Abstract
The Caudal-related homeobox genes Cdx1 and Cdx2 are intestine-specific transcription factors that regulate differentiation of intestinal cell types. Previously, we have shown Cdx1 to be antiproliferative and to promote cell differentiation. However, other studies have suggested that Cdx1 may be an oncogene. To test for oncogenic behavior, we used the murine villin promoter to ectopically express Cdx1 in the small intestinal villi and colonic surface epithelium. No changes in intestinal architecture, cell differentiation, or lineage selection were observed with expression of the transgene. Classic oncogenes enhance proliferation and induce tumors when ectopically expressed. However, the Cdx1 transgene neither altered intestinal proliferation nor induced spontaneous intestinal tumors. In a murine model for colitis-associated cancer, the Cdx1 transgene decreased, rather than increased, the number of adenomas that developed. In the polyps, the expression of the endogenous and the transgenic Cdx1 proteins was largely absent, whereas endogenous Villin expression was retained. This suggests that transgene silencing was specific and not due to a general Villin inactivation. In conclusion, neither the ectopic expression of Cdx1 was associated with changes in intestinal cell proliferation or differentiation nor was there increased intestinal cancer susceptibility. Our results therefore suggest that Cdx1 is not an oncogene in normal intestinal epithelium.
Introduction
The continuous renewal of intestinal epithelium provides many unique challenges. Rates of cell production must be precisely balanced by cell loss or destruction, otherwise the epithelial barrier function is compromised, or, alternatively, tumors and obstructing masses form, obliterating the normal lumen. Cell proliferation and differentiation are thus tightly controlled in the normal intestinal epithelium. Our current understanding of these processes is limited but improving. Many of the transcription and growth factors that regulate intestinal cell proliferation or differentiation have been identified [1–6].
The Caudal-related homeobox (Cdx) transcription factors play key roles in regulating intestinal epithelial differentiation and proliferation [7,8]. The Cdx homologues modulate a diverse set of processes including proliferation, apoptosis, cell adhesion, and the acquisition of a columnar morphology [7]. They are also necessary for the expression of an increasing number of intestine-specific genes [9–13]. By targeting these various processes and genes, the Cdx homologues contribute toward the maintenance of this balance between proliferation and differentiation.
Numerous studies have sought to define the role of the Cdx homologues in human colon carcinogenesis. Unfortunately, the question as to whether these transcription factors hinder or contribute to the process of carcinogenesis remains unsettled. Cdx2, the more intensively studied of the two, was historically thought of as a tumor suppressor [14,15], although recent studies have suggested that it may have oncogenic potential [16–18].
In contrast to Cdx2, little is known about Cdx1's role. Historically, Cdx1 was described as an oncogene due in part to reports that it promoted proliferation of IEC6 and Caco-2 cells [19,20]. Moreover, Wnt/β-catenin signaling is required for Cdx1 expression [21], suggesting that Cdx1 may promote Wnt-mediated proliferation. However, we have reported that restoring Cdx1 expression to colon cancer cells inhibited proliferation by blocking β-catenin/T cell factor transcriptional activity [22]. Thus, in our model, Cdx1 could negatively feedback on the Wnt/β-catenin signaling pathway to limit cell proliferation. The data from human colon cancer specimens do not completely clarify the issue. In the majority of human colon cancer specimens studied, CDX1 expression is lost due to active CDX1 gene silencing by promoter hypermethylation [23–25]. However, a subset of colon cancers may express increased levels of Cdx1 mRNA and protein [26,27].
Therefore, to directly test what the effects of Cdx1 overexpression are on intestinal oncogenesis, we generated transgenic mice with ectopic and overexpression of Cdx1 in the small intestinal and colonic epithelium using the murine Villin promoter. This expression did not alter endogenous Cdx1 mRNA levels, but there was a reciprocal reduction in Cdx2 mRNA and protein levels. The transgene had no effect on intestinal cell proliferation rates or differentiation of the four cell lineages. We observed the mice for up to 24 months and did not observe the development of any spontaneous intestinal polyps or cancers. Moreover, in a mouse model of inflammation-associated neoplasia, we found that the ectopic Cdx1 expression reduced neoplasia formation by nearly 50%. In addition, we noted the reduction of endogenous and absence of transgenic Cdx1 expression in the polyps that did form, whereas endogenous Villin expression remained robust. This suggests that loss of the Cdx1 transgene expression was a specific event and not simply due to a general loss of Villin gene expression. We conclude that ectopic overexpression of Cdx1 in normal intestinal epithelium does not have an oncogenic effect but may instead have significant antitumorigenic properties.
Materials and Methods
Transgenic Construct
To add a cMyc-tag to Cdx1, a full-length mouse Cdx1 cDNA was liberated from pRC-Cdx1 [28] and ligated into pCMV-Tag3c (Stratagene, La Jolla, CA). Then this cMyc-tagged Cdx1 cDNA, along with the SV40 polyA, were subcloned from pCMV-Tag3c into pBluescript KS to generate pCdx1-KS. The 12.4-kb mouse Villin promoter [29] was also subcloned into pBluescript to generate pVillin-KS. Then the cMyc-tag-Cdx1-SV40 polyA cassette was ligated into pVillin-KS to generate the final Villin-myc Cdx1 construct. TOPFLASH reporter was kindly provided by Ken Kinzler (Johns Hopkins University, Baltimore, MD).
Generation of Villin-Cdx1 Transgenic Mice
All animal experiments were performed under an animal use protocol approved by the University of Pennsylvania IACUC Committee. The final Villin-myc Cdx1 DNA construct was linearized and injected into the male pronuclei of fertilized eggs and implanted into pseudopregnant females by the Transgenic and Chimeric Mouse Core Facility at the University of Pennsylvania. Founder animals were identified by polymerase chain reaction (PCR) amplification of tail DNA with genotyping primers and confirmed by Southern blot with a villin intron probe [29]. Three founders were obtained from two separate injections. Transgene founders of the BGSJL/F1 strain (Jackson Laboratory, Bar Harbor, ME) were bred with normal CD-1 mice (Charles River, Wilmington, MA), and offspring were analyzed for the transgene by PCR. Therefore, mice used in these studies were on a mixed genetic background. There were no significant phenotypic differences between the three lines. The reported figures are therefore the combined results of the three lines.
Immunohistochemistry
Intestinal regions were isolated, rinsed in ice-cold PBS, fixed, and embedded as described [30]. Primary antibodies used include Cdx1 CPSP [30], bromodeoxyuridine (BrdU) (Upstate, Charlottesville, VA), monoclonal Cdx2 (Biogenex, San Ramon, CA), sucrase isomaltase (SI) (gift of K. Yeh, Louisiana State University, Shreveport, LA), α-chromagranin (Immunostar, Hudson, WI), lysozyme (Dako, Carpinteria, CA), and human cMyc-tag (BD Pharmingen, San Jose, CA). The primary antibodies were visualized using a biotinylated secondary antibody and an avidin-horseradish peroxidase conjugate (Vectastain ABC kit; Vector Laboratories, Burlingame, CA). The slides were developed with DAB (Vector Laboratories). For Alcian blue staining, slides were deparaffinized. After application of 3% aqueous acetic acid to the slides, 1% Alcian blue in 3% acetic acid, pH 2.5, was applied. Sections were washed and counterstained with 0.1% nuclear fast red, dehydrated, and mounted. Alkaline phosphatase activity was analyzed by deparaffinization of slides followed by incubation with 5-bromo-4-chloro-3-indolyl-phosphatase-4-nitro blue tetrazolium chloride. Crypt depth and villi length were measured in the ileumof transgenic and non-transgenic littermate controls using a software (IPLab; Scanalytics, Fairfax, VA). At least 10 crypts and 10 villi were measured in each mouse. Three transgenic and three littermate control mice were measured from each transgenic line for this study.
