Abstract
The RNA subunits of RNase Ps of Archaea and eukaryotes have been thought to depend fundamentally on protein for activity, unlike those of Bacteria that are capable of efficient catalysis in the absence of protein. Although the eukaryotic RNase P RNAs are quite different than those of Bacteria in both sequence and structure, the archaeal RNAs generally contain the sequences and structures of the bacterial, phylogenetically conserved catalytic core. A spectrum of archaeal RNase P RNAs were therefore tested for activity in a wide range of conditions. Many remain inactive in ionically extreme conditions, but catalytic activity could be detected from those of the methanobacteria, thermococci, and halobacteria. Chimeric holoenzymes, reconstituted from the Methanobacterium RNase P RNA and the Bacillus subtilis RNase P protein subunits, were functional at low ionic strength. The properties of the archaeal RNase P RNAs (high ionic-strength requirement, low affinity for substrate, and catalytic reconstitution by bacterial RNase P protein) are similar to synthetic RNase P RNAs that contain all of the catalytic core of the bacterial RNA but lack phylogenetically variable, stabilizing elements.
RNase P is an endoribonuclease best known for its role in tRNA biosynthesis, in which it is the enzyme responsible for the removal of 5′ leader sequences from transfer RNA precursors (for reviews see refs. 1 and 2). In Bacteria, RNase P consists of two subunits: a large (≈140-kDa, 400-nt) RNA and a small (≈14-kDa, 120-amino acid) protein. Both RNA and protein are required in vivo and for optimal activity in vitro in reactions at low ionic strength (3, 4). The RNA is the catalytic subunit of the bacterial enzyme; at elevated ionic strength in vitro, it is by itself capable of processing pre-tRNAs catalytically; i.e., it is a ribozyme (5). The protein component of the bacterial enzyme alters substrate recognition by directly contacting the leader region of the pre-tRNA (6). The RNase P enzymes of Archaea (formerly archaebacteria) and Eukarya (the nuclear/cytoplasmic portion of the eukaryotic cell) also contain RNA subunits, but these RNAs have not been shown to be catalytically active. Although it seems likely that the catalytic function of the archaeal and eukaryal enzymes resides in the RNA, the expression of this activity apparently requires the presence of the protein subunits.
RNase P enzymes have been characterized from only two archaeal species: the thermoacidophilic crenarchaeote Sulfolobus acidocaldarius (7) and the extremely halophilic euryarchaeote Haloferax volcanii (8). The S. acidocaldarius RNase P is resistant to micrococcal nuclease treatment and contains a 315-nt RNA that persists after nuclease treatment (9). The enzyme is large (≈400 kDa apparent molecular mass) and has a low density in Cs2SO4 (1.27 g/cm3), similar to the densities of the eukaryotic nuclear enzyme and implying a high protein:RNA content. The H. volcanii RNase P, on the other hand, resembles the bacterial enzyme in density in Cs2SO4 (1.61 g/cm3; ref. 10) and nuclease sensitivity. The H. volcanii enzyme contains a 435-nt RNA (8). Neither proteins associated with the archaeal enzymes nor sequences encoding polypeptides with recognizable similarity to eukaryal, mitochondrial, or bacterial RNase P proteins have been identified in archaeal genomes. The RNase P RNAs from S. acidocaldarius and H. volcanii are similar in size and, to some extent, sequence to those of Bacteria, but these RNAs by themselves could not be shown to be catalytically active after deproteinization or when synthesized in vitro (8, 9). This finding and the observation in subsequent tests that the RNase P RNAs from Methanosarcina barkeri (11) and Methanococcus jannaschii (E.S.H., J.K.H., and J.W.B., unpublished results) also lack activity led to the conclusion that archaeal RNase P RNAs in general, like those of eukaryotes, are not by themselves catalytically active.
