Abstract
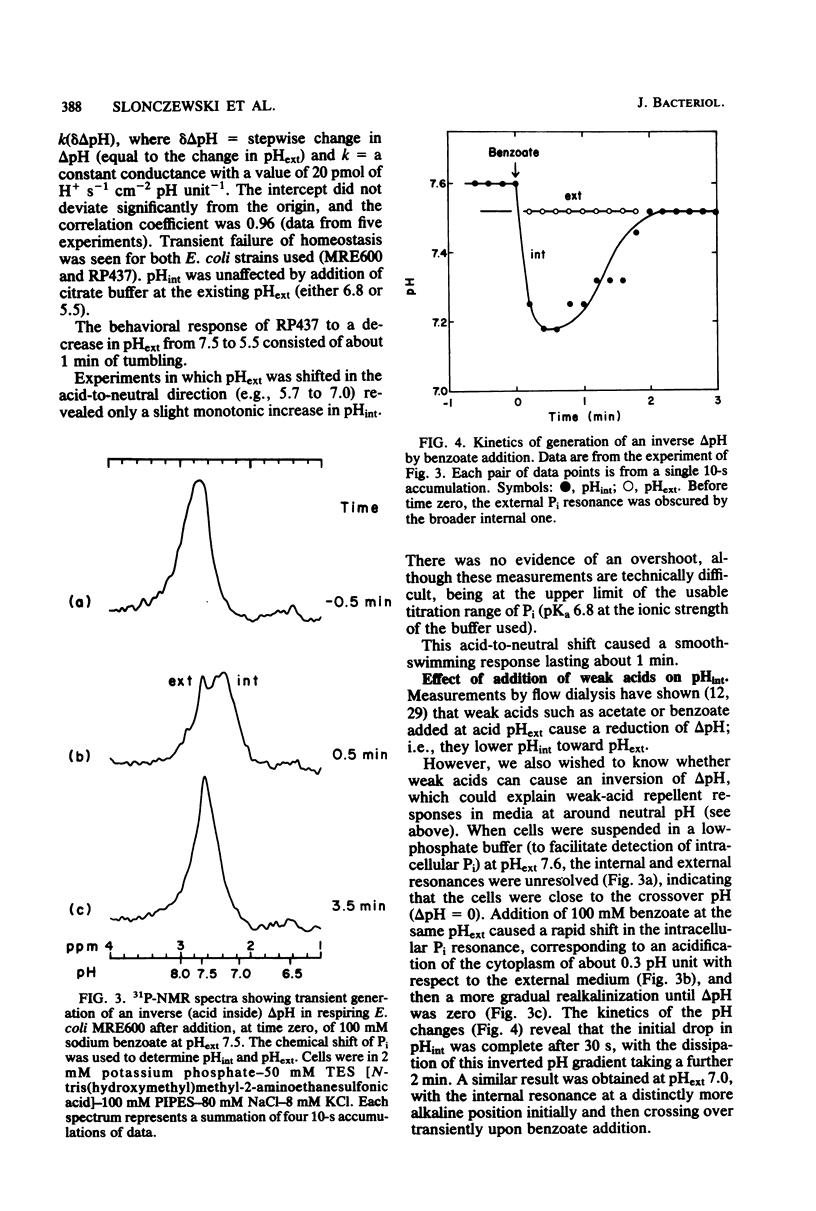
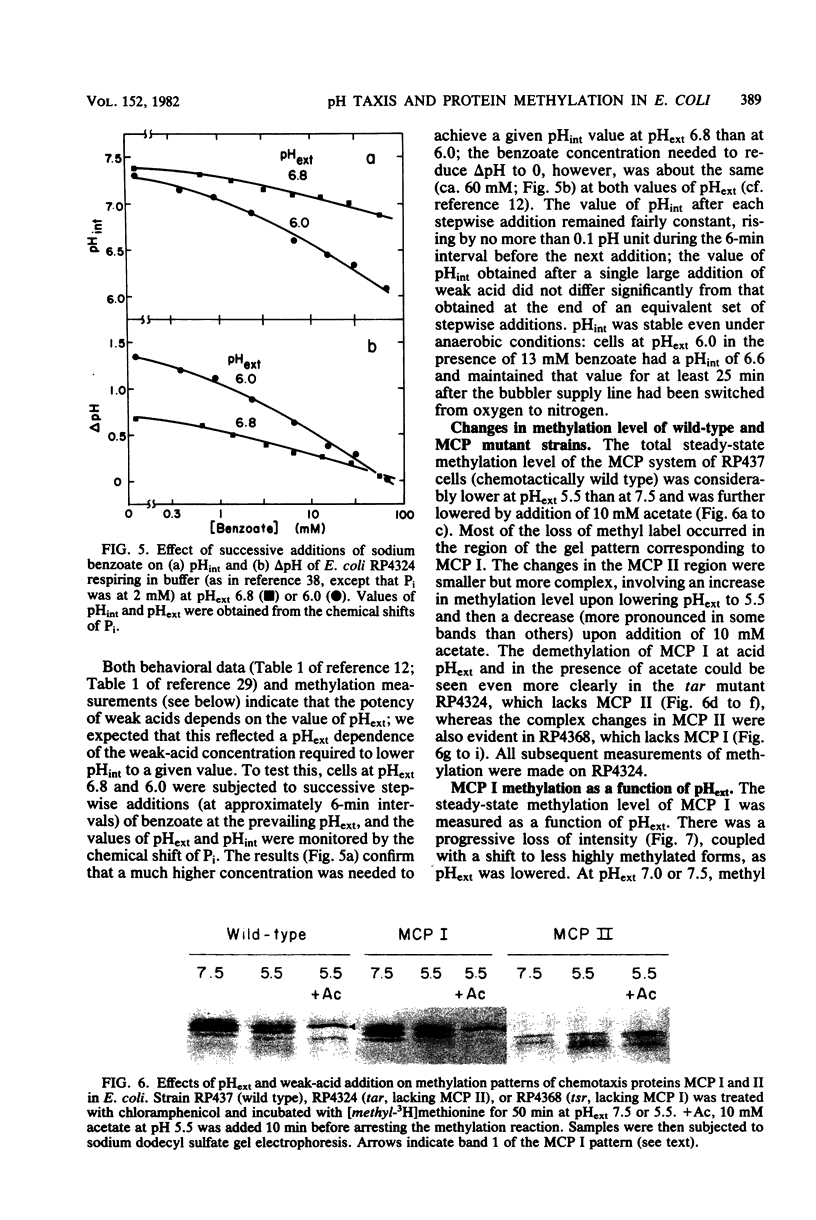
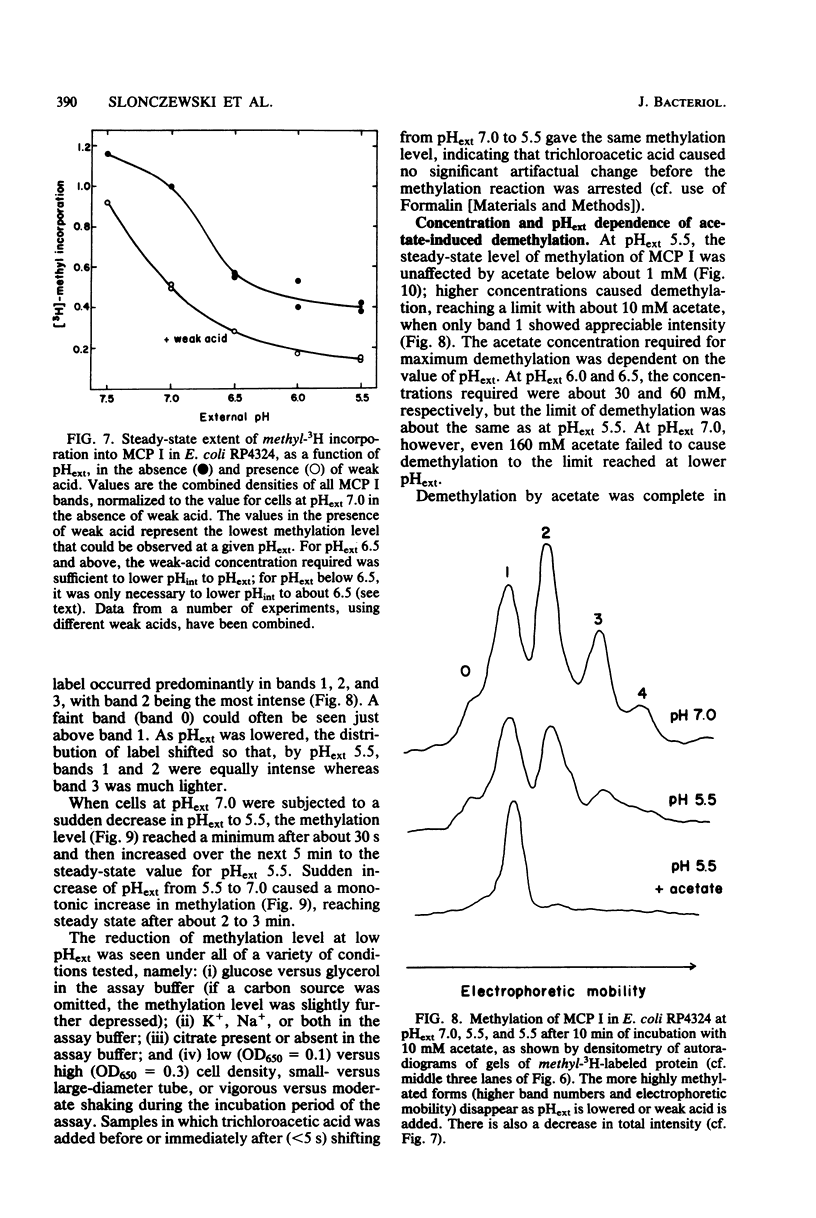
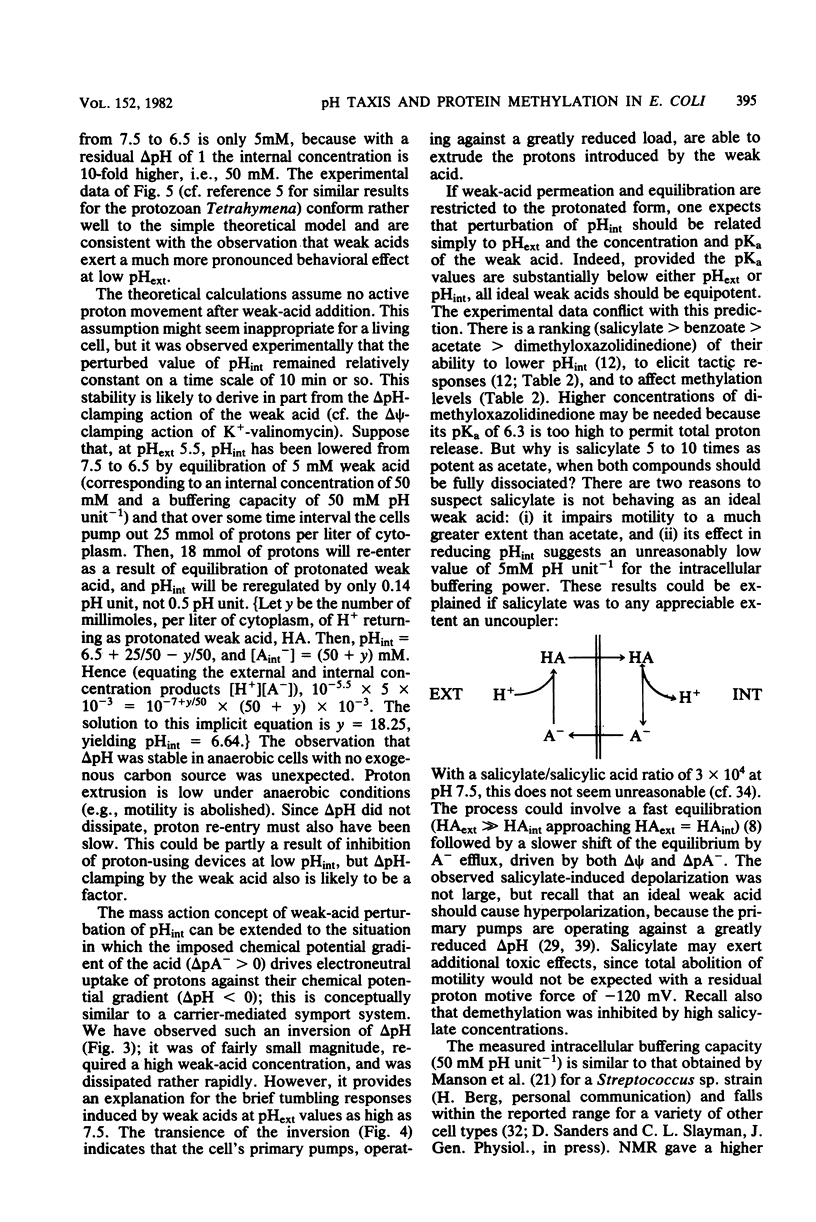
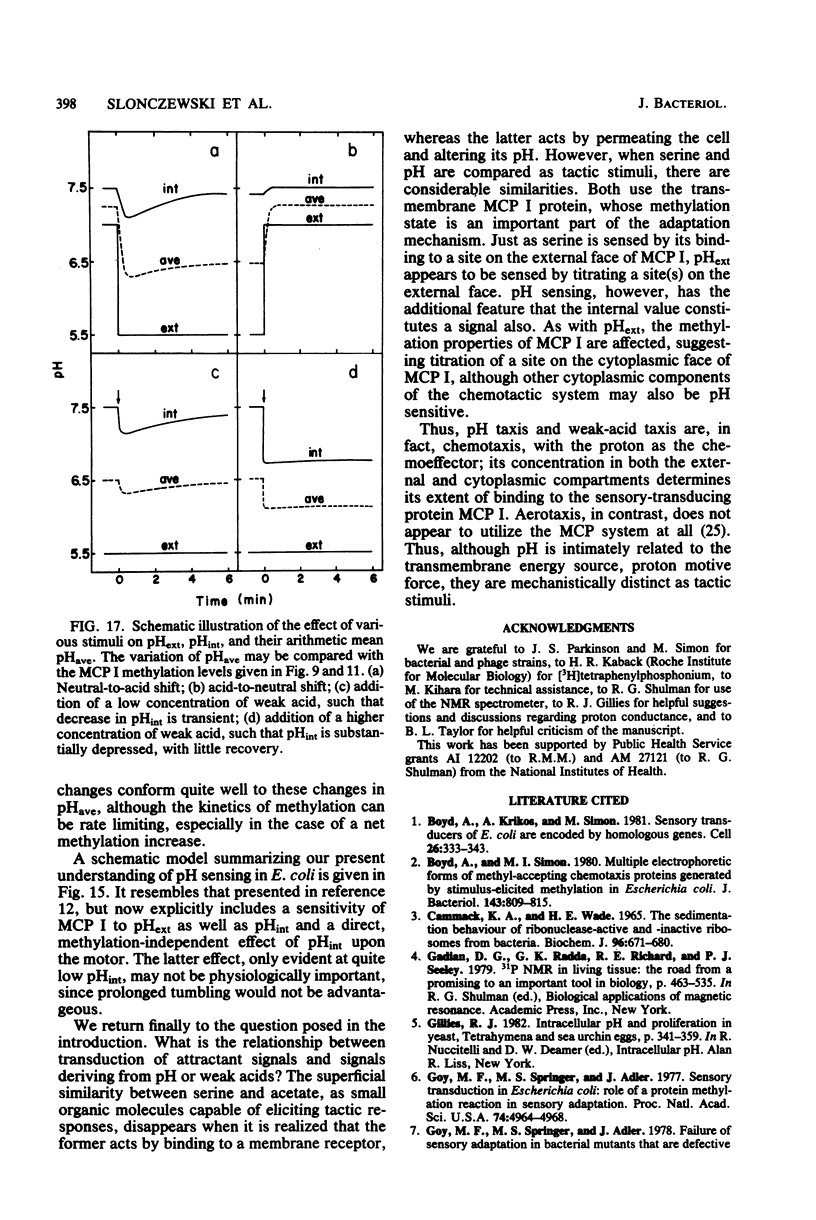
Intracellular pH (pHint) and extracellular pH (pHext) of Escherichia coli were measured at 12-s time resolution by 31P-nuclear magnetic resonance: a sudden neutral-to-acid shift in pHext (e.g., from 7.0 to 5.6) caused a transient failure of homeostasis, with pHint decreasing by about 0.4 unit in ca. 30 s and then returning to its original value (ca. 7.5) over a period of several minutes. Membrane proton conductance was estimated to be 20 pmol s−1 cm−2 pH unit−1. Addition of the membrane-permeant weak acid benzoate at constant pHext also caused a lowering of pHint; at high concentrations it generated an inverted transmembrane pH gradient (ΔpH). The buffering capacity of the cells was estimated by such experiments to be ca. 50 mM per pH unit. Effects of pH-related stimuli on the methyl-accepting chemotaxis proteins (MCPs) were examined: the steady-state methylation of MCP I was found to decrease when pHint was lowered by weak acid addition or when pHext was lowered. The extent of demethylation in the latter case was too great to be explained by imperfect steady-state homeostasis; a small but reproducible undershoot in methylation level correlated with the observed short-term homeostatic failure. MCP II underwent smaller and more complex changes than MCP I, in response to pH-related stimuli. The methylation level