Western Blot Analysis
Nuclear proteins were isolated from the intestinal epithelium of adult mice by an adaptation of a previously described method [9]. Western blot analysis was then conducted. The blot was incubated with rabbit polyclonal Cdx1 antibody and then visualized with secondary antibodies and electrochemiluminescence detection (ECL Plus kit; Bio-Rad). To verify equal loading of samples, the blots were stripped and reprobed with anti-YY1 (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:500.
RNA Detection By Quantitative Reverse Transcription-Polymerase Chain Reaction
Sections of the distal jejunum were stored in a nontoxic tissue storage reagent (RNAlater; Ambion, Austin, TX). Total RNA was isolated from the tissue with a RNeasy Midi kit (Qiagen, Valencia, CA). cDNA was prepared from 1 µg of total RNA using Superscript II (Invitrogen, Carlsbad, CA) and random hexamers.
Primers for regular reverse transcription-polymerase chain reaction (RT-PCR) detection of total Cdx1:
FCdx1 468 5′ GCGCCTACGAATGGATG 3′
RCdx1 613 5′ TCTTTACCTGCCGCTCTGTGAG 3′ (spans intron 1)
Transgene-specific RNA primers:
FCdx1 665 5′ CGGCAGGTAAAGATCTGGTTC 3′ (crosses exon 2/3 boundary)
RSV40 5′ GAGCCTTGGGACTGTGAATCT 3′
Quantitative RT-PCR was performed on a detection system (ABI 7000; Applied Biosystems, Foster City, CA), with SYBR green used as the fluorescent dye. Primers were designed using Primer Express software (Applied Biosystems).
Cdx1 primers that recognize both the transgene and the endogenous Cdx1
5′ AGACCGAACCAAGGACAAG 3′
5′ TGATGTACCGGCTGTAGTGAA 3′
Specific primers that only recognize endogenous Cdx1
qFCdx1UTR CTAGGACAAGTAGCTTGCCCTCTT 3′
qRCdx1UTR 5′ TCCAACAGGCTCACCACACA 3′
Specific primers that only recognize endogenous Cdx2
5′ CGATACATCACCATCAGGAGG 3′
5′ TGGCTCTGCCGGTTCTGAAA 3′
Transgene-specific quantitative PCR primers
FVMCex1 5′ TGGCTGCCTCTTCCAGACA 3′
qRCdx1ex1 5′ CCACGTAGGGCTTCAGATCC 3′.
Standard PCR conditions were as then used. A dissociation curve was run with each PCR to ensure that primer-dimer formation did not occur. PCR results were analyzed using the ΔΔCt analysis (User Bulletin 2, Applied Biosystems), with 36B4 used as the housekeeping gene.
Proliferation Studies
Mice were injected with BrdU (Zymed) 1 hour before sacrifice. After BrdU immunohistochemistry, BrdU-labeled nuclei were counted in 15 full-length crypts that touched the basement membranes from regions of the small intestine and colon. Positive cells were expressed as the number of BrdU(+) cells per crypt unit. Three transgenic and matched nontransgenic littermates from each of the three Villin-Cdx1 lines were used in this study.
Azoxymethane/Dextran Sodium Sulfate Studies
Animals were injected with a single 10 mg/kg dose of azoxymethane (AOM) (Sigma, St. Louis, MO) at 6 weeks of age. One week later, animals received 3% dextran sodium sulfate (DSS) (MP Biochemicals, Solon, OH) in their drinking water for 7 days. After 14 days on regular water, the treated animals received a second round of DSS. A subgroup of these mice received a third round. Control animals received AOM alone, or DSS alone, at the same time as the test groups. Animals were euthanized at 14 weeks after the AOM injection. The colons were opened longitudinally, stained with 1% methylene blue, and examined under a dissecting microscope and scored for polyps. The tissue was then fixed and sections with polyps were subdivided for microscopic analysis. Polyps were independently examined by two blinded investigators who scored them as adenoma, lymphoid aggregate, or normal colon. A board-certified pathologist blinded to the treatment scored each adenoma for its degree of dysplasia. Polyp diameter was measured using IPLab software (Scanalytics) and recorded during the histologic examination of the polyps. Student's t test was performed to compare findings in the AOM/DSS-treated transgenic mice with the AOM/DSS-treated controls. After these parameters were recorded, some polyps had additional sections immunohistochemically stained for Cdx1, the cMyc-tag, and Cdx2 as before.
Results
Development of a Transgenic Mouse Line Ectopically Expressing Cdx1 throughout the Mouse Intestine
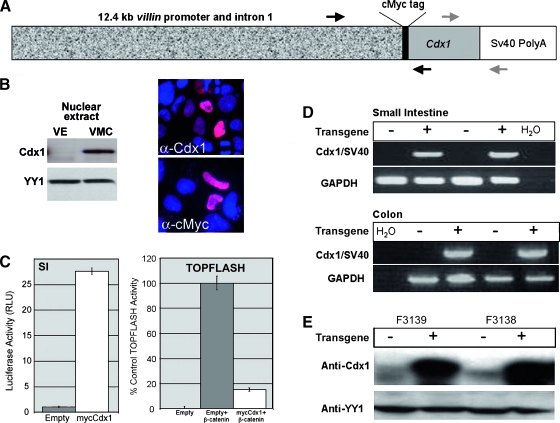
Transgenic mice were generated containing a construct with 12.4 kb mouse villin regulatory sequences (kindly provided by Dr. Deborah Gumucio, University of Michigan) driving expression of mouse Cdx1 cDNA (Figure 1A). An N-terminal cMyc-tag was added to differentiate transgenic from endogenous Cdx1. Before generating the transgenic mice, the villin-mycCdx1 construct was tested to confirm expression of active Cdx1 protein. The mycCdx1 protein was expressed and localized to the nucleus when the villin-mycCdx1 construct was transfected into Caco-2 cells (Figure 1B). In addition, we demonstrate that this construct can induce transcriptional activity from a canonical Cdx-responsive promoter, SI, when they are cotransfected into 293T cells. Lastly, the mycCdx1 protein can inhibit β-catenin/TCF transcriptional activity in 293T cells, as demonstrated by the suppression of TOPFLASH reporter activity (Figure 1C). Both results are consistent with wild-type Cdx1 activity, as previously published [22].
Figure 1.
Development of a transgenic mouse line ectopically expressing Cdx1 throughout the mouse intestine. (A) Mouse Cdx1 cDNA was cloned into pCMV-Tag3C to add an N-terminal human cMyc tag and SV40 PolyA, then placed behind 12.4 kb of mouse villin gene regulatory sequences, which included the promoter, untranslated exon 1, intron 1, and the start of exon 2. Genotyping primers, indicated by black arrows, amplified the junction of villin and Cdx1. RT-PCR primers, represented by gray arrows, amplified the junction of Cdx1 and SV40 PolyA. An N-terminal cMyc tag was added to differentiate transgenic from endogenous Cdx1. (B) The villin-mycCdx1 transgene expresses a functioning Cdx1 protein. mycCdx1 protein is detected in nuclear extracts from cells after transfection and is detected in the nucleus by immunofluorescence using anti-Cdx1 or anti-myc antibodies. Equal protein loading was confirmed by YY1 immunoblot analysis. VE, Villin-empty; VMC, Villin-mycCDX1. (C) Transient transfection of mycCdx1 into 293T induces luciferase activity from a canonical Cdx reporter, SI. Gray bar, empty vector control; white bar, mycCdx1. Cotransfection of the canonical β-catenin/TCF reporter TOPFLASH with mycCdx1 constructs demonstrates that the chimera can inhibit β-catenin/TCF transcriptional activity. Black bar, empty vector alone; gray bar, empty vector with β-catenin expression vector; white bar, mycCdx1 with β-catenin expression vector. (D) Transgene expression is shown by RT-PCR. RNA was isolated from the distal jejunum and colon of 1-month-old offspring from two different founders. RT-PCR with Cdx1/SV40 primers demonstrated expression of the transgene in both the small intestine and colon. (E) Overexpression of Cdx1 in the intestine in the Villin-Cdx1 transgenic mice. Jejunum tissue from offspring of two different founders. Nuclear extracts were isolated and loaded on sodium dodecyl sulfate-polyacrylamide gels. Western blot with Cdx1 antibody demonstrates overexpression of Cdx1 in transgenic animals. The same blot was stripped and reprobed with an antibody to nuclear protein YY1 to demonstrate even loading. F3138 and F3139 are the designations of the founder line serving as the source of the jejunal tissue.