We recently developed a well defined secondary structure model of RNase P RNAs from Archaea by using comparative sequence analysis (Fig. 1; ref. 11 and J.K.H., E.S.H., and J.W.B., unpublished data). Nearly all of the conserved sequences and secondary structures in the catalytically active bacterial RNAs (12) are also present in the archaeal RNAs. The structural distinctions between the bacterial and archaeal RNAs have largely disappeared as our understanding of their structures has improved; thus, the dramatic difference in their ability to catalyze pre-tRNA 5′ processing was puzzling. A phylogenetic spectrum of archaeal RNase P RNAs were therefore tested for pre-tRNA processing activity in a wide variety of conditions, including ionic conditions outside the range usually considered. We show here that RNase P RNAs from the methanobacteria, thermococci, and extreme halophiles are, like their bacterial homologs, able to process pre-tRNAs in the absence of protein at very high ionic strength. The biochemical properties of the archaeal RNAs are similar to those of synthetic minimal bacterial RNase P RNAs (RNAs that contain only the phylogenetically conserved “core” elements of the bacterial RNAs; refs. 13 and 14), suggesting that the archaeal RNAs contain all of the elements required for substrate recognition and catalysis but are structurally defective in the absence of protein.
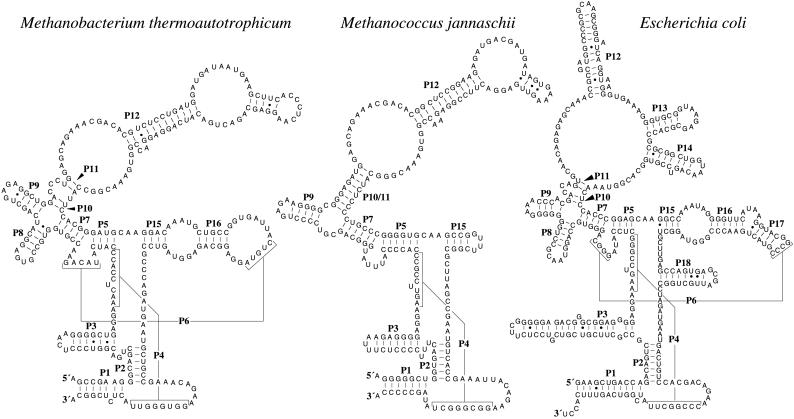
Figure 1.
Secondary structures of the M. thermoautotrophicum strain ΔH, M. jannaschii, and Escherichia coli RNase P RNAs (refs. 11 and 12, and J.K.H., E.S.H., and J.W.B., unpublished data). Helices are numbered P1–P18 as described (24). Helices P4 and P6 are shown with lines and brackets. The 5′ and 3′ ends of the native archaeal RNAs were determined by primer extension and nuclease S1 protection. Additional RNase P RNA sequences and structures are available on the Ribonuclease P database at www.mbio.ncsu.edu/RNaseP/home.html (25).
MATERIALS AND METHODS
RNase P Activity Assays.
RNase P RNA assays contained 1.5 nM 32P-labeled Bacillus subtilis or Methanobacterium thermoautotrophicum pre-tRNAAsp, 50 mM Tris (pH 8), 25–300 mM MgCl2, 0.1–5 M ammonium acetate, 0.1% SDS, and 0.05% Nonidet P-40. Incubations ranged from 2 to 16 h at 30–75°C (typically 45°C). Optimal pH was evaluated in activity assays that used Hepes in place of Tris. Reaction products were separated by electrophoresis on 8 or 12% polyacrylamide/7.5 M urea/TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) gels (SequaGel, National Diagnostics) and visualized by phosphorimagery (15).
For assays of synthetic RNAs, RNase P RNAs were synthesized from cloned genes by using T7 or T3 RNA polymerase, as described by the manufacturer (Promega). For assays of activity in total cellular nucleic acid, proteins were removed from archaeal cell lysates (prepared by using a French press and cells resuspended in 50 mM Tris⋅Cl, pH 8/25 mM MgCl2/0.1 M ammonium acetate/0.05% Nonidet P-40 as described in ref. 15) by repeated phenol extraction, followed by a phenol:chloroform extraction. Nucleic acids were precipitated with ethanol and resuspended in 10 mM Tris, pH 8/1 mM EDTA/0.1% SDS. Assays contained 10–40 μg/ml cellular nucleic acid.
Determination of Native 5′ and 3′ Ends.