of MCP I could not, by any condition tested, be driven below a limit of ca. 15% of the control level (unstimulated cells at pHext 7.0). The weak-acid concentration needed to reach that limit was dependent on pHext, as would be expected on the basis of ΔpH-driven concentrative effects. The potency ranking of weak acids was the same with respect to lowering pHint, demethylating MCP I, and causing repellent behavioral responses. The data are consistent with a model whereby MCP I and hence tactic behavior are sensitive to both pHint and pHext. Evidence is presented that pHint may also have a direct (non-MCP-related) effect on motor function. Comparison of methyl-3H- and 35S-labeled MCP I revealed that in both unstimulated and repellent-stimulated cells the major species did not carry methyl label, yet it had an electrophoretic mobility that indicated that it was more positively charged than the unmethylated form observed in methyltransferase mutants, and it was susceptible to base hydrolysis. This suggests that a substantial fraction of MCP I molecules is methylated or otherwise modified but neither exchanges methyl label nor undergoes reverse modification by repellent stimuli.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyd A., Krikos A., Simon M. Sensory transducers of E. coli are encoded by homologous genes. Cell. 1981 Nov;26(3 Pt 1):333–343. doi: 10.1016/0092-8674(81)90202-6. [DOI] [PubMed] [Google Scholar]
- Boyd A., Simon M. I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980 Aug;143(2):809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammack K. A., Wade H. E. The sedimentation behaviour of ribonuclease-active and -inactive ribosomes from bacteria. Biochem J. 1965 Sep;96(3):671–680. doi: 10.1042/bj0960671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Failure of sensory adaptation in bacterial mutants that are defective in a protein methylation reaction. Cell. 1978 Dec;15(4):1231–1240. doi: 10.1016/0092-8674(78)90049-1. [DOI] [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4964–4968. doi: 10.1073/pnas.74.11.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutknecht J., Tosteson D. C. Diffusion of weak acids across lipid bilayer membranes: effects of chemical reactions in the unstirred layers. Science. 1973 Dec 21;182(4118):1258–1261. doi: 10.1126/science.182.4118.1258. [DOI] [PubMed] [Google Scholar]
- Hedblom M. L., Adler J. Genetic and biochemical properties of Escherichia coli mutants with defects in serine chemotaxis. J Bacteriol. 1980 Dec;144(3):1048–1060. doi: 10.1128/jb.144.3.1048-1060.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskunas S. R., Burgess R. R., Nomura M. Identification of a gene for the alpha-subunit of RNA polymerase at the str-spc region of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5036–5040. doi: 10.1073/pnas.72.12.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskunas S. R., Lindahl L., Nomura M. Identification of two copies of the gene for the elongation factor EF-Tu in E. coli. Nature. 1975 Oct 9;257(5526):458–462. doi: 10.1038/257458a0. [DOI] [PubMed] [Google Scholar]