Three distinct transgenic lines were established and studied using the villin-mycCdx1 construct. All three lines were viable and yielded transgenic pups in the expected Mendelian ratios. Transgenic pups appeared healthy and grew at rates similar to their wild-type littermates, with indistinguishable weight gains (data not shown). All three lines expressed the transgene mRNA and protein (Figure 1, D and E, and data not shown). Levels of transgenic protein expression appeared similar in the three lines, and transgenic Cdx1 levels were uniformly much higher than that of Cdx1 in wild-type littermates.
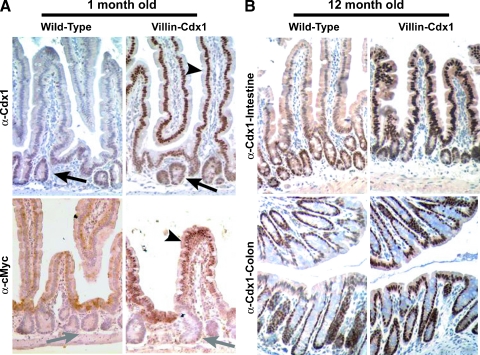
Transgene expression was detected at 1 month of life and was maintained in the animal for at least 12 months for all three founders studied (Figure 2). Expression earlier than 1 month was not evaluated for this study, but others have reported expression as early as embryonic day 15 (E15) when using this promoter as a transgene [29]. We have in fact studied several animals as old as 24 months, and transgene expression was still maintained (data not shown). Expression of the transgene Cdx1 protein, rather than enhancement of endogenous Cdx1 gene expression, was further confirmed by the detection of cMyc-tagged nuclear protein expression in the villus epithelial cells of the small intestine (Figure 2A). In summary, the 12.4 kb mouse villin regulatory sequences successfully directed high levels of Cdx1 transgene expression to the villi and superficial epithelium of the small intestine and colon in three lines of transgenic mice.
Figure 2.
Cdx1 ectopic expression in the small intestine villi and colon surface epithelium of Villin-Cdx1 transgenic mice. The small intestine and colon from transgenic or littermate control mice were excised at 1 and 12 months, fixed overnight in 4% paraformaldehyde, then processed for immunohistochemical studies. (A) Anti-Cdx1 antibody (CPSP) demonstrating that Cdx1 protein is limited to the crypt epithelial cells in 1-month-old wild-type mice (arrow). In contrast, in villin-Cdx1 transgenic mice, we see darkly stained nuclei in the upper crypt and in the villus epithelial cells (arrowhead). A mouse monoclonal antibody against the human cMyc-tag does not recognize endogenous mouse cMyc in the crypt cells of 1-month-old wild-type offspring (gray arrow) but detects nuclear-localized transgene expression in the small intestinal villi of transgenic mice (arrowhead). (B) These patterns of Cdx1 expression persist in 1-year-old wild-type and transgenic animals. Cdx1 protein is limited to the colon crypts of 1-year-old wild-type mice but is highly expressed in the colon surface epithelium of transgenic mice.
Ectopic Cdx1 Expression Does Not Alter Intestinal Epithelial Architecture, Cell Differentiation, or Cell Lineage Decisions
The villin-Cdx1 transgene directs ectopic Cdx1 expression to the villi and surface epithelium of the small intestine and colon (Figure 2). Transgene expression was somewhat variable in the duodenum and highest in the ileum. Colonic expression was maintained throughout the colon to the rectum. Ectopic Cdx1 expression did not appear to alter the architecture of the crypts and villi of the small intestine and colon at 1 month or 12 months of age (Figure 2). Crypt depth and villus length were no different in the villin-Cdx1 mice (100 ± 26 and 240 ± 25 µM, respectively) when compared to nontransgenic littermate controls (109 ± 26 and 230 ± 35 µM, respectively).
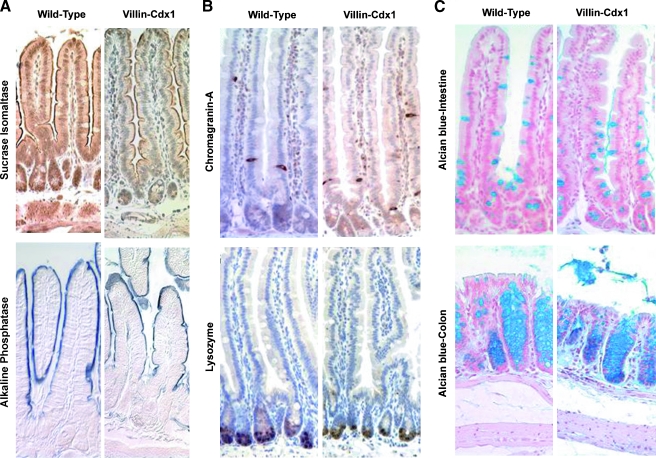
Cell differentiation in the intestinal epithelium is assessed by cell shape and morphology, cell lineage determination (absorptive enterocyte, goblet cell, enteroendocrine cell, or paneth cell), as well as the expression of markers for terminal differentiation. We stained small intestinal tissue for two enzymes associated with intestinal differentiation, SI, and alkaline phosphatase. The expression levels of both enzymes are regulated in part by the transcriptional activity of Cdx1 or Cdx2. Whereas SI expression is believed to require Cdx2 transcriptional activity for its expression, alkaline phosphatase expression is inhibited by Cdx2 and enhanced by Cdx1 [9,31]. The pattern of SI expression appeared to be unchanged in the villin-Cdx1 mice compared to their littermate controls (Figure 3A). Extensive brush border staining was detected in both the transgenic and the control mice. In addition, robust alkaline phosphatase enzymatic activity was similarly detected along the small intestine brush border surface of Villin-Cdx1 and control mice. Together, these findings suggest that ectopic Cdx1 expression in the villus epithelial cells did not hinder or disrupt the normal patterns of intestinal epithelial cell differentiation.
Figure 3.
No change in markers for enterocyte differentiation or cell lineages due to ectopic Cdx1 expression. (A) Immunohistochemistry with anti-sucrase isomaltase antibody shows no change in pattern of sucrase isomaltase gene expression in the small intestine brush border between 7-month-old wild-type and villin-Cdx1 transgenic mice. No difference in activity of brush border enzyme alkaline phosphatase noted in 7-month-old wild-type and transgenic mice. (C) Seven-month-old wild-type and villin-Cdx1 transgenic mice show no difference in the distribution of immunohistochemical staining for an enteroendocrine cell marker (chromagranin-A) or a paneth cell marker (lysozyme). (B) Similarly, staining with Alcian blue, a marker for goblet cells in the ileum of the small intestine and colon, was unchanged by ectopic Cdx1 expression in wild-type and transgenic small intestine.