Identification of the native 5′ and 3′ ends of the M. thermoautotrophicum ΔH RNase P RNA was performed as described (8). The location of the 5′ end was determined by primer extension by using oligonucleotide Mfo145R (CGTGTCGTTTCTGCTCC). The location of the 3′ end was determined by S1 nuclease mapping by using a probe DNA fragment that overlapped the 3′ end of the RNase P RNA, produced by PCR amplification from genomic DNA by using oligonucleotide primers ΔH138F (GATAATGAAGCTTCACCCTCAAG) and ΔH908RXba (GCTCTAGAGCTCCATCGCGCCTGCG). The downstream priming-site sequence was taken from the complete genome sequence (16).
RNase P RNA-Encoding Genes.
RNase P RNA-encoding genes were cloned after PCR amplification from genomic DNAs by using previously described methods (11). The oligonucleotide primers used were ΔH5′Bam (CGGGATCCACCGGGCAAGCCGAAGGGC) and ΔH3′Xba (GCTCTAGACCGGGCATGCCGAGAG) for obtaining genes from Methanobacterium spp., Pyro5′Xba (GCTCTAGATAGGCGAGGGGGCTGGGG) and Pyro3′Bam (CGGGATCCTAGGCGAGGGGGCTATAG) for obtaining genes from Pyrococcus and Thermococcus spp., Mja5′Xba (GCTCTAGAGGGTAAGGGGGCTGGTG) and Mja3′Bam (CGGGATCCGGTATGGGGGCTATAGC) for obtaining genes from Methanococcus spp., and Sac5′Bam (CGGGATCCTAGGGGAGCCTAACAGG) and Sac3′Xba (GCTCTAGAGGGGAGCCTAACAATAACC) for obtaining the gene from Metallosphaera sedula. Primer sequences are based on data from the complete genome sequences of M. thermoautotrophicum ΔH (16), Pyrococcus furiosus (Utah Genome Center; www.genome.utah.edu), and M. jannaschii (17), respectively. The Sac primers were designed based on the published gene sequence of S. acidocaldarius RNase P RNA (9).
RNase H Depletions.
Reactions (10 μl) containing 100 mM KCl, 10 mM MgCl2, 20 mM Hepes⋅KOH (pH 7.4), 1 mM DTT, 0.2 μg of RNase P RNA, and 2 μg of oligonucleotide Mfo145R or Eco145R (GCGGTTTGCTCTCTGTTGC) were heated to 80°C for 3 min and cooled to 30°C over 30 min; RNase H (2 units; Life Technologies, Grand Island, NY) was added and incubated for 1 h at 30°C. The remaining RNase P activity was assessed as described above.
Unimolecular Enzyme:Substrate RNAs.
Unimolecular enzyme:substrate RNAs were constructed by using RNase P RNA sequences from Methanobacterium formicicum and M. barkeri and the B. subtilis tRNAAsp. Full-length RNase P RNA-encoding sequences were amplified from cloned genes by PCR by using primers ΔH5′Bam and ΔH3′Xba for M. formicicum or Msb5′Bam (CGGGATCCATGCGAGAGAGGCTGG) and Msb3′RBam (CGGGATCCATGCGAGTGAGGCACG) for M. barkeri; this DNA was phosphorylated with ATP and T4 polynucleotide kinase and treated with T4 DNA ligase to generate circular genes. Secondary amplifications from these DNAs with primers Mfo261FBglIITP (GAAGATCTAGGTAACTCGCATAGATG) and Mfo260RHindTP (CCCAAGCTTCTGCCTCATACAGGATTC) for M. formicicum or MsbTP2505′ (CGTCTAGAAACGCATAGCCGAATG) and MsbTP2503′ (AACTGCAGAACAACCGGGAGAGTCCG) for M. barkeri generated circularly permuted RNase P RNA genes. Circularly permuted DNAs were cloned immediately downstream of the tRNAAsp from B. subtilis in pDW128 (18). Radiolabeled enzyme:substrate RNAs were generated from linearized plasmids by using T7 RNA polymerase and [α-32P]GTP.
Reconstitution of Chimeric Holoenzymes.
RNase P reconstitution assays were conducted in 50 mM Tris⋅Cl (pH 8), 100 mM ammonium acetate, 25 mM MgCl2, 1.5 nM B. subtilis pre-tRNAAsp, 20–200 μg/ml RNase P RNA, 18 μg/ml nonsense transcript of the M. thermoautotrophicum RNase P RNA gene (included as a “decoy” for traces of nuclease activity in the RNase P protein preparation), and 0–10 μg/ml B. subtilis RNase P protein (a gift from C. Fierke, Duke University, Durham, NC). Reactions containing archaeal RNase P RNA and negative controls containing no RNase P RNA were incubated at 37°C for 16 h. Reactions containing E. coli RNase P RNA were incubated at 37°C for 5 min. Cleavage products were separated by electrophoresis in denaturing 8% acrylamide gels followed by phosphorimagery.