- Kihara M., Macnab R. M. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol. 1981 Mar;145(3):1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene S. J., Toews M. L., Adler J. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J Biol Chem. 1977 May 25;252(10):3214–3218. [PubMed] [Google Scholar]
- Koiwai O., Hayashi H. Studies on bacterial chemotaxis. IV. Interaction of maltose receptor with a membrane-bound chemosensing component. J Biochem. 1979 Jul;86(1):27–34. [PubMed] [Google Scholar]
- Kort E. N., Goy M. F., Larsen S. H., Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr Biochemistry of sensing and adaptation in a simple bacterial system. Annu Rev Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Letellier L., Shechter E. Cyanine dye as monitor of membrane potentials in Escherichia coli cells and membrane vesicles. Eur J Biochem. 1979 Dec 17;102(2):441–447. doi: 10.1111/j.1432-1033.1979.tb04259.x. [DOI] [PubMed] [Google Scholar]
- Maloney P. C. Membrane H+ conductance of Streptococcus lactis. J Bacteriol. 1979 Oct;140(1):197–205. doi: 10.1128/jb.140.1.197-205.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson M. D., Tedesco P., Berg H. C., Harold F. M., Van der Drift C. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci U S A. 1977 Jul;74(7):3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskavitch M. A., Kort E. N., Springer M. S., Goy M. F., Adler J. Attraction by repellents: an error in sensory information processing by bacterial mutants. Science. 1978 Jul 7;201(4350):63–65. doi: 10.1126/science.351803. [DOI] [PubMed] [Google Scholar]