We then examined for changes in the different lineages of our villin-Cdx1 mice by staining for specific markers. Chromagranin A, a component of secretory vesicles, has long been used as a general marker for the enteroendocrine cell lineage. Similarly, the enzyme lysozyme, a component of the paneth cell secretory granules, has been used to identify paneth cells in the intestinal epithelium. We stained for these markers in our villin-Cdx1 and control mice and found no significant differences in the numbers of paneth and enteroendocrine cells (Figure 3B). Alcian blue avidly stains intestinal mucins present in goblet cells. Alcian blue staining suggested that there might be a small increase in goblet cell numbers in our villin-Cdx1 mice. However, formal counting of Alcian blue-stained cells found only a small, statistically insignificant increase in ileal goblet cell numbers in two of the transgenic lines (Figure 3C and data not shown). In summary, ectopic expression of Cdx1 in the murine small intestine did not alter epithelial cell morphology, differentiation, or cell lineage selection.
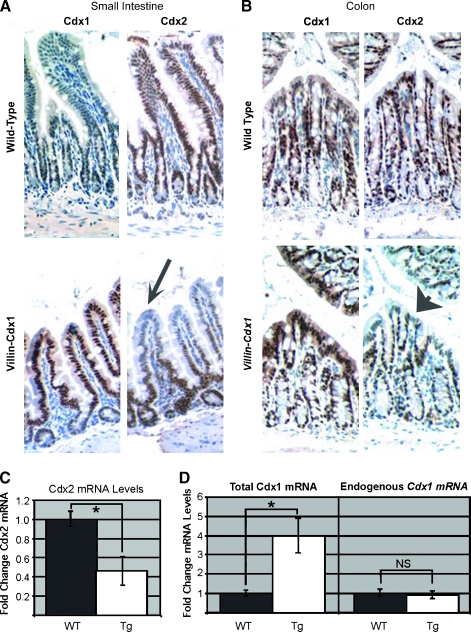
Expression of the Homologue Cdx2 Is Diminished By Ectopic Cdx1 Expression in Transgenic Mice
The Caudal homologue Cdx2 is a phosphoprotein that is expressed throughout the entire crypt-villus axis [7,32,33]. Its expression overlaps with Cdx1 in the crypts. In our transgenic mice, we found that Cdx2 staining was considerably reduced in both the small intestine and the colon in the regions where the Cdx1 transgene is expressed (Figure 4, A and B). This finding was somewhat unexpected, as SI and alkaline phosphatase levels were not altered in the villin-Cdx1 mice (Figure 2A). We confirmed the reduction in Cdx2 using quantitative real-time SYBR green RT-PCR. Cdx2 RNA is decreased two-fold in the villin-Cdx1 mouse small intestines when compared to nontransgenic littermates (Figure 4C).
Figure 4.
Ectopic expression of Cdx1 reduces Cdx2 expression in the small intestine villi and colon surface epithelium. Immunohistochemistry with either Cdx1 or Cdx2 antibodies on adjacent sections from 9-month-old nontransgenic and Villin-Cdx1 littermates. (A) Small intestine of nontransgenic mice with normal Cdx1 and Cdx2 expressions. Arrow points to reduced Cdx2 staining. (B) Cdx1 and Cdx2 expression pattern in the colon of transgenic and nontransgenic mice. Arrowhead points to reduced Cdx2 staining. (C) Real-time RT-PCR quantification of Cdx2 mRNA levels in the small intestine of transgenic and nontransgenic littermates. (D) Endogenous Cdx1 and total mRNA levels in the small intestine determined by quantitative RT-PCR. WT, wild-type; Tg, transgenic; NS, nonsignificant differences.
We had previously demonstrated that Cdx1 expression could reduce the β-catenin-dependent activity of its own promoter [22]. To determine if transgenic Cdx1 altered the expression of the endogenous Cdx1 gene, we measured both total and endogenous Cdx1 mRNA levels using real-time quantitative RT-PCR. We used two primer pairs, one that recognizes both the transgenic and endogenous Cdx1 mRNA, and one primer pair specific for the endogenous Cdx1 gene transcript (Figure 1A). Total Cdx1 mRNA levels were increased three- to five-fold in the small intestine of transgenic mice (Figure 4D). This increase was entirely due to the villin-Cdx1 transgene, as mRNA levels from the endogenous Cdx1 gene were unchanged. In summary, ectopic expression of Cdx1 induced a reciprocal reduction in the expression of the Caudal homologue Cdx2 without affecting epithelial cell differentiation.
Proliferation Rates in the Small Intestine and Colon Were Not Altered By Ectopic Expression of Cdx1
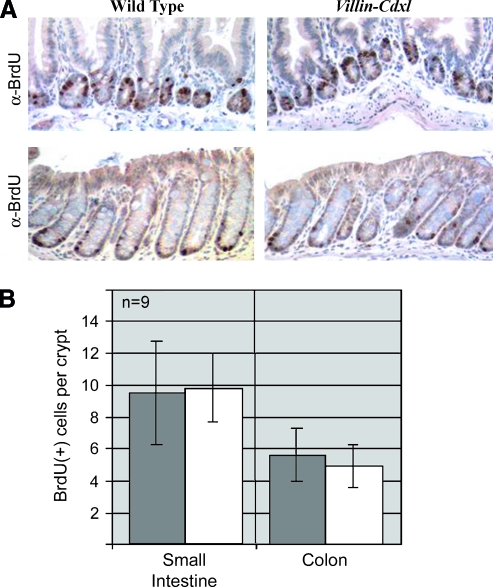
In prior studies of colon cancer cells, we demonstrated that restoration of Cdx1 expression significantly reduced rates of cell proliferation [34–36]. We therefore tested for changes in intestinal epithelial proliferation in our transgenic villin-Cdx1 mice by measuring BrdU labeling rates. As in nontransgenic controls, BrdU (US Biological, Swampscott,MA) incorporation was localized to the crypt compartment and appeared unaltered by ectopic Cdx1 expression (Figure 5A and data not shown). When extensive cell counts were performed on 6-month-old mice, no significant differences emerged in either the small intestine or the colon (Figure 5B). A subgroup analysis with each of the transgenic lines yielded similar findings (data not shown). We concluded that ectopic expression of Cdx1 by the murine villin promoter did not alter intestinal epithelial or colonic rates of proliferation or its distribution along the crypt-villus axis.
Figure 5.
No change in crypt proliferation pattern due to ectopic expression of Cdx1. (A) Immunohistochemistry with anti-BrdU antibody shows that proliferating cells remain restricted to the crypts in both 6-month-old wild-type and transgenic mice small intestine and colon. (B) Quantification of epithelial proliferation based on BrdU incorporation in the ileum and colon. BrdU-positive cells per crypt were counted in 15 well-shaped crypts from three 6-month-old mice for each founder. Matched wild-type littermates were used as controls. Data for the combined analysis from all three transgenic lines, from the ileum and colon (n = 9) are presented. Dark gray bar, wild-type mice; white bar, Villin-Cdx1 transgenic mice.
Ectopic Cdx1 Expression in the Surface Epithelium of the Colon Did Not Cause or Accelerate Tumor Formation
One important question these transgenic mice are designed to address was whether the Cdx1 transcription factor had oncogenic properties in intestinal epithelium. Gene products classically thought of as oncogenes typically induce tumors when ectopically expressed or overexpressed [37]. However, despite examining more than 150 mice from each of the three lines, including ages 1 month to 24 months, no intestinal tumors or polyps were observed in the transgenic mice (data not shown). It therefore seems unlikely that Cdx1 is an intestinal oncogene in the classic sense.
