Abstract
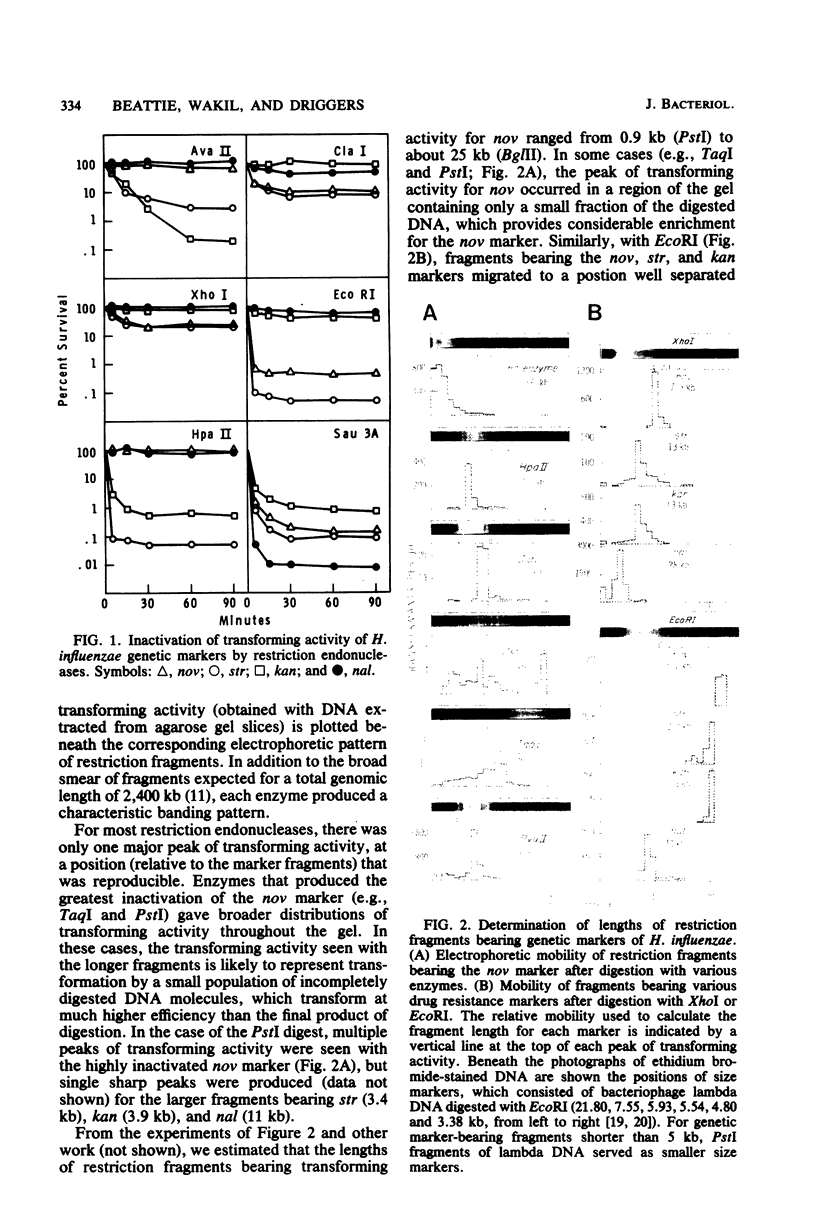
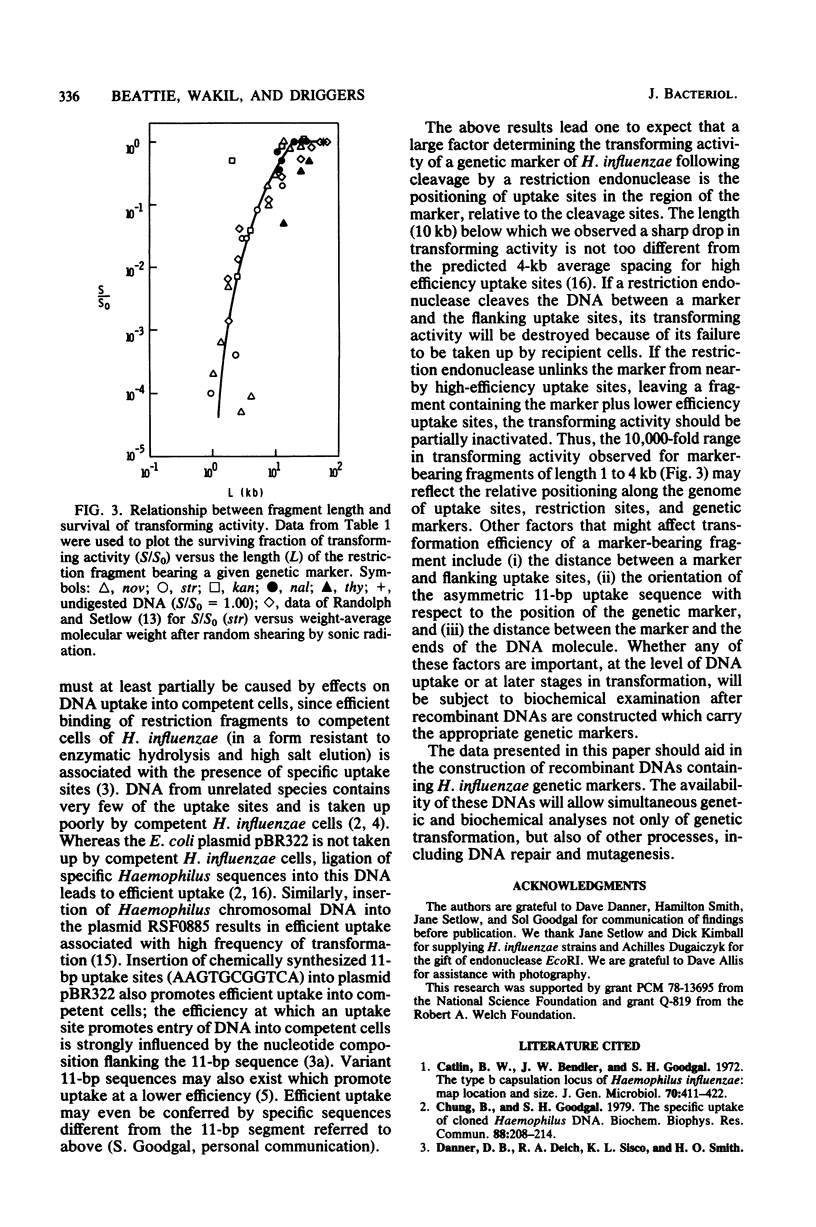
Cleavage of DNA from Haemophilus influenzae with restriction endonucleases caused inactivation of transforming ability to an extent that depended on the genetic marker and the enzyme. The rate of inactivation, but not the final level of survival, depended on the concentration of enzyme in the restriction digest. In general, the greatest extent of inactivation of transforming activity was obtained with endonucleases that are known to produce the shortest fragments. We electrophoresed restriction digests of H. influenzae DNA in agarose gels and assayed transforming activity of DNA extracted from gel slices. In this way, we determined the lengths of restriction fragments that contain genetic markers of H. influenzae. For the marker that we studied most thoroughly (nov), the shortest restriction fragment that possessed detectable transforming activity was a 0.9-kilobase pair fragment produced by endonuclease R . PstI. The shortest marker-bearing restriction fragment that retained substantial transforming activity (50% of value for undigested DNA) was a 2.1-kilobase pair EcoRI fragment bearing the kan marker. Among marker-bearing restriction fragments 1 to 4 kilobase pairs in length, survival of transforming activity varied 10,000-fold. We relate these observations to the recent findings by Sisco and Smith (Proc. Natl. Acad. Sci. U.S.A. 76:972-976, 1979) that efficient entry of DNA into competent H. influenzae cells appears to require the presence of a recognition sequence that is scattered throughout the Haemophilus genome in many more copies than in unrelated genomes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Catlin B. W., Bendler J. W., 3rd, Goodgal S. H. The type b capsulation locus of Haemophilus influenzae: map location and size. J Gen Microbiol. 1972 May;70(3):411–422. doi: 10.1099/00221287-70-3-411. [DOI] [PubMed] [Google Scholar]
