid replication appears to lie between the Tn1 insertion at coordinate 6.3 kb and a second Tn1 insertion at coordinate 6.5 kb. All R1162 sequences between position 6.5 kb and the EcoRI site at coordinate 8.7/0 kb were dispensible for replication in Escherichia coli and Pseudomonas putida. Plasmids carrying insertions in a variety of restriction sites in an R1162::Tn1 derivative were unstable in P. putida but stable in E. coli. Tn5 insertions in R1162 showed a hot spot at coordinate 7.5 kb. A Tn5 insertion at coordinate 8.2 kb appeared to mark the 3' end of the streptomycin phosphotransferase coding sequence. All R1162::Tn5 derivatives showed specific instability in Pseudomonas strains but not in E. coli. The instability could be relieved by internal deletions of Tn5 sequences. In the haloaromatic-degrading Pseudomonas sp. strain B13, introduction of an unstable R1162::Tn5 plasmid led to loss of ability to utilize m-chlorobenzoate as a growth substrate. Our results showed that alteration of plasmid sequence organization in nonessential regions can result in restriction of plasmid host range.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedik M., Fennewald M., Shapiro J. Transposition of a beta-lactamase locus from RP1 into Pseudomonas putida degradative plasmids. J Bacteriol. 1977 Feb;129(2):809–814. doi: 10.1128/jb.129.2.809-814.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S., Shapiro J. TOL is a broad-host-range plasmid. J Bacteriol. 1978 Jul;135(1):278–280. doi: 10.1128/jb.135.1.278-280.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggins L. W., McCluskey E. Characterization of the ColE1-Km plasmids pCR1 and pCR11 by electron microscope and restriction endonuclease mapping. Plasmid. 1979 Jul;2(3):446–453. doi: 10.1016/0147-619x(79)90028-3. [DOI] [PubMed] [Google Scholar]
- Covey C., Richardson D., Carbon J. A method for the deletion of restriction sites in bacterial plasmid deoxyribonucleic acid. Mol Gen Genet. 1976 May 7;145(2):155–158. doi: 10.1007/BF00269587. [DOI] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Meyer R. J., Helinski D. R. Suppression of Co1E1 replication properties by the Inc P-1 plasmid RK2 in hybrid plasmids constructed in vitro. J Mol Biol. 1979 Sep 25;133(3):295–318. doi: 10.1016/0022-2836(79)90395-4. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bonewald R. The use of plasmid R1162 and derivatives for gene cloning in the methanol-utilizing Pseudomonas AM1. Mol Gen Genet. 1980;178(2):375–380. doi: 10.1007/BF00270487. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Kelley W. S. Effects of different alleles of the E. coli K12 pol A gene on the replication of non-transferring plasmids. Mol Gen Genet. 1976 Feb 2;143(3):311–318. doi: 10.1007/BF00269409. [DOI] [PubMed] [Google Scholar]
- Guerry P., van Embden J., Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974 Feb;117(2):619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Yanofsky C., Lovett M. A., Helinski D. R. Plasmid ColEl as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3455–3459. doi: 10.1073/pnas.71.9.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kretschmer F. J., Chang A. C., Cohen S. N. Indirect selection of bacterial plasmids lacking identifiable phenotypic properties. J Bacteriol. 1975 Oct;124(1):225–231. doi: 10.1128/jb.124.1.225-231.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F'kn plasmid. J Bacteriol. 1976 Aug;127(2):982–987. doi: 10.1128/jb.127.2.982-987.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. J., Shapiro J. A. Genetic organization of the broad-host-range IncP-1 plasmid R751. J Bacteriol. 1980 Sep;143(3):1362–1373. doi: 10.1128/jb.143.3.1362-1373.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R., Boch G., Shapiro J. Transposition of DNA inserted into deletions of the Tn5 kanamycin resistance element. Mol Gen Genet. 1979 Mar 9;171(1):7–13. doi: 10.1007/BF00274009. [DOI] [PubMed] [Google Scholar]
- Meyer R., Figurski D., Helinski D. R. Molecular vehicle properties of the broad host range plasmid RK2. Science. 1975 Dec 19;190(4220):1226–1228. doi: 10.1126/science.1060178. [DOI] [PubMed] [Google Scholar]
- Meyer R., Hinds M., Brasch M. Properties of R1162, a broad-host-range, high-copy-number plasmid. J Bacteriol. 1982 May;150(2):552–562. doi: 10.1128/jb.150.2.552-562.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahari K. Deletion plasmids from transformants of Pseudomonas aeruginosa trp cells with the RSF1010-trp hybrid plasmid and high levels of enzyme activity from the gene on the plasmid. J Bacteriol. 1978 Oct;136(1):312–317. doi: 10.1128/jb.136.1.312-317.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Construction of haloaromatics utilising bacteria. Nature. 1979 Feb 1;277(5695):385–386. doi: 10.1038/277385a0. [DOI] [PubMed] [Google Scholar]
- Riley M., Anilionis A. Evolution of the bacterial genome. Annu Rev Microbiol. 1978;32:519–560. doi: 10.1146/annurev.mi.32.100178.002511. [DOI] [PubMed] [Google Scholar]
- Rubens C., Heffron F., Falkow S. Transposition of a plasmid deoxyribonucleic acid sequence that mediates ampicillin resistance: independence from host rec functions and orientation of insertion. J Bacteriol. 1976 Oct;128(1):425–434. doi: 10.1128/jb.128.1.425-434.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Meyer R., Helinski D. R. Regions of broad-host-range plasmid RK2 which are essential for replication and maintenance. J Bacteriol. 1980 Jan;141(1):213–222. doi: 10.1128/jb.141.1.213-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windass J. D., Worsey M. J., Pioli E. M., Pioli D., Barth P. T., Atherton K. T., Dart E. C., Byrom D., Powell K., Senior P. J. Improved conversion of methanol to single-cell protein by Methylophilus methylotrophus. Nature. 1980 Oct 2;287(5781):396–401. doi: 10.1038/287396a0. [DOI] [PubMed] [Google Scholar]