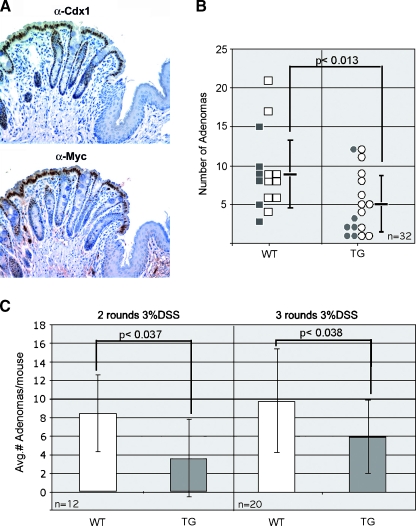
To further test for oncogenic potential of Cdx1, we investigated whether the ectopic Cdx1 expression would increase rates of colonic neoplasia in a mouse model for inducible colon cancer [38,39]. We treated mice with AOM (10 mg/ml) and either two rounds or three rounds of 3% DSS in their drinking water. As this treatment generally produces tumors in the rectum, we confirmed expression of our transgene to the squamo-columnar border (Figure 6A). With these doses of DSS, we obtained neoplastic polyps in all the mice included in the study (Table 1). Polyps were located in the distal colon, and in general, the nontransgenic mice appeared to have bulkier disease. None of the control or transgenic mice treated with either AOM or DSS alone developed histologically confirmed adenomas. Only the combination of these agents elicited a true neoplastic response.
Figure 6.
Ectopic Cdx1 expression directed by the villin-Cdx1 transgene reduced the number of adenomas elicited by AOM/DSS treatment. (A) Significant transgene expression demonstrated in rectum to squamo-columnar junction. Immunohistochemical staining for Cdx1 (CPSP) and the myc-tagged transgene protein (anti-myc). (B) Combined analysis of both treatment groups. Black squares, wild-type littermates receiving two rounds of 3% DSS; white squares, wild-type littermates receiving three rounds of 3% DSS; black circles, villin-Cdx1 transgenic mice receiving two rounds of 3% DSS; white circles, villin-Cdx1 transgenic mice receiving three rounds of 3% DSS. Statistical significance was determined by Student's t test using a two-tailed distribution. (C) Subgroup analysis of treatment groups using Student's t test using a single-tailed distribution. TG, transgenic; WT, wild-type.
Table 1.
Polyp Formation in Transgenic and Control Mice with AOM and DSS Treatment.
| Villin-Cdx1 | Nontransgenic Littermate | |
| AOM and two rounds of 3% DSS | 20 adenomas and 1 carcinoma in six mice (100% mice with polyps) | 50 adenomas in six mice (100% mice with polyps) |
| AOM and two rounds of water | 0 adenomas | 0 adenomas |
| Vehicle and two rounds of 3% DSS | 0 adenomas | 0 adenomas |
| AOM and three rounds of 3% DSS | 64 Adenomas in 11 mice (100% mice with polyps) | 88 adenomas in nine mice (100% mice with polyps) |
| AOM and three rounds of water | 0 adenomas | 0 adenomas |
| Vehicle and three rounds of 3% DSS | 0 adenomas | 0 adenomas |
Six-week-old villin-Cdx1 mice and nontransgenic littermate controls were given intraperitoneal injections of AOM (10 mg/kg) or vehicle. One week later, they received 3% DSS in their drinking water for 7 days. Mice received one or two additional rounds of 3% DSS for 7 days. Fourteen weeks after the initial AOM injection, mice were sacrificed; the number of colonic polyps was scored and adenomas were confirmed histologically.
Villin-Cdx1 transgenic mice had about half as many neoplastic polyps as their similarly treated nontransgenic littermates (5 vs 9.2; P < .013) (Figure 6B). Mice treated with two rounds of 3% DSS developed 3.5 adenomas/mouse compared to their nontransgenic littermates who develop 8.3 neoplastic polyps on average (P < .037) (Figure 6C). Treatment of the mice with three rounds of DSS led to an increase in the average number of polyps in all mice. However, the villin-Cdx1 transgenic mice still had fewer adenomas than controls (5.8 polyps/mouse vs 9.8 polyps/mouse; P < .038).
Other tumor parameters were compared in this study, including polyp diameter, degree of dysplasia, and expression of Cdx1 and Cdx2. A single carcinoma in situ was identified among the more than 200 polyps examined, and it developed in a villin-Cdx1 transgenic mouse. The remaining polyps were all classified as adenomas, with either low-grade or high-grade dysplasia. Despite the carcinoma, there was no significant difference between the groups in degree of polyp dysplasia (data not shown). The diameter of the adenomatous polyps was quite variable, ranging from 0.30 to 7.9 mm in the villin-Cdx1 mice and from 0.34 to 8.2 mm in the control mice. Despite the great variability in polyp sizes, there was a trend for smaller polyps in the transgenic mice in both treatment groups. However, statistical significance was only achieved in the group receiving the three rounds of DSS treatment (1.6 vs 2.1 mm; P = .019) (data not shown).
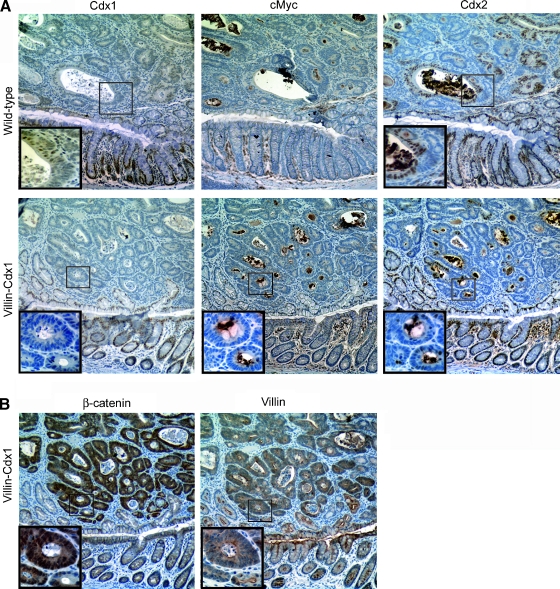
Immunohistochemical levels of Cdx1 and Cdx2 were significantly diminished in all polyps tested. Nuclear Cdx1 staining was infrequently detected in the 10 transgenic and 10 nontransgenic polyps studied (Figure 7A). Patchy Cdx2 expression persisted in many of the polyps, but overall Cdx2 levels were reduced compared to adjacent normal colon. Of interest, in the transgenic mice, the Myc-tag antibody failed to detect transgenic Cdx1 protein in the polyps, indicating a loss of villin-Cdx1 transgene expression.
Figure 7.
Expression of the transgene villin-Cdx1 transgene was lost in the adenomas elicited by AOM/DSS treatment. (A) Sections were obtained from the AOM/DSS polyps and were immunohistochemically stained for Cdx1, the Myc-tagged Cdx1, or Cdx2 proteins. A region of normal epithelium was included for comparison. (B) Staining for β-catenin and villin expression in the polyps from transgenic mice. Insets: higher magnification photo of neoplastic cells.
In conjunction with the absence of transgene expression, we noted elevated levels of nuclear β-catenin staining, and retention of villin protein expression, although the villin protein does not localize to the apical surface in adenomas as efficiently as it does in normal epithelial cells (Figure 7B). Thus the loss of villin-Cdx1 expression is not due to a general loss of villin gene expression or that the neoplastic polyp cells are in an undifferentiated state. Lastly, the distribution of β-catenin protein in the normal intestine appeared unaltered by the expression of the transgene. Together, these findings suggest that the villin-Cdx1 transgene might limit tumorigenesis at an early stage, possibly in the aberrant crypt foci, by inhibiting proliferation or neoplastic progression. Dysregulated Wnt/β-catenin signaling occurs frequently in aberrant crypt foci and could be the target of the transgene effect [40,41]. Determining the molecular mechanism for this interesting observation is a focus for future studies.