RESULTS AND DISCUSSION
In preliminary experiments, lysates from a variety of archaeal cultures were capable of 5′ processing either the B. subtilis or M. thermoautotrophicum pre-tRNAAsp (15). Removal of protein from these lysates in many cases eliminated catalytic activity, but those of the methanobacteria and the thermococci (P. furiosus and Thermococcus celer) retained RNase P activity (data not shown). This activity was resistant to exhaustive phenol extraction and the inclusion of SDS (1%) in the reaction buffer, supporting the belief that catalysis was independent of protein. RNAs from the extreme halophiles H. volcanii and Natronobacterium gregoryi purified by organic extraction failed to process pre-tRNA, but the activity present in cell lysates was resistant to the inclusion of SDS in the reaction buffer, suggesting that the RNA was responsible for this activity.
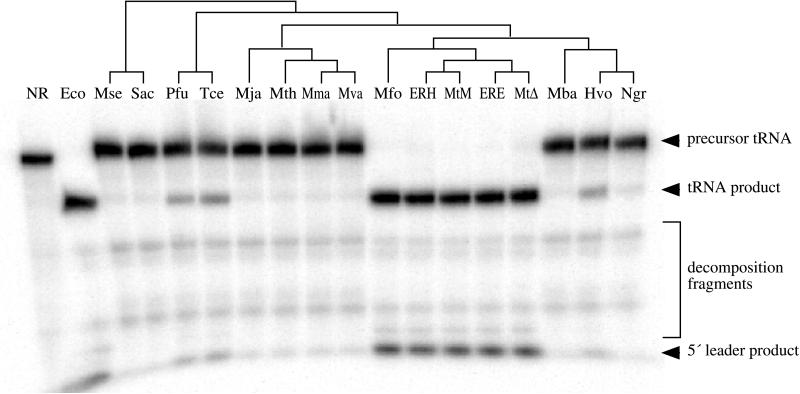
The convincing proof that ribozymes, including bacterial RNase P RNA, are catalytically proficient in the absence of associated proteins came from experiments with RNAs transcribed in vitro (19). Synthetic RNAs, transcribed from cloned RNase P RNA-encoding genes, from a wide phylogenetic range of archaeal species were therefore tested for pre-tRNA 5′-processing activity; those of the methanobacteria, the extreme halophiles, and thermococci, but not other Archaea, were found to contain RNase P activity (Fig. 2). The catalytic proficiency of these synthetic RNAs indicates that they do not depend on protein for activity, nor are posttranscriptional modifications such as pseudouridylation or 2′-O-methylation essential for activity.
Figure 2.
Catalytic activity of synthetic archaeal RNase P RNAs. Assays contained 4 M ammonium acetate, 300 mM MgCl2, 50 mM Tris⋅Cl (pH 8), 0.1% SDS, 0.05% Nonidet P-40, 1.5 nM uniformly labeled B. subtilis pre-tRNAAsp, and ≈300 nM RNase P RNA (synthesized from cloned genes by transcription in vitro) and were incubated for 3.5 h at 45°C. The location of substrate and product bands are indicated. RNase P RNAs tested were no RNA (NR), E. coli (Eco), M. sedula (Mse), S. acidocaldarius (Sac), P. furiosus (Pfu), T. celer (Tce), M. jannaschii (Mja), M. thermolithotrophicus (Mth), Methanococcus marapaludis (Mma), Methanococcus vannielii (Mva), M. formicicum (Mfo), M. thermoautotrophicum ER-H (ERH), M. thermoautotrophicum Marburg (MtM), M. thermoautotrophicum ER-E (ERE), M. thermoautotrophicum ΔH (MtΔ), M. barkeri (Mba), H. volcanii (Hvo), and Natronobacterium gregoryi (Ngr). Activity by the N. gregoryi RNA is evident but weak. The phylogenetic relationships between these organisms based on small subunit ribosomal RNA sequences (26) are shown as a cladogram above the gel. Placement of the ER-E and ER-H RNAs, which were cloned from enrichment cultures inoculated with wastewater sludge, are based on similarity to the M. thermoautotrophicum strains ΔH and Marburg RNase P RNA sequences.
