Abstract
Recombinant plasmids were constructed which carry defined regions of the threonine (thr) operon regulatory region of Escherichia coli. In vitro transcription experiments utilizing plasmid or restriction fragment templates showed that two major RNA transcripts, which differ in length by one to a few bases, are transcribed from this region. The approximate length of the transcripts is 150 to 170 bases, and the site(s) of termination is near or within the thr attenuator. The efficiency of termination at the thr operon attenuator in vitro is approximately 90%. A regulatory mutation, thr79-20, which is a G-C insertion in the attenuator, reduces the frequency of transcription termination to 75%. In addition, in vivo RNA transcripts were identified which hybridize to the thr operon regulatory region. These transcripts appeared to be identical to the two major in vitro transcripts as judged by their mobilities on 8% polyacrylamide-8 M urea gels. This result indicates that the thr operon regulatory region is transcribed in vivo and that termination occurs near or within the thr attenuator.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes W. M. DNA sequence from the histidine operon control region: seven histidine codons in a row. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4281–4285. doi: 10.1073/pnas.75.9.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes W. M. Plasmid detection and sizing in single colony lysates. Science. 1977 Jan 28;195(4276):393–394. doi: 10.1126/science.318764. [DOI] [PubMed] [Google Scholar]
- Bertrand K., Squires C., Yanofsky C. Transcription termination in vivo in the leader region of the tryptophan operon of Escherichia coli. J Mol Biol. 1976 May 15;103(2):319–337. doi: 10.1016/0022-2836(76)90315-6. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakesley R. W., Wells R. D. 'Single-stranded' DNA from phiX174 and M13 is cleaved by certain restriction endonucleases. Nature. 1975 Oct 2;257(5525):421–422. doi: 10.1038/257421a0. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chapman J., Gardner J. F. Secondary lambda attachment site in the threonine operon attenuator of Escherichia coli. J Bacteriol. 1981 Jun;146(3):1046–1054. doi: 10.1128/jb.146.3.1046-1054.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G. E., Farnham P. J., Platt T. Synthetic sites for transcription termination and a functional comparison with tryptophan operon termination sites in vitro. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4180–4184. doi: 10.1073/pnas.78.7.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Blasi F., Di Lauro R., Frunzio R., Bruni C. B. Nucleotide sequence of the attenuator region of the histidine operon of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4276–4280. doi: 10.1073/pnas.75.9.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREUNDLICH M. Multivalent repression in the biosynthesis of threonine in Salmonella typhimurium and Escherichia coli. Biochem Biophys Res Commun. 1963 Feb 6;10:277–282. doi: 10.1016/0006-291x(63)90430-3. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. A model for transcription termination suggested by studies on the trp attenuator in vitro using base analogs. Cell. 1980 Jul;20(3):739–748. doi: 10.1016/0092-8674(80)90320-7. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981 Feb 11;9(3):563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frunzio R., Bruni C. B., Blasi F. In vivo and in vitro detection of the leader RNA of the histidine operon of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1981 May;78(5):2767–2771. doi: 10.1073/pnas.78.5.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. F. Regulation of the threonine operon: tandem threonine and isoleucine codons in the control region and translational control of transcription termination. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1706–1710. doi: 10.1073/pnas.76.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
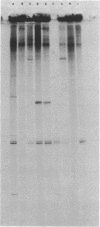
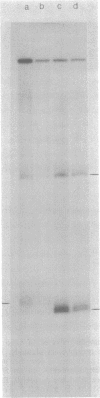
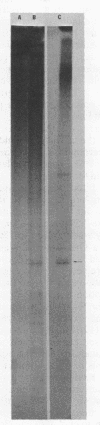
- Gardner J. F., Reznikoff W. S. Identification and restriction endonuclease mapping of the threonine operon regulatory region. J Mol Biol. 1978 Dec 5;126(2):241–258. doi: 10.1016/0022-2836(78)90361-3. [DOI] [PubMed] [Google Scholar]
- Gardner J. F., Smith O. H. Operator-promoter functions in the threonine operon of Escherichia coli. J Bacteriol. 1975 Oct;124(1):161–166. doi: 10.1128/jb.124.1.161-166.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill R. M., Wessler S. R., Keller E. B., Calvo J. M. leu operon of Salmonella typhimurium is controlled by an attenuation mechanism. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4941–4945. doi: 10.1073/pnas.76.10.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Chow L., Helinski D. R. Characterization of a mini-ColC1 plasmid. J Bacteriol. 1976 Apr;126(1):447–453. doi: 10.1128/jb.126.1.447-453.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. A., Walseth T. F. The enzymatic preparation of [alpha-32P]ATP, [alpha-32P]GTP, [32P]cAMP, and [32P]cGMP, and their use in the assay of adenylate and guanylate cyclases and cyclic nucleotide phosphodiesterases. Adv Cyclic Nucleotide Res. 1979;10:135–167. [PubMed] [Google Scholar]