We conclude that ectopic Cdx1 expression in the surface epithelium of the colon does not increase cell proliferation nor induce tumor formation or accelerate tumor growth. It does not behave as an oncogene in the normal intestinal epithelium. Instead, it is able to reduce polyp formation in a mouse model for colitis-associated cancer and may therefore act to suppress colon carcinogenesis.
Discussion
Cdx1 and Cdx2 Are Caudal Homologues with a Conserved Structure and Overlapping Function
The Caudal-related homeodomain transcription factors Cdx1 and Cdx2 are highly conserved. As Caudal homologues, they are highly conserved structurally, with a high degree of homology throughout, and they share a 90% amino acid identity in the all-important DNA binding domain. This degree of structural conservation would suggest that these homologues might share a number of important transcriptional targets and thereby have shared functions. Indeed, this has been borne out in at least one study, where it was established that Cdx1 and Cdx2 have compensatory roles in anteroposterior patterning and posterior axis elongation [42]. Despite this shared structure and function, Cdx1 and Cdx2 have long been thought of as having opposing effects on proliferation and gene expression in the intestinal epithelium. This is perhaps best evidenced by the fact that Cdx1 has been called an oncogene by some investigators, whereas Cdx2 has long been thought of as a tumor suppressor [12,14,15,20,27,31,43].
We have been interested in the role played by the Cdx factors in normal intestinal cell proliferation and colon tumorigenesis. We found that both Cdx1 and Cdx2 could limit colon cancer cell proliferation by inhibiting Wnt/β-catenin transcriptional activity [22,34,35]. Our findings, therefore, did not agree with the accepted description of Cdx1. To directly test whether Cdx1 was an intestinal oncogene, we carried out the present study.
Ectopic Expression of Cdx1 Had No Effect on Intestinal Epithelial Architecture, Differentiation, or Proliferation
We generated three transgenic lines with robust transgene expression. Of importance for the interpretation of these results, Cdx1 was both overexpressed and ectopically expressed. Thus, if Cdx1 had oncogenic properties in the intestinal epithelium, we would expect to see intestinal polyps and cancers develop, as was seen when K-ras was expressed in the intestine using a villin promoter [44]. However, no spontaneous tumors or cancers were observed in our transgenic mice, even in mice up to 24 months, nor was there any effect on proliferation in the crypt compartment. Together this is strong evidence that Cdx1 does not act as classic oncogene in intestinal epithelial cells.
Villin-Cdx1 Mice Had Reduced Adenoma Formation in a Model for Colitis-Associated Cancer
As an additional test for Cdx1 oncogenic potential, we looked for synergy between our transgene and a mouse model for colon cancer. If Cdx1 possesses oncogenic properties in the colon, we would expect it to enhance tumor production or growth in a mouse model for colon carcinogenesis. We chose to use a combination of AOM with DSS because it was an established model for inducible colon cancer in CD-1 mice [45]. CD-1 is an outbred strain of mice, which may account for the large variability in polyp numbers that we observed in our study. Nonetheless, our findings were very significant. The transgenic mice had half as many rectal adenomas as their wild-type littermates with this treatment. This difference was highly significant and confirmed by statistical testing (P < .013). In addition, the polyps that developed in the villin-Cdx1 mice tended to be physically smaller than those in their nontransgenic littermates. These observations are exactly the opposite of what would be expected from Cdx1 if it behaved as an oncogene in intestinal epithelium. In summary, whereas this single study may be insufficient to designate Cdx1 as a tumor suppressor in the intestinal epithelium, it is clear from this work that Cdx1 does not behave as an oncogene in the intestinal epithelium.
Of great importance was the finding that Cdx1 expression, including that of the Cdx1 transgene, was lost in the neoplastic polyps. This confirms earlier observations that Cdx1 expression is lost in most human and murine polyps and cancers [22,23,25,30]. This finding was somewhat unexpected, as the transgene was expressed under the control of a villin promoter, a gene whose expression is often maintained colon cancers [46]. The endogenous villin gene remained active and was expressed in all polyps, thus the loss of the villin-Cdx1 transgene expression was specific to the transgene and not due to a general loss of villin expression or a poorly differentiated state of the neoplasm. It suggests that there must be a powerful advantage for neoplastic intestinal cells to lose their Cdx1 expression.
We do not yet know the mechanism by which the expression of the transgene is silenced, whether by loss of heterozygosity, epigenetic effects, or another mechanism. Whereas loss of endogenous Cdx1 gene expression in most colon cancers appears to be from epigenetic silencing of the Cdx1 promoter [23–26], there are no reports in the literature describing epigenetic regulation or loss of heterozygosity of the villin gene. It is therefore not clear which mechanism is involved here. Given that the villin-Cdx1 transgene consists of more than 12 Kb of mouse villin promoter and intronic sequence, inserted randomly in the mouse genome, differentiating it from the endogenous villin gene and determining the mechanism by which the transgene is silenced would be a difficult task and is beyond the focus of this investigation into whether Cdx1 behaves in vivo as a classic oncogene. It remains, however, an interest for future investigations. In conclusion, these observations together strongly argue against an oncogenic role for Cdx1 in intestinal carcinogenesis.
Cdx1 Expression Can Modulate Cdx2 Expression in Murine Intestinal Epithelium
One unexpected finding was the reduction in Cdx2 expression noted in the intestinal villi and colonic surface epithelium. Now, we do not know if this is a direct or indirect effect by Cdx1 on the Cdx2 gene. Although there have been reports describing a regulatory effects of Cdx1 or Cdx2 on Cdx1 gene expression [47], the reverse has not been described. In transient transfection studies with human colon cancer DLD1 cells, we have noted a reduction in CDX2 mRNA levels when Cdx1 is expressed (data not shown). However, in studies with two different murine Cdx2 promoter-luciferase reporter constructs, including one with 8.8 kb of promoter sequence, Cdx1 expression had no effect on Cdx2 promoter activity. Thus, this observation will require additional experimentation to establish that Cdx1 regulates Cdx2 expression. However, this remains an important observation, because intestinal epithelial differentiation and gene expression patterns were unaltered with the loss of Cdx2 protein. It suggests that the ectopically expressed Cdx1 can compensate for Cdx2 and support normal intestinal cell differentiation and gene expression. This observation therefore further undermines the hypothesis that Cdx1 and Cdx2 have opposing effects on the intestinal epithelial proliferation and differentiation and supports our hypothesis that these factors share overlapping targets and functions.
One disappointment was that we did not see a reduction in intestinal cell proliferation with Cdx1 overexpression. There are several potential explanations for this. One possibility is that we did not get enough Cdx1 overexpression in the proliferative crypt compartment. Our immunohistochemical staining seems to support this possibility (Figure 2), although other investigators using the same villin promoter construct report significant transgene expression in crypt epithelial cells [29]. Another possible explanation could be that the compensatory reduction in Cdx2 expression might have removed an important anti-proliferative stimulus and, therefore, the Cdx1 transgene, whereas substituting for the lost Cdx2 could not provide any additional anti-proliferative effects. One final possibility is that, despite the in vitro evidence to the contrary, Cdx1 has no antiproliferative effect on normal intestinal epithelial cells. We favor the first two explanations and are considering future studies to address these concerns directly.