- Navon G., Ogawa S., Shulman R. G., Yamane T. High-resolution 31P nuclear magnetic resonance studies of metabolism in aerobic Escherichia coli cells. Proc Natl Acad Sci U S A. 1977 Mar;74(3):888–891. doi: 10.1073/pnas.74.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J. W., Deamer D. W. Net proton-hydroxyl permeability of large unilamellar liposomes measured by an acid-base titration technique. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2038–2042. doi: 10.1073/pnas.77.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwano M., Taylor B. L. Novel sensory adaptation mechanism in bacterial chemotaxis to oxygen and phosphotransferase substrates. Proc Natl Acad Sci U S A. 1982 Jan;79(1):11–15. doi: 10.1073/pnas.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki Y., Tanford C. Proton and hydroxide ion permeability of phospholipid vesicles. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4324–4328. doi: 10.1073/pnas.78.7.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978 Jul;135(1):45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos S., Schuldiner S., Kaback H. R. The use of flow dialysis for determinations of deltapH and active transport. Methods Enzymol. 1979;55:680–688. doi: 10.1016/0076-6879(79)55076-9. [DOI] [PubMed] [Google Scholar]
- Repaske D. R., Adler J. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J Bacteriol. 1981 Mar;145(3):1196–1208. doi: 10.1128/jb.145.3.1196-1208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins C., Dahlquist F. W. The methyl-accepting chemotaxis proteins of E. coli: a repellent-stimulated, covalent modification, distinct from methylation. Cell. 1981 Aug;25(2):333–340. doi: 10.1016/0092-8674(81)90051-9. [DOI] [PubMed] [Google Scholar]
- Roos A., Boron W. F. Intracellular pH. Physiol Rev. 1981 Apr;61(2):296–434. doi: 10.1152/physrev.1981.61.2.296. [DOI] [PubMed] [Google Scholar]
- Schuldiner S., Kaback H. R. Membrane potential and active transport in membrane vesicles from Escherichia coli. Biochemistry. 1975 Dec 16;14(25):5451–5461. doi: 10.1021/bi00696a011. [DOI] [PubMed] [Google Scholar]
- Sherris D., Parkinson J. S. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silerman M., Matsumura P., Draper R., Edwards S., Simon M. I. Expression of flagellar genes carried by bacteriophage lambda. Nature. 1976 May 20;261(5557):248–250. doi: 10.1038/261248a0. [DOI] [PubMed] [Google Scholar]
- Silverman M., Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonczewski J. L., Rosen B. P., Alger J. R., Macnab R. M. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6271–6275. doi: 10.1073/pnas.78.10.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. A., Stock J. B., Koshland D. E., Jr Role of membrane potential and calcium in chemotactic sensing by bacteria. J Mol Biol. 1981 Jun 25;149(2):241–257. doi: 10.1016/0022-2836(81)90300-4. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Maderis A. M., Koshland D. E., Jr Bacterial chemotaxis in the absence of receptor carboxylmethylation. Cell. 1981 Nov;27(1 Pt 2):37–44. doi: 10.1016/0092-8674(81)90358-5. [DOI] [PubMed] [Google Scholar]