- Johnston H. M., Barnes W. M., Chumley F. G., Bossi L., Roth J. R. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci U S A. 1980 Jan;77(1):508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston H. M., Roth J. R. DNA sequence changes of mutations altering attenuation control of the histidine operon of Salmonella typhimurium. J Mol Biol. 1981 Feb 5;145(4):735–756. doi: 10.1016/0022-2836(81)90312-0. [DOI] [PubMed] [Google Scholar]
- Kasai T. Regulation of the expression of the histidine operon in Salmonella typhimurium. Nature. 1974 Jun 7;249(457):523–527. doi: 10.1038/249523a0. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Calvo J. M. Alternative secondary structures of leader RNAs and the regulation of the trp, phe, his, thr, and leu operons. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6186–6190. doi: 10.1073/pnas.76.12.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Hatfield G. W. Multivalent translational control of transcription termination at attenuator of ilvGEDA operon of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1862–1866. doi: 10.1073/pnas.77.4.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman I. R. T4 DNA polymerase. Methods Enzymol. 1974;29:46–53. [PubMed] [Google Scholar]
- Lowe P. A., Hager D. A., Burgess R. R. Purification and properties of the sigma subunit of Escherichia coli DNA-dependent RNA polymerase. Biochemistry. 1979 Apr 3;18(7):1344–1352. doi: 10.1021/bi00574a034. [DOI] [PubMed] [Google Scholar]
- Lynn S. P., Cohen L. K., Kaplan S., Gardner J. F. RsaI: a new sequence-specific endonuclease activity from Rhodopseudomonas sphaeroides. J Bacteriol. 1980 May;142(2):380–383. doi: 10.1128/jb.142.2.380-383.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E., Reznikoff W. S. In vitro analysis of the Escherichia coli RNA polymerase interaction with wild-type and mutant lactose promoters. J Mol Biol. 1978 Nov 15;125(4):467–490. doi: 10.1016/0022-2836(78)90311-x. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., Thornton K., Reznikoff W. S. lac Promoter mutations located downstream from the transcription start site. J Mol Biol. 1980 May 25;139(3):537–549. doi: 10.1016/0022-2836(80)90145-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nargang F. E., Subrahmanyam C. S., Umbarger H. E. Nucleotide sequence of ilvGEDA operon attenuator region of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1823–1827. doi: 10.1073/pnas.77.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxender D. L., Zurawski G., Yanofsky C. Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5524–5528. doi: 10.1073/pnas.76.11.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Myers P. A., Morrison A., Murray K. A specific endonuclease from Arthrobacter luteus. J Mol Biol. 1976 Mar 25;102(1):157–165. doi: 10.1016/0022-2836(76)90079-6. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Myers P. A., Morrison A., Murray K. A specific endonuclease from Haemophilus haemolyticus. J Mol Biol. 1976 May 5;103(1):199–208. doi: 10.1016/0022-2836(76)90060-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Saint-Girons I., Margarita D. Operator-constitutive mutants in the threonine operon of Escherichia coli K-12. J Bacteriol. 1975 Dec;124(3):1137–1141. doi: 10.1128/jb.124.3.1137-1141.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Hutchinson C. A., 3rd, Harris J. I. A thermostable sequence-specific endonuclease from Thermus aquaticus. Proc Natl Acad Sci U S A. 1977 Feb;74(2):542–546. doi: 10.1073/pnas.74.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lund E., Smithies O., Blattner F. R. Hybridization of labeled RNA to DNA in agarose gels. Nucleic Acids Res. 1975 Oct;2(10):1911–1929. doi: 10.1093/nar/2.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C., Lee F., Bertrand K., Squires C. L., Bronson M. J., Yanofsky C. Nucleotide sequence of the 5' end of tryptophan messenger RNA of Escherichia coli. J Mol Biol. 1976 May 15;103(2):351–381. doi: 10.1016/0022-2836(76)90317-x. [DOI] [PubMed] [Google Scholar]
- Stauffer G. V., Zurawski G., Yanofsky C. Single base-pair alterations in the Escherichia coli trp operon leader region that relieve transcription termination at the trp attenuator. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4833–4837. doi: 10.1073/pnas.75.10.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Cameron J. R., Davis R. W. Functional genetic expression of eukaryotic DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1976 May;73(5):1471–1475. doi: 10.1073/pnas.73.5.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thèze J., Saint-Girons I. Threonine locus of Escherichia coli K-12: genetic structure and evidence for an operon. J Bacteriol. 1974 Jun;118(3):990–998. doi: 10.1128/jb.118.3.990-998.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Jacquemin-Sablon A., Live T. R., Fareed G. C., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. VI. Further purification and properties of polynucleotide ligase from Escherichia coli infected with bacteriophage T4. J Biol Chem. 1968 Sep 10;243(17):4543–4555. [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Zurawski G., Brown K., Killingly D., Yanofsky C. Nucleotide sequence of the leader region of the phenylalanine operon of Escherichia coli. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4271–4275. doi: 10.1073/pnas.75.9.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Elseviers D., Stauffer G. V., Yanofsky C. Translational control of transcription termination at the attenuator of the Escherichia coli tryptophan operon. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5988–5992. doi: 10.1073/pnas.75.12.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]