The extent of activity for all of the active archaeal RNase P RNAs was quite low; initial assays included large molar excesses (in the range of 100:1) of RNase P RNA over substrate and incubation for several hours, but often resulted in only partial processing of the substrate. The potential for contamination by catalytically active bacterial RNase P RNAs was therefore a significant concern. Previous descriptions of catalytic activity by the H. volcanii RNA (8) and reconstitution of chimeric RNase P holoenzymes from archaeal and bacterial components (8, 10) were previously discounted as the result of contamination, because the amount of activity recovered was very small and the results could not be reproduced (9). Three observations argued against the possibility that the catalytic activity reported here might result from contamination by bacterial RNase P RNA rather than from the archaeal RNA: (i) correlation of the presence or absence of activity from RNA extracted from cells and RNAs synthesized in vitro from genes cloned from the same species (data not shown), (ii) the unusual biochemical properties of the activity (see below), and (iii) the precise electrophoretic comigration of activity with the archaeal RNA (data not shown). The direct experimental evidence that the observed activity resides in the archaeal RNase P RNA comes from oligonucleotide-directed RNase H depletion experiments (Fig. 3) and intramolecular cleavage of unimolecular enzyme:substrate RNAs (Fig. 4). In the first instance, pretreatment of the M. thermoautotrophicum ΔH RNase P RNA with RNase H in the presence of an M. thermoautotrophicum-specific oligonucleotide, but not an E. coli-specific oligonucleotide, reduces RNase P activity. However, the E. coli-specific oligonucleotide and RNase H reduced the activity of the E. coli, but not M. thermoautotrophicum, RNase P RNA. In the second case, cleavage of the tethered M. formicicum RNase P RNA:B. subtilis pre-tRNAAsp was independent of concentration, indicating cleavage was in cis. The M. formicicum RNase P RNA:B. subtilis pre-tRNAAsp required conditions similar to the native M. thermoautotrophicum RNA (see below) for optimal activity. The analogous enzyme:substrate RNA based on the M. barkeri RNase P RNA, an RNase P RNA that is not catalytically active, does not self-cleave (data not shown).
Figure 3.
Oligonucleotide-directed RNase H depletion of RNase P activity. RNase P RNAs were annealed with oligonucleotides complementary to J11/12 in either the M. thermoautotrophicum (MtΔ) or the E. coli (Eco) RNase P RNA and then digested with ribonuclease H. Reagents included in each reaction are indicated above each lane. Activity by the M. thermoautotrophicum RNase P RNA was reduced only in reactions containing the Methanobacterium-specific oligonucleotide; reactions lacking the oligonucleotide or containing the E. coli-specific oligonucleotide were not inhibited. Activity by the E. coli RNase P RNA was reduced only in reactions containing the E. coli-specific oligonucleotide; reactions lacking the oligonucleotide or containing the Methanobacterium-specific oligonucleotide were not inhibited.
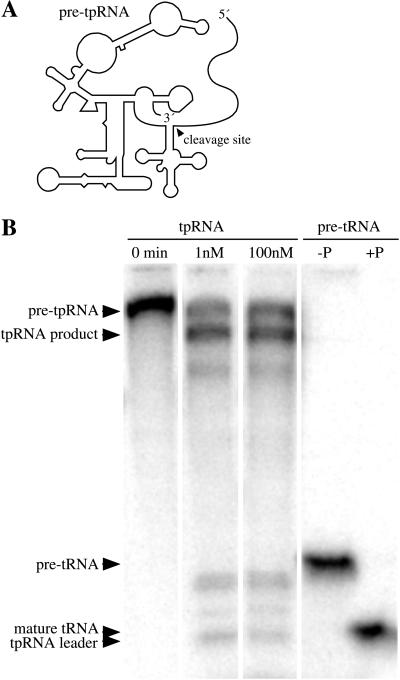
Figure 4.