Tissue and Genetic Context of Expression Likely Explains the Opposing Observations Regarding Cdx1 and Cdx2 in Carcinogenesis
Cdx1 and Cdx2 are potent transcription factors with a diverse array of target genes and reported effects on proliferation and apoptosis [7,8]. We believe the context in which the expression occurs defines the cell's response. In the context of normal intestinal epithelium, where Cdx1 and Cdx2 expression occur naturally, these factors serve to limit proliferation and promote differentiation and predominantly act to inhibit tumorigenesis. However, in other contexts, including other tissue contexts, the response to Cdx1 expression can be quite different. When Cdx1 or Cdx2 are expressed in some nonintestinal tissues, such as gastric epithelium or bone marrow stem cells, Cdx expression may support carcinogenesis [17,48]. This may also be true in intestinal cells lacking normal retinoblastoma function, either due to experimental manipulations [49] or to natural occurrence, such as colon cancer cells in which the retinoblastoma protein has been inactivated by hyperphosphorylation. In summary, our observations support the conclusion that the homeodomain transcription factor Cdx1 is not an oncogene and does not have oncogenic properties in the context of normal intestinal epithelium. Furthermore, overexpressed Cdx1 does not oppose Cdx2 actions in intestinal epithelium but can instead substitute for Cdx2 when Cdx2 expression is lost.
Acknowledgments
We thank Deborah Gumucio for the Villin promoter and Andrew Midzak and Xianglin Wang for technical assistance.
Abbreviations
- AOM
azoxymethane
- BrdU
bromodeoxyuridine
- CDX1, CDX2
human genes
- Cdx1, Cdx2
mouse genes or species not specified
- DSS
dextran sodium sulfate
- RT-PCR
reverse transcription-polymerase chain reaction
- SI
sucrase isomaltase
- TCF
T cell factor
Footnotes
This work was supported by the National Institutes of Health (NIH)/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants DK047437 and DK068366 (to J. Lynch) and DK49210 and DK59539 (to D. G. Silberg), by the Pilot Project award from the Center for Molecular Studies in Digestive and Liver Disease (to D. Silberg and E. R. Suh), and by the Morphology, Cell Culture, Transgenic Animal, and Molecular Biology Core Facilities of the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Disease at the University of Pennsylvania (P30-DK50306). We also thank the University of Pennsylvania Transgenic and Chimeric Mouse Facility for their expertise.
References
- 1.Crosnier C, Stamataki D, Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet. 2006;7:349–359. doi: 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- 2.van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T, et al. Wnt signalling induces maturation of paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 3.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 5.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 6.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 7.Guo RJ, Suh ER, Lynch JP. The role of Cdx proteins in intestinal development and cancer. Cancer Biol Ther. 2004;3:593–601. doi: 10.4161/cbt.3.7.913. [DOI] [PubMed] [Google Scholar]
- 8.Beck F. The role of Cdx genes in the mammalian gut. Gut. 2004;53:1394–1396. doi: 10.1136/gut.2003.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudreau F, Rings EH, van Wering HM, Kim RK, Swain GP, Krasinski SD, Moffett J, Grand RJ, Suh ER, Traber PG. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J Biol Chem. 2002;277:31909–31917. doi: 10.1074/jbc.M204622200. [DOI] [PubMed] [Google Scholar]
- 10.Malakooti J, Dahdal RY, Dudeja PK, Layden TJ, Ramaswamy K. The human Na(+)/H(+) exchanger NHE2 gene: genomic organization and promoter characterization. Am J Physiol Gastrointest Liver Physiol. 2001;280:G763–G773. doi: 10.1152/ajpgi.2001.280.4.G763. [DOI] [PubMed] [Google Scholar]
- 11.Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. Expression of the gut-enriched Kruppel-like factor (Kruppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto H, Bai YQ, Yuasa Y. Homeodomain protein CDX2 regulates goblet-specific MUC2 gene expression. Biochem Biophys Res Commun. 2003;300:813–818. doi: 10.1016/s0006-291x(02)02935-2. [DOI] [PubMed] [Google Scholar]
- 13.Troelsen JT, Mitchelmore C, Spodsberg N, Jensen AM, Noren O, Sjostrom H. Regulation of lactase-phlorizin hydrolase gene expression by the caudal-related homeodomain protein Cdx-2. Biochem J. 1997;322:833–838. doi: 10.1042/bj3220833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aoki K, Tamai Y, Horiike S, Oshima M, Taketo MM. Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Delta716 Cdx2+/- compound mutant mice. Nat Genet. 2003;35:323–330. doi: 10.1038/ng1265. [DOI] [PubMed] [Google Scholar]
- 15.Bonhomme C, Duluc I, Martin E, Chawengsaksophak K, Chenard MP, Kedinger M, Beck F, Freund JN, Domon-Dell C. The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut. 2003;52:1465–1471. doi: 10.1136/gut.52.10.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witek ME, Nielsen K, Walters R, Hyslop T, Palazzo J, Schulz S, Waldman SA. The putative tumor suppressor Cdx2 is overexpressed by human colorectal adenocarcinomas. Clin Cancer Res. 2005;11:8549–8556. doi: 10.1158/1078-0432.CCR-05-1624. [DOI] [PubMed] [Google Scholar]
- 17.Rawat VP, Cusan M, Deshpande A, Hiddemann W, Quintanilla-Martinez L, Humphries RK, Bohlander SK, Feuring-Buske M, Buske C. Ectopic expression of the homeobox gene Cdx2 is the transforming event in a mouse model of t(12;13)(p13;q12) acute myeloid leukemia. Proc Natl Acad Sci USA. 2004;101:817–822. doi: 10.1073/pnas.0305555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang LH, Chen F, Ying C, Chun SY, Knock SA, Appelman HD, Dang DT. CDX2 has tumorigenic potential in the human colon cancer cell lines LOVO and SW48. Oncogene. 2006;25:2264–2272. doi: 10.1038/sj.onc.1209247. [DOI] [PubMed] [Google Scholar]
- 19.Lorentz O, Duluc I, Arcangelis AD, Simon-Assmann P, Kedinger M, Freund JN. Key role of the Cdx2 homeobox gene in extracellular matrix-mediated intestinal cell differentiation. J Cell Biol. 1997;139:1553–1565. doi: 10.1083/jcb.139.6.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soubeyran P, Haglund K, Garcia S, Barth BU, Iovanna J, Dikic I. Homeobox gene Cdx1 regulates Ras, Rho and PI3 kinase pathways leading to transformation and tumorigenesis of intestinal epithelial cells. Oncogene. 2001;20:4180–4187. doi: 10.1038/sj.onc.1204551. [DOI] [PubMed] [Google Scholar]
- 21.Lickert H, Domon C, Huls G, Wehrle C, Duluc I, Clevers H, Meyer BI, Freund JN, Kemler R. Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development. 2000;127:3805–3813. doi: 10.1242/dev.127.17.3805. [DOI] [PubMed] [Google Scholar]