- Tsang N., Macnab R., Koshland D. E., Jr Common mechanism for repellents and attractants in bacterial chemotaxis. Science. 1973 Jul 6;181(4094):60–63. doi: 10.1126/science.181.4094.60. [DOI] [PubMed] [Google Scholar]
- Tso W. W., Adler J. Negative chemotaxis in Escherichia coli. J Bacteriol. 1974 May;118(2):560–576. doi: 10.1128/jb.118.2.560-576.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Werf P., Koshland D. E., Jr Identification of a gamma-glutamyl methyl ester in bacterial membrane protein involved in chemotaxis. J Biol Chem. 1977 Apr 25;252(8):2793–2795. [PubMed] [Google Scholar]
- Wang E. A., Koshland D. E., Jr Receptor structure in the bacterial sensing system. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. A., Mowry K. L., Clegg D. O., Koshland D. E., Jr Tandem duplication and multiple functions of a receptor gene in bacterial chemotaxis. J Biol Chem. 1982 May 10;257(9):4673–4676. [PubMed] [Google Scholar]
- Zaritsky A., Kihara M., Macnab R. M. Measurement of membrane potential in Bacillus subtilis: a comparison of lipophilic cations, rubidium ion, and a cyanine dye as probes. J Membr Biol. 1981;63(3):215–231. doi: 10.1007/BF01870983. [DOI] [PubMed] [Google Scholar]
- Zilberstein D., Schuldiner S., Padan E. Proton electrochemical gradient in Escherichia coli cells and its relation to active transport of lactose. Biochemistry. 1979 Feb 20;18(4):669–673. doi: 10.1021/bi00571a018. [DOI] [PubMed] [Google Scholar]