Intramolecular cleavage by a unimolecular substrate:enzyme RNA. (A) The secondary structure of the conjugated B. subtilis pre-tRNAAsp:M. formicicum (circularly permuted) RNase P RNA (pre-tpRNA) is shown diagrammatically. (B) Unreacted pre-tpRNA is shown in the first lane. Lanes 2 and 3 are the same RNA, incubated in parallel, at 1 and 100 nM; the extent of cleavage is ≈65% in both cases. The three other weaker cleavage products representing either Mg2+-sensitive sites or miscleavages were not characterized. Lanes 4 and 5 are the trans-cleavage analogs of lanes 1 and 3, in which the B. subtilis pre-tRNAAsp, but not the M. formicicum RNase P RNA, is radiolabeled.
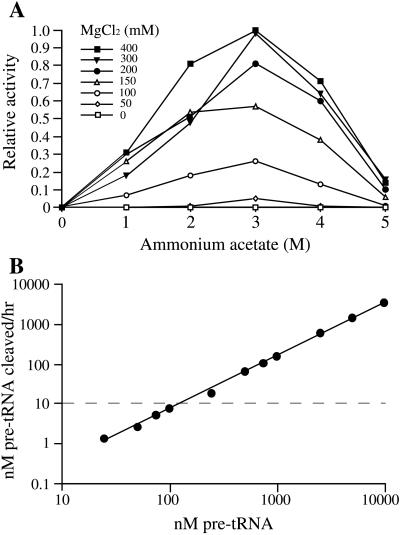
The reaction conditions required by the archaeal RNase P RNA activity are unusual. Both the native and synthetic M. thermoautotrophicum RNase P RNAs were found to require 300 mM MgCl2 and 3 M ammonium acetate for maximal activity (Fig. 5A), well above the requirements of any characterized bacterial RNase P RNA. Mn2+ could efficiently replace Mg2+ and was effective at lower concentrations (100 mM) but at the expense of a dramatic increase in nonspecific hydrolysis of substrate, product, and presumably enzyme. Ca2+, Zn2+, and Cu2+ were unable to promote cleavage. The Mg2+ requirement was not reduced by increases in ammonium acetate concentration nor visa versa. The high ionic strength may be required for stabilization of the RNA structure, and the high Mg2+ may be required for further specific stabilization and/or to overcome poor binding of catalytically involved Mg2+. The RNase P RNA synthetic transcript from H. volcanii also required very high ionic strength for activity (4 M ammonium acetate and 300 mM MgCl2) for maximal activity. The optimal temperatures and pH for these RNase P RNAs was 45–50°C and 8.0, respectively. However, the upper bounds of these assays are limited by the extent of nonspecific hydrolysis in these reactions at elevated temperatures and pH, although high temperature or pH alone did not allow optimal catalysis at lower ionic strength or Mg2+ concentrations.
Figure 5.
Activity of the M. thermoautotrophicum RNase P RNA as functions of ammonium acetate, MgCl2, and substrate concentration. (A) Activity of the M. thermoautotrophicum RNase P RNA as functions of ammonium acetate and MgCl2 concentration. Reactions contained 20 nM substrate RNA and 1 nM RNase P RNA. Maximal activity in this assay represents 47% cleavage. (B) Activity of the M. thermoautotrophicum RNase P RNA as a function of substrate concentration. Reactions were performed under optimal conditions (see Materials and Methods and A). Reactions contained 10 nM RNase P RNA and were incubated for 2.5 h; points above the dashed horizontal line represent multiple turnovers over the course of the incubation. All data points in both A and B are represented by reactions with less than 50% cleavage, within the linear range of enzyme concentration and time for the reaction (data not shown).
Poor catalysis by the archaeal RNase P RNAs, even in optimal conditions, seems to result at least in part from poor affinity for substrate. Substrate cleavage rates increase linearly with substrate concentration up to at least 10 μM (Fig. 5B), implying that the Km of the reaction is in excess of ≈40 μM. This result may occur in part because of the extreme conditions required to obtain activity; the Km of the E. coli RNase P RNA-catalyzed reaction is also quite high under these far-from-optimal conditions (2.2 μM, as opposed to 74 nM in reactions containing 1 M ammonium acetate and 25 mM MgCl2). The Vmax of the reaction catalyzed by the archaeal RNA could not be determined, but given the high Km and respectable catalytic rate at high substrate concentrations, there is no evidence for a defect in Vmax or kcat. At the highest substrate concentrations, each mole of archaeal RNase P RNA processed more than 100 mol of substrate during the 2.5-h reaction (2.2 min−1).