- 22.Guo RJ, Huang E, Ezaki T, Patel N, Sinclair K, Wu J, Klein PS, Suh E, Lynch JP. Cdx1 inhibits human colon cancer cell proliferation by reducing β-catenin/TCF transcriptional activity. J Biol Chem. 2004;279:36865–36875. doi: 10.1074/jbc.M405213200. [DOI] [PubMed] [Google Scholar]
- 23.Suh ER, Ha CS, Rankin EB, Toyota M, Traber PG. DNA methylation down-regulates CDX1 gene expression in colorectal cancer cell lines. J Biol Chem. 2002;227:35795–35800. doi: 10.1074/jbc.M205567200. [DOI] [PubMed] [Google Scholar]
- 24.Xu XL, Yu J, Zhang HY, Sun MH, Gu J, Du X, Shi DR, Wang P, Yang ZH, Zhu JD. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J Gastroenterol. 2004;10:3441–3454. doi: 10.3748/wjg.v10.i23.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong NA, Britton MP, Choi GS, Stanton TK, Bicknell DC, Wilding JL, Bodmer WF. Loss of CDX1 expression in colorectal carcinoma: promoter methylation, mutation, and loss of heterozygosity analyses of 37 cell lines. Proc Natl Acad Sci USA. 2004;101:574–579. doi: 10.1073/pnas.0307190101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilozzi E, Onelli MR, Ziparo V, Mercantini P, Ruco L. CDX1 expression is reduced in colorectal carcinoma and is associated with promoter hypermethylation. J Pathol. 2004;204:289–295. doi: 10.1002/path.1641. [DOI] [PubMed] [Google Scholar]
- 27.Domon-Dell C, Schneider A, Moucadel V, Guerin E, Guenot D, Aguillon S, Duluc I, Martin E, Iovanna J, Launay JF, et al. Cdx1 homeobox gene during human colon cancer progression. Oncogene. 2003;22:5969–5977. doi: 10.1038/sj.onc.1206756. [DOI] [PubMed] [Google Scholar]
- 28.Taylor JK, Boll W, Levy T, Suh E, Siang S, Mantei N, Traber PG. Comparison of intestinal phospholipase A/lysophospholipase and sucrase-isomaltase genes suggest a common structure for enterocyte-specific promoters. DNA Cell Biol. 1997;16:1419–1428. doi: 10.1089/dna.1997.16.1419. [DOI] [PubMed] [Google Scholar]
- 29.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 30.Silberg DG, Furth EE, Taylor JK, Schuck T, Chiou T, Traber PG. CDX1 protein expression in normal, metaplastic, and neoplastic human alimentary tract epithelium. Gastroenterology. 1997;113:478–486. doi: 10.1053/gast.1997.v113.pm9247467. [DOI] [PubMed] [Google Scholar]
- 31.Alkhoury F, Malo MS, Mozumder M, Mostafa G, Hodin RA. Differential regulation of intestinal alkaline phosphatase gene expression by Cdx1 and Cdx2. Am J Physiol Gastrointest Liver Physiol. 2005;289:G285–G290. doi: 10.1152/ajpgi.00037.2005. [DOI] [PubMed] [Google Scholar]
- 32.Rings EH, Boudreau F, Taylor JK, Moffett J, Suh ER, Traber PG. Phosphorylation of the serine 60 residue within the cdx2 activation domain mediates its transactivation capacity. Gastroenterology. 2001;121:1437–1450. doi: 10.1053/gast.2001.29618. [DOI] [PubMed] [Google Scholar]
- 33.Boulanger J, Vezina A, Mongrain S, Boudreau F, Perreault N, Auclair BA, Laine J, Asselin C, Rivard N. Cdk2-dependent phosphorylation of homeobox transcription factor CDX2 regulates its nuclear translocation and proteasome-mediated degradation in human intestinal epithelial cells. J Biol Chem. 2005;280:18095–18107. doi: 10.1074/jbc.M502184200. [DOI] [PubMed] [Google Scholar]
- 34.Lynch J, Suh ER, Silberg DG, Rulyak S, Blanchard N, Traber PG. The caudal-related homeodomain protein Cdx1 inhibits proliferation of intestinal epithelial cells by down-regulation of d-type cyclins. J Biol Chem. 2000;275:4499–4506. doi: 10.1074/jbc.275.6.4499. [DOI] [PubMed] [Google Scholar]
- 35.Lynch J, Keller M, Guo R, Yang D, Traber PG. Cdx1 inhibits the proliferation of human colon cancer cells by reducing cyclin D1 gene expression. Oncogene. 2003;22:6395–6407. doi: 10.1038/sj.onc.1206770. [DOI] [PubMed] [Google Scholar]
- 36.Keller MS, Ezaki T, Guo RJ, Lynch JP. Cdx1 or Cdx2 expression activates E-cadherin-mediated cell-cell adhesion and compaction in human COLO 205 cells. Am J Physiol Gastrointest Liver Physiol. 2004;287:G104–G114. doi: 10.1152/ajpgi.00484.2003. [DOI] [PubMed] [Google Scholar]
- 37.Lynch JP, Rustgi AK. Mechanisms of GI malignancies. In: Johnson L, editor. Physiology of the Gastrointestinal Tract, Vol. 1. 4th ed. San Diego, CA: Elsevier; 2004. [Google Scholar]
- 38.Tanaka T, Kohno H, Suzuki R, Yamada Y, Sugie S, Mori H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003;94:965–973. doi: 10.1111/j.1349-7006.2003.tb01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat Protoc. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Dong Chen W, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi M, Mutoh M, Kawamori T, Sugimura T, Wakabayashi K. Altered expression of beta-catenin, inducible nitric oxide synthase and cyclooxygenase-2 in azoxymethane-induced rat colon carcinogenesis. Carcinogenesis. 2000;21:1319–1327. [PubMed] [Google Scholar]
- 42.van den Akker E, Forlani S, Chawengsaksophak K, de Graaff W, Beck F, Meyer BI, Deschamps J. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 2002;129:2181–2193. doi: 10.1242/dev.129.9.2181. [DOI] [PubMed] [Google Scholar]
- 43.Moucadel V, Totaro MS, Dell CD, Soubeyran P, Dagorn JC, Freund JN, Iovanna JL. The homeobox gene Cdx1 belongs to the p53-p21(WAF)-Bcl-2 network in intestinal epithelial cells. Biochem Biophys Res Commun. 2002;297:607–615. doi: 10.1016/s0006-291x(02)02250-7. [DOI] [PubMed] [Google Scholar]
- 44.Janssen KP, el-Marjou F, Pinto D, Sastre X, Rouillard D, Fouquet C, Soussi T, Louvard D, Robine S. Targeted expression of oncogenic K-ras in intestinal epithelium causes spontaneous tumorigenesis in mice. Gastroenterology. 2002;123:492–504. doi: 10.1053/gast.2002.34786. [DOI] [PubMed] [Google Scholar]
- 45.Bissahoyo A, Pearsall RS, Hanlon K, Amann V, Hicks D, Godfrey VL, Threadgill DW. Azoxymethane is a genetic background-dependent colorectal tumor initiator and promoter in mice: effects of dose, route, and diet. Toxicol Sci. 2005;88:340–345. doi: 10.1093/toxsci/kfi313. [DOI] [PubMed] [Google Scholar]
- 46.West AB, Isaac CA, Carboni JM, Morrow JS, Mooseker MS, Barwick KW. Localization of villin, a cytoskeletal protein specific to microvilli, in human ileum and colon and in colonic neoplasms. Gastroenterology. 1988;94:343–352. doi: 10.1016/0016-5085(88)90421-0. [DOI] [PubMed] [Google Scholar]
- 47.Domon-Dell C, Freund JN. Stimulation of Cdx1 by oncogenic betacatenin/TCF4 in colon cancer cells; opposite effect of the CDX2 homeoprotein. FEBS Lett. 2002;518:83–87. doi: 10.1016/s0014-5793(02)02650-9. [DOI] [PubMed] [Google Scholar]
- 48.Mutoh H, Sakurai S, Satoh K, Tamada K, Kita H, Osawa H, Tomiyama T, Sato Y, Yamamoto H, Isoda N, et al. Development of gastric carcinoma from intestinal metaplasia in Cdx2-transgenic mice. Cancer Res. 2004;64:7740–7747. doi: 10.1158/0008-5472.CAN-04-1617. [DOI] [PubMed] [Google Scholar]
- 49.Haigis K, Sage J, Glickman J, Shafer S, Jacks T. The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J Biol Chem. 2006;281:638–647. doi: 10.1074/jbc.M509053200. [DOI] [PubMed] [Google Scholar]