- Chung B. C., Goodgal S. H. The specific uptake of cloned Haemophilus DNA. Biochem Biophys Res Commun. 1979 May 14;88(1):208–214. doi: 10.1016/0006-291x(79)91717-0. [DOI] [PubMed] [Google Scholar]
- Danner D. B., Smith H. O., Narang S. A. Construction of DNA recognition sites active in Haemophilus transformation. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2393–2397. doi: 10.1073/pnas.79.7.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deich R. A., Smith H. O. Mechanism of homospecific DNA uptake in Haemophilus influenzae transformation. Mol Gen Genet. 1980 Feb;177(3):369–374. doi: 10.1007/BF00271475. [DOI] [PubMed] [Google Scholar]
- Goodgal S. H., Gromkova R. Separation of specific segments of transforming DNA after treatment with endodeoxyribonuclease. Proc Natl Acad Sci U S A. 1973 Feb;70(2):503–506. doi: 10.1073/pnas.70.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriott R. M., Meyer E. M., Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970 Feb;101(2):517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriott R. M., Meyer E. Y., Vogt M., Modan M. Defined medium for growth of Haemophilus influenzae. J Bacteriol. 1970 Feb;101(2):513–516. doi: 10.1128/jb.101.2.513-516.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball R. F. Reversions of two proline-requiring auxotrophs of Haemophilus influenzae by n-methyl-n'-nitro-n-nitrosoguanidine and hydrazine. Mutat Res. 1976 Jul;36(1):29–38. doi: 10.1016/0027-5107(76)90018-x. [DOI] [PubMed] [Google Scholar]
- MACHATTIE L. A., BERNE K. I., THOMAS C. A., Jr ELECTRON MICROSCOPY OF DNA FROM HEMOPHILUS INFLUENZAE. J Mol Biol. 1965 Mar;11:648–649. doi: 10.1016/s0022-2836(65)80019-5. [DOI] [PubMed] [Google Scholar]