The protein component or components of the archaeal RNase P have yet to be identified despite the availability of several complete archaeal genome sequences and RNase P protein sequences from a wide variety of Bacteria and the yeast nucleus and mitochondrion (as well as some of the human sequences; refs. 20–23). Because of the similarity of the archaeal and bacterial RNase P RNAs, we attempted to reconstitute chimeric holoenzymes from archaeal RNase P RNA and bacterial RNase P protein (Fig. 6). Catalytic activity by the Methanobacterium RNase P RNAs was reconstituted at low ionic strength and Mg2+ concentration by using the B. subtilis RNase P protein, indicating the functional interaction of these heterologous subunits. It seems likely, therefore, that the structure and function of the bacterial and archaeal protein components are, at least in part, similar in the archaeal and bacterial enzymes. RNase P activity at moderate ionic strength absolutely depended on the presence of the protein, unlike the E. coli or B. subtilis RNase P RNAs, which can process this substrate at rates of ≈20% of that of the reconstituted enzymes in these conditions. We did not detect enhanced activity of the M. sedula, H. volcanii, M. barkeri, or M. vannielii RNAs in the presence of the bacterial protein under these conditions, although analogous reconstitution of H. volcanii RNA and B. subtilis protein has been reported (8).
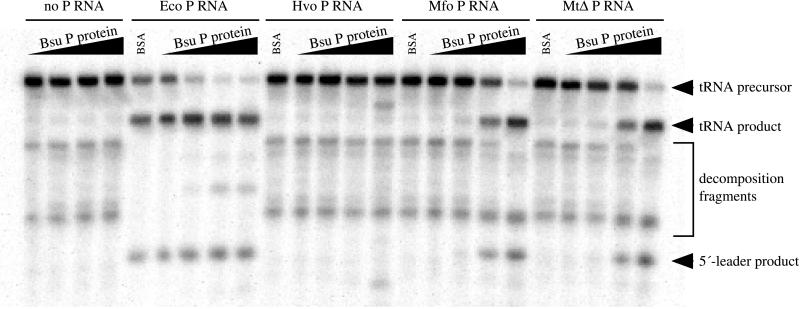
Figure 6.
Reconstitution of active chimeric holoenzymes. RNase P RNAs from E. coli (Eco, 38 μg/ml), H. volcanii (Hvo, 170 μg/ml), M. formicicum (Mfo, 60 μg/ml), or M. thermoautotrophicum ΔH (MtΔ, 33 μg/ml) were assayed for RNase P activity in the presence of 0, 0.1, 1, or 10 μg/ml B. subtilis RNase P protein (increasing protein concentration indicated by black wedges above the reaction lanes). Specific RNase P cleavage products are indicated by black arrowheads. The E. coli RNase P RNA is somewhat active by itself under these conditions, but activity is enhanced by the inclusion of the B. subtilis RNase P protein. The M. thermoautotrophicum and M. formicicum RNase P RNAs are not active in the absence of protein under these conditions but were activated by inclusion of the B. subtilis RNase P protein. Correct processing by the H. volcanii RNase P RNA was not observed under these conditions in either the presence or absence of the B. subtilis RNase P protein. However, a specific inappropriate cleavage was generated at the highest concentration of protein; the nature of these products and the apparent miscleavage have not been determined.
The catalytic proficiency of archaeal RNase P RNAs is much lower than their bacterial homologs, but their ability to process pre-tRNA at any rate implies that they contain all of the sequences and structures necessary for substrate recognition and catalysis. The catalytic competence of the archaeal RNAs is consistent with our understanding of the conserved features of the archaeal and bacterial RNase P RNAs; these RNAs are remarkably similar in both sequence and structure, and the invariably present sequences and structures of the catalytically active bacterial RNAs are present in the archaeal RNAs as well (except those of Methanococcus and Archaeoglobus). Synthetic RNase P RNAs that contain only these core bacterial sequences and structures are catalytically active; however, these RNAs are structurally deficient, with biochemical properties similar to those of the active archaeal RNAs (13, 14). The archaeal RNAs apparently depend on the protein component(s) of the holoenzyme for structural integrity but not for essential catalytic function. The exception to this generality is the RNase P RNAs of Methanococcus and Archaeoglobus, both of which lack core structural features (P8 and L15) that are directly involved in substrate recognition in Bacteria. These RNAs are not catalytically active, even in the presence of the bacterial protein. How the holoenzymes compensate for the lack of core RNA structures is not known.