- MASTER R. W. POSSIBLE SYNTHESIS OF POLYRIBONUCLEOTIDES OF KNOWN BASE-TRIPLET SEQUENCES. Nature. 1965 Apr 3;206:93–93. doi: 10.1038/206093b0. [DOI] [PubMed] [Google Scholar]
- Randolph M. L., Setlow J. K. Mechanism of inactivation of Haemophilus influenzae transforming deoxyribonucleic acid by sonic radiation. J Bacteriol. 1972 Jul;111(1):186–191. doi: 10.1128/jb.111.1.186-191.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow J. K., Brown D. C., Boling M. E., Mattingly A., Gordon M. P. Repair of deoxyribonucleic acid in Haemophilus influenzae. I. X-ray sensitivity of ultraviolet-sensitive mutants and their behavior as hosts to ultraviolet-irradiated bacteriophage and transforming deoxyribonucleic acid. J Bacteriol. 1968 Feb;95(2):546–558. doi: 10.1128/jb.95.2.546-558.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow J. K., Notani N. K., McCarthy D., Clayton N. L. Transformation of Haemophilus influenzae by plasmid RSF0885 containing a cloned segment of chromosomal deoxyribonucleic acid. J Bacteriol. 1981 Dec;148(3):804–811. doi: 10.1128/jb.148.3.804-811.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisco K. L., Smith H. O. Sequence-specific DNA uptake in Haemophilus transformation. Proc Natl Acad Sci U S A. 1979 Feb;76(2):972–976. doi: 10.1073/pnas.76.2.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. I., Blattner F. R., Davies J. The isolation and partial characterization of a new restriction endonuclease from Providencia stuartii. Nucleic Acids Res. 1976 Feb;3(2):343–353. doi: 10.1093/nar/3.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O. Recovery of DNA from gels. Methods Enzymol. 1980;65(1):371–380. doi: 10.1016/s0076-6879(80)65048-4. [DOI] [PubMed] [Google Scholar]
- Szybalski E. H., Szybalski W. A comprehensive molecular map of bacteriophage lambda. Gene. 1979 Nov;7(3-4):217–270. doi: 10.1016/0378-1119(79)90047-7. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]