Acknowledgments
We thank J. Reeve for cultures of M. thermoautotrophicum; W. Whitman for cultures of Methanococcus spp.; C. Fierke for the gift of B. subtilis RNase P protein; R. Kelley for cultures of various Crenarchaea and thermococci; A. Andrews for assistance with biochemical assays and construction of the unimolecular enzyme:substrate RNAs; J. Perez, L. Rudd, and B. Vucson for their work in the cloning and DNA sequence determination of RNase P RNA encoding genes; and N. Pace and C. Daniels for valuable discussions on this work. This work was supported by National Institutes of Health Grant GM52894.
Footnotes
References
- 1.Pace N R, Brown J W. J Bacteriol. 1995;177:1919–1928. doi: 10.1128/jb.177.8.1919-1928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karwan R, Pluk H, van Venrooij W J, editors. Molecular Biology Reports Special Issue: RNase MRP/RNase P Systems. Vol. 22. Nijmegen: Kluwer; 1996. [Google Scholar]
- 3.Reich C, Olsen G J, Pace B, Pace N R. Science. 1988;239:178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- 4.Kurz J C, Niranjanakumari S, Fierke C A. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 5.Guerrier-Takada C, Gardner K, Marsh T, Pace N R, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 6.Niranjanakumari S, Stams T, Crary S M, Christianson D W, Fierke C A. Proc Natl Acad Sci USA. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darr S C, Pace B, Pace N R. J Biol Chem. 1990;265:12927–12932. [PubMed] [Google Scholar]
- 8.Nieuwlandt D T, Haas E S, Daniels C J. J Biol Chem. 1991;266:5689–5695. [PubMed] [Google Scholar]
- 9.LaGrandeur T E, Darr S C, Haas E S, Pace N R. J Bacteriol. 1993;175:5043–5048. doi: 10.1128/jb.175.16.5043-5048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence N, Wesolowski D, Gold H, Bartkiewicz M, Guerrier-Takada C, McCain W H, Altman S. Cold Spring Harbor Symp Quant Biol. 1987;52:233–238. doi: 10.1101/sqb.1987.052.01.028. [DOI] [PubMed] [Google Scholar]
- 11.Haas E S, Armbruster D W, Vucson B M, Daniels C J, Brown J W. Nucleic Acids Res. 1996;24:1252–1259. doi: 10.1093/nar/24.7.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas E S, Brown J W. Nucleic Acids Res. 1998;26:4093–4099. doi: 10.1093/nar/26.18.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waugh D S, Green C J, Pace N R. Science. 1989;244:1569–1570. doi: 10.1126/science.2472671. [DOI] [PubMed] [Google Scholar]
- 14.Siegel R W, Banta A B, Haas E S, Brown J W, Pace N R. RNA. 1996;2:452–462. [PMC free article] [PubMed] [Google Scholar]
- 15.Pannucci J A, Haas E S, Brown J W. Nucleic Acids Symp Series. 1997;36:90–92. [Google Scholar]
- 16.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 18.Waugh D S. Ph.D. thesis. Bloomington, IN: Indiana University; 1989. [Google Scholar]
- 19.Guerrier-Takada C, Altman S. Science. 1984;223:285–286. doi: 10.1126/science.6199841. [DOI] [PubMed] [Google Scholar]
- 20.Chamberlain J R, Lee Y, Lane W S, Engelke D R. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eder P, Kekuda R, Stolc V, Altman S. Proc Natl Acad Sci USA. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stolc V, Altman S. Genes Dev. 1997;11:2414–2425. doi: 10.1101/gad.11.18.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morales M J, Dang Y L, Lou Y C, Sulo P, Martin N C. Proc Natl Acad Sci USA. 1992;89:9875–9879. doi: 10.1073/pnas.89.20.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas E S, Brown J W, Pitulle C, Pace N R. Proc Natl Acad Sci USA. 1994;91:2527–2531. doi: 10.1073/pnas.91.7.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown J W. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]